Abstract
In Drosophila melanogaster, the genetic and molecular bases of post-mating changes in the female's behavior and physiology are poorly understood. However, DNA microarray studies have demonstrated that, shortly after mating, transcript abundance of >1700 genes is altered in the female's reproductive tract as well as in other tissues. Many of these changes are elicited by sperm and seminal fluid proteins (Acps) that males transfer to females. To further dissect the transcript-level changes that occur following mating, we examined gene expression profiles of whole female flies at four time points following copulation. We found that, soon after copulation ends, a large number of small-magnitude transcriptional changes occurred in the mated female. At later time points, larger magnitude changes were seen, although these occurred in a smaller number of genes. We then explored how four individual Acps (ovulin, Acp36DE, Acp29AB, and Acp62F) with unique functions independently affected gene expression in females shortly after mating. Consistent with their early and possibly local action within the female, ovulin and Acp36DE caused relatively few gene expression changes in whole bodies of mated females. In contrast, Acp29AB and Acp62F modulated a large number of transcriptional changes shortly after mating.
IN insects, as in many mammals and other vertebrates, copulation brings male and female cells and molecules together in the female's reproductive tract. There, they can interact to facilitate post-copulatory events such as sperm transport and storage, sperm maintenance, egg release, and ultimately, fertilization. While much is known about the genes, cells, and molecules that males contribute to post-copulatory processes (reviewed in Chapman 2001; Wolfner 2002; Chapman and Davies 2004; Wolfner et al. 2005; Poiani 2006; Ravi Ram and Wolfner 2007a,b), technical issues have made it more difficult to dissect how females respond molecularly and genetically to mating.
Much of our understanding of the molecular and genetic bases of post-copulatory processes has come from studies of the genus Drosophila, particularly the species Drosophila melanogaster. Mating in D. melanogaster induces a series of changes in the female's behavior and physiology, including decreasing her attractiveness to other males, decreasing her receptivity to future matings, increasing her food consumption, elevating her egg-laying rate, facilitating storage and utilization of sperm, and decreasing her life span (reviewed in Chapman 2001; Swanson and Vacquier 2002b; Wolfner 2002; Kubli 2003; Chapman and Davies 2004; Wolfner et al. 2005; Poiani 2006; Wong and Wolfner 2006; Ravi Ram and Wolfner 2007a). The proximate causes of many of these changes are the seminal fluid proteins (Acps) that males transfer to females along with sperm. Acps are synthesized in and secreted from the male's accessory glands. To date, a total of 112 potential Acps have been identified in D. melanogaster (reviewed in Ravi Ram and Wolfner 2007a) and many of these exhibit structural homology to classes of proteins found in mammalian seminal fluid (Mueller et al. 2004).
Although the specific functions of most Acps are unknown, null mutations, RNA interference knockdowns, and ectopic expression of several Acp genes have facilitated characterization of their functions. For example, females who do not receive the 36-amino-acid sex peptide (SP; Acp70A; reviewed in Kubli 2003; Swanson 2003) have reduced rates of egg production, are more likely to remate (Chen et al. 1988; Aigaki et al. 1991; Soller et al. 1999; Chapman et al. 2003; Liu and Kubli 2003), eat less (Carvalho et al. 2006), and have a longer life span (Wigby and Chapman 2005). SP also causes females to increase transcript levels of some genes encoding antimicrobial peptides (Peng et al. 2005b). Another Acp, ovulin (Acp26Aa), is a 264-amino-acid male-derived prohormone that stimulates ovulation by females on the first day following mating (Herndon and Wolfner 1995; Heifetz et al. 2000). Acp36DE is a 912-amino-acid glycoprotein, and females mated to males who lack Acp36DE store 10% of the number of sperm normally stored by mated females (Neubaum and Wolfner 1999; Bloch Qazi and Wolfner 2003). Acp29AB is a 234-amino-acid predicted lectin. Association studies (Clark et al. 1995; Fiumera et al. 2005) and genetic data (A. Wong, S. N. Albright, J. D. Giebel, K. Ravi Ram, S. Ji, A. C. Fiumera and M. F. Wolfner, unpublished results) suggest that Acp29AB may play a role in sperm competition. Finally, Acp62F, a 115-amino-acid trypsin inhibitor, one of four Acps that have been shown to be toxic upon ectopic expression (Lung et al. 2002; Mueller et al. 2007) and was suggested to contribute to shortening the life span of mated females, plays a role in sperm competition (Mueller et al. 2008). Despite what we know about the functions of these Acps, it is not known how they act within the female to mediate these important changes.
To begin to explore the genetic changes that occur in the mated female, three separate microarray and proteomic studies (Lawniczak and Begun 2004; McGraw et al. 2004; Mack et al. 2006) compared gene expression profiles between virgin and mated females. Those studies demonstrated that within 1–3 hr after mating, when the physiological effects of Acps on the mated female are first observed, transcript abundance of >1700 genes is altered in whole female flies. However, the magnitude of most of these changes is small (less than twofold), suggesting that the early physiological and behavioral alterations in the mated female do not rely on major transcriptional changes. In addition, consistent with the fact that many Acps pass through the uterus and enter the hemolymph of the female where they may act to modulate her behavior by acting on neuroendocrine targets (Lung and Wolfner 1999), mating-responsive transcript-level changes are not confined to the reproductive tract. Many of the transcript-level changes following mating in whole female flies (Lawniczak and Begun 2004; McGraw et al. 2004; Mack et al. 2006) were not detected when only the lower reproductive tracts were examined (Mack et al. 2006). These data suggest that many transcriptional changes likely occur in other parts of the female fly.
Although comparison of gene expression profiles of virgin and mated females provided valuable information regarding the numbers, classes, and time frame of mating-induced transcript-level changes, these studies cannot detect the specific factors that trigger gene expression changes resulting from mating. To disentangle how sperm, Acps, or non-sperm/non-Acp components of mating contribute to these gene expression changes, McGraw et al. (2004) mated females to males that either do not transfer sperm or do not transfer Acps and sperm. This allowed for differentiation of the effects of sperm and Acps on female gene expression. While sperm alone were found to modulate the expression of 549 genes, Acps alone were found to regulate the transcript abundance of up to 160 genes in mated females (McGraw et al. 2004). For example, receipt of Acps by females resulted in upregulation of 12 genes involved in immune/defense response, and 5 of these were upregulated twofold or more. In addition, Acps were shown to cause upregulation of cytochrome P450 genes, transporter genes, and proteases in mated females, but downregulation of several genes involved in ligand binding or metabolism.
In this study, we further dissected the transcript-level changes that occur in mated females using DNA microarrays representing ∼88% of D. melanogaster version 4.1 annotated genes. First, although it has been observed that in the reproductive tract transcriptional changes are small shortly after mating but at later time points post-mating larger-scale changes occur (Mack et al. 2006), it was unknown whether these patterns would also hold in whole female flies. To test this, we compared transcriptome profiles in whole bodies of females at four different times post-mating. Second, we predicted that shortly after mating, when Acps have been shown to have their first effects on the female, individual Acps would generate unique changes to the mated female's transcriptome profile. By mating females to one of four strains of males with a null mutation for an individual Acp (ovulin, Acp36DE, Acp29AB, or Acp62F), we compared transcriptome profiles between females who did or did not receive a specific Acp from a male.
MATERIALS AND METHODS
Drosophila stocks and crosses:
D. melanogaster were reared on standard yeast/glucose medium at 25° under a 12:12-hr light:dark cycle. All females used in these studies were from a highly inbred Canton-S laboratory strain. Virgin flies were collected within 4 hr of eclosion and aged in single-sex groups in 25 × 95-mm vials on standard yeast/dextrose media until they were 3 or 4 days old. Within 3 hr of the beginning of their light cycle, single, aged virgin females were paired individually in a fresh vial with single, aged virgin males of the appropriate genotype and allowed to mate. All matings were observed and females that mated for <∼18–20 min were discarded. Flies were flash frozen in liquid nitrogen after copulation ended at the appropriate time point (see below). Virgin females used as a reference sample for the time-course study were reared as above and flash frozen alongside experimental females.
To determine the time course of the females' post-mating transcriptome changes, wild-type Canton-S males were mated to wild-type Canton-S females. Randomly chosen subsets of females were flash frozen at four time points (1–3, 6–8, 12, and 24 hr) following the termination of copulation. Labeled cDNA derived from mated females from each time point was competitively hybridized on microarrays to differentially labeled cDNA from virgin females as described below.
To determine the effect of individual Acps on the females' post-mating transcriptome, cDNAs with a different fluorescent label—from wild-type Canton-S females mated to null mutant males and from wild-type Canton-S females mated to genetically matched control males—were competitively hybridized to microarrays. To ensure that females differed only in whether they did or did not receive an Acp from her mate, the genetic background of control males was made to be as identical as possible to that of mutant males. Acp null mutant and genetically matched control males were derived from the following stocks: (1a) stock 3211—Acp36DE1/Df(2L)H20 that transfer normal quantities of sperm and seminal fluid, but no Acp36DE (Neubaum and Wolfner 1999) and (1b) stock 3191—Acp36DE+/Df(2L)H20 males controlled for the background of the line listed above as well as for effects of other loci uncovered by the deficiency (see Neubaum and Wolfner 1999); (2a) Acp26Aa1/Df(2L)PM101 males (see Herndon and Wolfner 1995) that do not transfer ovulin or sperm and (2b) Acp26Aa+/CyO males that transfer both sperm and ovulin; (3a) Acp62F1b/Acp62F1b males that transfer normal amounts of sperm and seminal fluid, but no Acp62F and (3b) Acp62F1b/TM3,Sb control males (Mueller 2005; Mueller et al. 2008); (4a) Acp29AB1/Df(2L)ED611 males that transfer sperm and seminal fluid proteins, but no Acp29AB (A. Wong, S. N. Albright, J. D. Giebel, K. Ravi Ram, S. Ji, A. C. Fiumera and M. F. Wolfner, unpublished results) and (4b) Df(2L)ED611/CyO control males.
RNA preparation and probing of microarrays:
Since matings were performed over several days, frozen females were pooled and 25 females from each treatment group were randomly chosen for each RNA extraction to minimize the effects of day of mating. Total RNA was extracted from previously frozen females using Trizol (Invitrogen) according to manufacturer protocols and purified with RNeasy mini kits (QIAGEN). RNA quality was assessed using the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) and quantified using a Nanodrop spectrophotometer. Direct labeling of cDNA was conducted by the Duke University Microarray Facility. Primer annealing was performed by incubating 10 μg of total RNA in 10 μl of RNAase-free water with 1 μl of oligo(dT) primer for 10 min at 65° and 5 min at 25°. First-strand labeled cDNA synthesis was performed by combining 5 μl 5× reaction buffer, 2.5 μl 0.1 m DTT, 1.25 μl Cy (3 or 5) dCTP, 1.25 μl Cy (3 or 5) dUTP, and 2.0 μl of Superscript III (Invitrogen) to the primer annealing reaction, incubated at 42° for 2 hr, denatured at 95° for 5 min, hydrolyzed with 6.25 μl 1 m NaOH at 37° for 10 min and neutralized with 6.26 μl 1 m Tris–HCl and 6.25 μl 1 m HCl. Samples were then purified with QiaQuick PCR purification kits (QIAGEN) and dried with a speed vacuum for ∼38 min.
Microarray slides (DGRC-1 transcriptome microarrays; Johnston et al. 2004) were prehybridized in filtered buffer containing 5× SSC, 0.1% SDS, and 0.5% BSA for 1 hr at 42°. Samples were then resuspended in 85 μl of hybridization buffer (42.5 μl formamide, 20.4 μl RNase-free water, 21.25 μl 20× SSC, and 0.85 μl 10% SDS) and 1.85 μl of COT-1 DNA and 1.9 μl of poly(dA) was added. The hybridization mixture was then denatured at 95° for 2 min. Arrays were hybridized at 42° overnight. Slides were then washed in 1× SSC, 0.2% SDS at 42° briefly to remove coverslips, 1× SSC, 0.2% SDS at 42° for 1 min, 0.2 SSC, 0.1% SDS at room temperature for 1 min, 0.2× SSC at room temperature for 2 min, 0.05× SSC at room temperature for 1 min and centrifuged at 1000–1500 rpm for 2 min. Axon GenePix 4000B and GenePixPro 5.0 software were used to scan and analyze fluorescent signals.
For each time point analyzed (1–3, 6–8, 12, and 24 hr) and for each Acp analyzed (ovulin, Acp36DE, Acp29AB, and Acp62F), four separate DGRC-1 transcriptome microarrays were hybridized. The labeling of Cy3 and Cy5 dyes was reversed (dye flip) for two of the four replicate arrays to control for dye bias associated with unequal incorporation of the two Cy dyes into cDNA (Jin et al. 2001). Each labeled cDNA sample was derived from an independent RNA extraction.
Data analysis:
Raw fluorescence intensities from GenePixPro 5.0 were range adjusted on the basis of the maximum expression value of all elements on the array and log2 transformed to obtain approximately normal distribution of data points (Gibson and Wolfinger 2004). Data from spots whose fluorescence was lower than the mean background intensity level of the array were removed from the analysis. In the time-course study, removal of these spots resulted in a remaining total of 6795 spots for the 1- to 3-hr time point, 7412 spots for the 6- to 8-hr time point, 6995 spots for the 12-hr time point, and 7009 spots for the 24-hr time point. A total of 6095 genes were represented in all four time points. In the experiment with Acp null mutants, removal of aberrant spots resulted in a remaining total of 6301 spots for ovulin, 5156 spots for Acp36DE, 6582 spots for Acp29AB, and 8404 spots for Acp62F. A mixed model ANOVA procedure performed in SAS was then used to compute relative intensity values for each spot relative to the sample mean and to adjust for overall array and dye effects (Chu et al. 2002; Hsieh et al. 2003; Gibson and Wolfinger 2004). Residuals of this model, or relative fluorescence intensities, were then compared on a gene-by-gene basis using the following model:
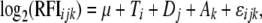 |
where log2(RFIijk) is the relative intensity of the spot, T is the fixed effect of the ith treatment (i = 1, 2), D is the fixed effect of jth dye (Cy3 or Cy5), and A is the random effect of the kth array (k = 1, … , 4). Random effects were presumed to be normally distributed with a mean of zero and a variance of σ2. The mean and unexplained error is represented by μ and ɛ, respectively. Once expression values were generated for each gene over all treatments, the DIFFS procedure in PROC MIXED was used to estimate the magnitude and significance in expression levels between treatments. Statistical significance was assessed at the test-wise threshold of P ≤ 0.05 and P ≤ 0.01. To control for multiple testing, we employed the approach of Storey and Tibshirani (2003) and calculated the 5% false discovery rate (FDR) and Q-value cutoff for each analysis. Overrepresentation of gene classes was assessed with EASE (Hosack et al. 2003). Hierarchical clustering analysis was performed with Cluster (Eisen et al. 1998) and visualized using Maple Tree (http://mapletree.sourceforge.net/).
RESULTS AND DISCUSSION
Time-course analysis of post-mating transcript-level changes in female D.melanogaster:
Although we detected many mating-responsive genes in females, different significance cutoff values gave quite different sets of genes (Table 1). For the remainder of this discussion, we will focus on the genes in the stringent 5% FDR (Q-value) set, and we will primarily discuss genes that exhibited transcript-level changes of twofold or more in mated vs. virgin females. Identities of all other genes can be accessed in the supplemental material.
TABLE 1.
Differentially expressed genes at different time points post-mating relative to virgin females
| Significance cutoff | No. of genes upregulated | No. of genes downregulated |
|---|---|---|
| 1–3 hr post-mating | ||
| P ≤ 0.05 | 1698 | 713 |
| P ≤ 0.01 | 875 | 272 |
| Q-value | 1563 | 616 |
| 6–8 hr post-mating | ||
| P ≤ 0.05 | 483 | 379 |
| P ≤ 0.01 | 408 | 317 |
| Q-value | 74 | 39 |
| 12 hr post-mating | ||
| P ≤ 0.05 | 869 | 399 |
| P ≤ 0.01 | 354 | 108 |
| Q-value | 128 | 41 |
| 24 hr post-mating | ||
| P ≤ 0.05 | 611 | 502 |
| P ≤ 0.01 | 164 | 169 |
| Q-value | 55 | 56 |
The number of differentially expressed genes at four time points post-mating at three different significance cutoffs. “Upregulated” designates the number of genes with higher transcript abundance than in virgin females, while “downregulated” designates the number of genes with lower transcript abundance than in virgin females.
In comparisons of females from each post-mating time point to virgin females, we detected 2179 differentially expressed genes (32.1% of 6795 genes) at 1–3 hr, 113 differentially expressed genes (1.5% of 7412 genes) at 6–8 hr, 170 differentially expressed genes (2.4% of 6995 genes) at 12 hr, and 110 differentially expressed genes (1.6% of 7009 genes) at 24 hr.
At 1–3 hr post-mating, 1563 genes had higher transcript levels in mated females than in virgin females (a fourfold or greater increase in transcript levels): CG3739, CG4476, CG6910, Odorant binding protein 99a (Obp99a), and female-specific independent of transformer (fit). Another 64 genes exhibited between two- and fourfold increases in transcript levels; because they are too numerous to list here, these genes can be accessed in the supplemental material. A total of 616 genes had lower transcript abundance relative to virgin females (fourfold or greater decrease in transcript levels: CG15351, Actin88F, Chorion protein 36, Chorion protein 38, flightin, Neuropeptide-like precursor 3, and Troponin C4) compared to virgin females. Of the genes exhibiting a decreased transcript abundance of twofold or more, those involved in eggshell formation (six genes), muscle contraction (five genes), and proteolysis (eight genes) were significantly overrepresented (EASE score = 0.001 for all cases). At 6–8 hr post-mating, transcript abundance of 74 genes was greater than in virgin females [twofold or greater increase in transcript levels: CG16743, CG17234, CG18067, Immune molecule 2, Jonah 25Bi, and lethal (2) essential for life] relative to virgin flies while 39 genes had lower transcript levels relative to virgin females (twofold or greater decrease in transcript levels: CG7953, CG16758, and CG16775). At 12 hr post-mating, transcript levels of 128 genes were greater than in virgin females (twofold or greater increase in transcript levels: CG15351, CG31661, Odorant binding protein 99a, Vitelline membrane 26Aa, Vitelline membrane 29Ab, Vitelline membrane 34Ca, yellow-g, Yolk protein 1, and Yolk protein 2) while 41 genes had lower transcript levels relative to virgin females (twofold or greater decrease in transcript levels: CG1213, CG1648, CG16758, and upheld). Finally, at 24 hr post-mating, transcript levels of 55 genes were greater than in virgin females (twofold or greater increase in transcript levels: CG9050, CG15351, yellow-g, Vitelline membrane 26Aa, Vitelline membrane 32E, Vitelline membrane 34Ca, Yolk protein 1, Yolk protein 2, and Yolk protein 3) and 56 genes had lower transcript levels (twofold or greater decrease in transcript levels: transient receptor potential) relative to virgin females. The genes mentioned in the results and discussion are presented in Table 2.
TABLE 2.
Genes with high fold-change in transcript levels in mated females relative to virgin females
| Gene | Log2 fold change |
|---|---|
| Genes regulated at 1–3 hr post mating | |
| CG3739 | 1.20 |
| CG4476 | 1.46 |
| CG6910 | 1.13 |
| Odorant binding protein 99a | 1.05 |
| female-specific independent of transformer | 1.26 |
| CG15351 | −2.23 |
| Actin88F | −2.90 |
| Chorion protein 36 | −2.75 |
| Chorion protein 38 | −2.93 |
| flightin | −2.08 |
| Neuropeptide-like precursor 3 | −2.04 |
| Troponin C4 | −2.05 |
| Genes regulated at 6–8 hr post-mating | |
| CG16743 | 1.36 |
| CG17234 | 1.54 |
| CG18067 | 1.21 |
| Immune molecule 2 | 1.26 |
| Jonah 25Bi | 2.15 |
| Lethal (2) essential for life | 1.00 |
| CG7953 | −1.16 |
| CG16758 | −1.01 |
| CG16775 | −1.07 |
| Genes regulated at 12 hr post-mating | |
| CG15351 | 1.09 |
| CG31661 | 1.23 |
| Odorant binding protein 99a | 1.27 |
| Vitelline membrane 26Aa | 1.34 |
| Vitelline membrane 26Ab | 1.59 |
| Vitelline membrane 34Ca | 1.44 |
| yellow-g | 1.75 |
| Yolk protein 1 | 1.60 |
| Yolk protein 2 | 1.74 |
| CG1213 | −1.30 |
| CG1648 | −1.07 |
| CG16758 | −1.26 |
| upheld | −1.26 |
| Genes regulated at 24 hr post-mating | |
| CG9050 | 1.70 |
| CG15351 | 2.56 |
| yellow-g | 3.15 |
| Vitelline membrane 26Aa | 1.88 |
| Vitelline membrane 32E | 2.12 |
| Vitelline membrane 34Ca | 2.19 |
| Yolk protein 1 | 1.62 |
| Yolk protein 2 | 1.87 |
| Yolk protein 3 | 1.87 |
| transient receptor potential | −1.21 |
Only genes within the Q-value set that exhibit fourfold or greater changes in expression levels are shown for the 1- to 3-hr time point. Genes exhibiting twofold or greater differences in transcript levels are given for all other time points. Data for all other genes can be accessed in the supplemental material.
Transcript-level changes are greater in magnitude (but fewer in number) by 6 hr post-mating:
Although at 1–3 hr post-mating, the number of genes whose transcript levels were altered by mating was significantly greater than at later time points, the overall magnitude of these changes was significantly lower (one-way ANOVA of log2 fold change for all genes within the Q-value set each time point: F3, 2571 = 3.07, P = 0.03). Only 2.9% of the genes whose expression levels were altered relative to virgin females at this time point exhibited twofold or greater changes in transcript levels compared to virgin females. At later post-mating time points, we find a smaller number of differentially expressed genes relative to virgin females, although the proportion of genes exhibiting twofold or greater changes is significantly higher than at 1–3 hr post-mating (Table 3). No differences in the proportion of genes with twofold or greater changes in transcript abundances were detected in pairwise comparisons of any of the later time points.
TABLE 3.
Proportion of transcriptional changes twofold in magnitude or greater in mated females relative to virgin females
| Time point (hr) | No. of twofold or greater changes | % total changes | Probability of being different from 1 to 3 hr |
|---|---|---|---|
| 1–3 | 64/2179 | 2.94 | — |
| 6–8 | 9/113 | 7.96 | 0.008 |
| 12 | 13/169 | 7.69 | 0.003 |
| 24 | 10/111 | 9.01 | 0.002 |
The total number of genes exhibiting twofold or larger changes in magnitude is shown over the total number of significantly differentially regulated genes (using Q-value cutoff) at each time point and the percentage of each of these fractions is given. The probability of this value being significantly different from the 1- to 3-hr time point (from Fisher's exact test).
These data lend further support to our previous hypothesis that sexually mature females are molecularly “poised” to respond to mating (Heifetz and Wolfner 2004; McGraw et al. 2004; Mack et al. 2006). In other words, large transcript-level changes do not appear to be required to activate important and immediate post-mating events such as sperm transport and initiation of egg laying. These results are further supported by a recent report. Mack et al. (2006) assessed gene expression profiles in the lower reproductive tracts of D. melanogaster females at four time points following copulation. At 3 hr following copulation, females had altered transcript levels of 55 genes in the lower reproductive tract and most of these were small-magnitude changes in expression levels. However, at 6 hr following copulation, females exhibited peak expression differences. This suggests that, like our analysis of the whole-body transcriptome, transcript-level changes in specific tissues over time may also exhibit analogous post-mating expression patterns.
Transcript abundance of follicle-cell-expressed eggshell formation genes is decreased shortly after mating:
Virgin, sexually mature D. melanogaster females rarely oviposit; however, once mating occurs, females ovulate and lay between 40 and 80 eggs per day (Manning 1967; Herndon and Wolfner 1995; Kubli 1996). In virgin females, once a maximum number of mature oocytes (stage 14) have accumulated within the ovary, the remaining immature oocytes are arrested at stage 9 (Soller et al. 1997). Thus, upon mating, the first eggs that are laid are the late- or mature-stage oocytes present in the female prior to mating (Mahowald and Kambysellis 1980). These oocytes have likely already acquired necessary egg formation proteins. However, subsequently laid eggs must be released from stage 9 arrest and continue through oogenesis where synthesis and deposition of eggshell proteins from follicle cells is required (reviewed in Orr-Weaver 1991).
We observed that several genes expressed in follicle cells and required for eggshell formation during oogenesis (four vitelline membrane genes, two chorion protein genes, and yellow-g and yellow-g2) have lower transcript levels at 1–3 hr after mating than in virgin females. However, at later time points, transcript levels of these genes are greater than or similar to virgin levels. Thus, the initial deficit in abundance of these RNAs likely reflects the death of follicle cells once eggshell proteins have been deposited onto the first eggs released following mating. The high levels of these transcripts observed at 12 and 24 hr post-mating likely reflects the production of new oocytes being synthesized once the mature oocytes are cleared from the ovaries, after stage 9 arrest of oogenesis has been released.
Currently little is known of the mechanisms that trigger the mated female to begin synthesizing the proteins required for eggshell formation. However, at least one seminal fluid protein, SP, stimulates egg laying for up to 6 hr in mated females (reviewed in Kubli 2003) and longer if sperm are also present in the female (Peng et al. 2005a). SP is suggested to modulate juvenile hormone (JH) levels (Moshitzky et al. 1996) and JH-dependent pathways that subsequently affect synthesis of yolk proteins in the mated female (Soller et al. 1997, 1999). Furthermore, the release of oocytes from stage 9 arrest appears to be dependent on receipt of SP (Chen et al. 1988; Aigaki et al. 1991; Soller et al. 1997, 1999). In addition to SP, other components of the seminal fluid and sperm may also contribute to these events. Another seminal fluid protein, ovulin (Acp26Aa), stimulates the female's ovulation rate between 1.5 and 6 hr following mating and is thought to function through interactions with neuromuscular targets near the lateral oviduct or by acting on neuroendocrine targets (Heifetz and Wolfner 2004; Heifetz et al. 2005). In the absence of sperm transfer, oocyte development and egg-laying rates are substantially decreased (Xue and Noll 2000; Heifetz et al. 2001). Even when sperm are transferred, if they are not properly stored, egg-laying rates are less than half of wild-type levels (Neubaum and Wolfner 1999).
Comparison of these genes to other post-mating gene expression studies:
We compared the genes identified in this time-course analysis to genes that were previously shown using Affymetrix GeneChips to be regulated by sperm, by Acps, or by other non-sperm/non-Acp aspects of mating at 1–3 hr after the end of copulation (McGraw et al. 2004). These results are summarized in Table 4. Surprisingly, of the genes that were assayed in both experiments, only 36.47% of the genes that we found to be regulated by mating in the previous study were also identified in this study. However, of the genes that appeared in both studies, over half of the transcriptional changes (55.80%) were in the same direction. Not surprisingly, at 6–8, 12, and 24 hr post-mating, only a small percentage of the changes that we previously detected at 1–3 hr post-mating continue to exhibit transcriptional changes at later time points.
TABLE 4.
Comparison of genes identified in this study to genes previously shown to be regulated by Acps, sperm, or non-sperm/non-Acp components of mating
| Regulated by | Total overlap (%) | Consistent (%) |
|---|---|---|
| 1–3 hr post mating | ||
| Acps | 36/83 (43.37) | 13/36 (36.11) |
| Sperm | 118/316 (37.34) | 86/118 (72.88) |
| Non-sperm/non-Acp | 122/365 (33.42) | 55/122 (45.08) |
| Total | 276/764 (36.13) | 154/276 (55.80) |
| 6–8 hr post-mating | ||
| Acps | 10/96 (10.42) | 8/10 (80) |
| Sperm | 15/338 (4.44) | 12/15 (72.88) |
| Non-sperm/non-Acp | 12/410 (2.93) | 12/12 (100) |
| Total | 37/844 (4.38) | 32/37 (86.49) |
| 12 hr post-mating | ||
| Acps | 2/86 (2.33) | 0/2 |
| Sperm | 10/23 (3.10) | 8/10 (80) |
| Non-sperm/non-Acp | 8/377 (2.12) | 4/8 (50) |
| Total | 20/786 (2.54) | 12/20 (60) |
| 24 hr post-mating | ||
| Acps | 1/87 (1.15) | 1/1 (100) |
| Sperm | 5/321 (1.56) | 4/5 (80) |
| Non-sperm/non-Acp | 2/380 (0.53) | 2/2 (100) |
| Total | 8/788 (1.02) | 7/8 (87.5) |
For each time point, the total number of genes found to be significantly differentially regulated (at the Q-value significance cutoff) in this study is given over the total number of genes that were significantly differentially regulated in McGraw et al. (2004). Only genes that were represented on both microarray platforms were considered. The percentage of this total is given in parentheses. “Consistent” indicates the total number of genes that exhibited consistency in the directionality of changes (e.g., increased or decreased transcript levels relative to virgin females) in both studies over the total number of genes significantly differentially expressed in both studies. The percentage of this total is also given in parentheses.
The discrepancies that we observed between the 1- and 3-hr post-mating time points in the two experiments may be the result of differences in the sensitivities of the microarray platforms used or of small variations in conditions (e.g., media) that may have slightly differed between the times that the two experiments were performed (see Anholt and Mackay 2004) although the flies and manipulations in this experiment were carefully controlled to be as similar to the previous study as possible. Alternatively, we postulate that many of the small-magnitude fluctuations in transcript-level changes that are observed at 1–3 hr post-mating are akin to a stress response as the female adjusts to the presence of foreign sperm and seminal fluid proteins within her body. These small fluctuations may vary as a result of complex interactions between the genotypes of the sexual partners (L. A. McGraw, A. G. Clark and M. F. Wolfner, unpublished results) as well as from subtle environmental influences (Amitin and Pitnick 2006; McGraw et al. 2007).
Post-mating transcriptional changes are not limited to the female's reproductive tract:
Many of the seminal fluid proteins that are responsible for much of the female's dramatic behavioral and physiological responses to mating do not remain within the reproductive tract, but instead pass through the uterus and enter the hemolymph where they may target other tissues (Monsma et al. 1990; Lung and Wolfner 1999; Ravi Ram et al. 2005). In this study and a previous study (McGraw et al. 2004), we assayed the whole-body transcriptome of mated female D. melanogaster to avoid potentially missing transcriptional changes that may be occurring elsewhere in the body. However, because sampling whole bodies of mated females may dilute tissue-specific transcriptome changes, we compared the mating-regulated genes from this study to genes that were found to differ in transcript abundance 3 hr post-mating in the lower reproductive tract of female D. melanogaster (Mack et al. 2006). Although numerous differences are likely to be observed between data sets from time points that differ by a range of 2 hr, of the 2179 genes that we found to be differentially expressed at 1–3 hr post-mating, 12 of these were also regulated at 3 hr post-mating in the lower reproductive tract (CG11880, CG18067, CG2852, CG2918, CG4825, CG5484, CG5791, CG6084, T-complex Chaperonin 5, Chorion protein 1, Sec61β, and Tcp1-like). While 8 of these genes were modulated in the same direction in both data sets, transcript levels of 3 (CG2918, CG5791, and CG18067) were greater in the reproductive tract, but lower when examining the whole female fly. One gene, Sec61β, had lower transcript levels in the reproductive tract but higher transcript levels when examining the whole fly. Comparing our 6- to 8-hr time point to genes that were differentially expressed in the female reproductive tract at 6 hr post-mating revealed 8 genes (CG3756, CG9080, CG18067, Arginine methyltransferase 8, lethal (2) essential for life, selR, sister of feo, and Tim8) in common with both data sets. Transcript abundance of 7 of these genes was in the same direction in both tissue samples, but one gene, sister of feo, had higher transcript levels in the reproductive tract but lower transcript levels in tissue from whole flies. None of the genes that we identified at 24 hr post-mating in whole female flies was expressed in the lower reproductive tract at this same time point.
Comparing our data to those of Mack et al. (2006) demonstrates two important properties of post-mating transcriptional profiles in females. First, we identified a large number of genes whose transcript levels were altered in whole female flies at several time points post-mating; however, only a small portion of these genes are differentially expressed within the lower reproductive tract of the mated female. This suggests that many of the genes involved in the female's response to mating are not limited to expression in the lower reproductive tract. Although many of the genes that we identified, such as those involved in egg formation, are primarily expressed in ovaries (and thus not detected by Mack et al. 2006), it is likely that transcriptional changes that occur in the brain, fat body, or other tissues also make important contributions to the changes in behavior and physiology of mated females. Second, a few genes detected in this study and in the study by Mack et al. (2006) were modulated in one direction in the female's lower reproductive tract, but in the opposite direction when assaying whole female bodies. This suggests that the same gene could be differentially regulated in different tissues. Future studies examining tissue specificity of the gene expression changes that we have identified in this study will be necessary for understanding how these genes contribute to the mating-induced behavioral and physiological changes that occur in females after mating.
Gene expression consequences of four individual Acps:
In comparisons of females mated to either control or Acp null males, at the test-wise threshold of P ≤ 0.05 in the 1- to 3-hr post-mating tests, we detected 776 (12.32% of 6301) genes whose transcript levels are altered by the receipt of ovulin, 514 genes (9.97% of 5156 genes) by the receipt of Acp36DE, 2106 genes (32.0% of 6582 genes) by the receipt of Acp29AB, and 2849 genes (33.9% of 8404 genes) by the receipt of Acp62F. The 5% FDR set of genes contained only 1 gene for ovulin contrasts, 0 genes for Acp36DE contrasts, 413 genes (6.27%) for Acp29AB contrasts, and 2151 genes (25.59% of 6582 genes) for Acp62F contrasts. Here, we focus our detailed description of the genes that we identified using a moderately conservative significance level of P ≤ 0.001. However, because many of the genes that we identified at the 5% significance cutoff level were previously shown to be mating regulated (Lawniczak and Begun 2004; McGraw et al. 2004), in some places in the text we also report genes that fall into the P ≤ 0.05 cutoffs and note this in the text. These significance thresholds were chosen so as not to omit genes with small changes that, although they did not meet the most stringent statistical cutoff, may nonetheless be biologically significant. Details of all genes can be accessed in the supplemental material.
Two Acps have little or no detectable effect on the female's transcriptional profile at 1–3 hr post-mating:
Because both ovulin and Acp36DE elicit their effects in females almost immediately following copulation, we chose a 1- to 3-hr time frame to measure post-mating transcriptional responses. Our data (described below) suggest, however, that neither ovulin nor Acp36DE contribute extensively to the female's transcriptome changes at this time point. Instead, as has been previously suggested, ovulin may act by binding to or interacting with previously existing neuromuscular or neuroendocrine targets (Heifetz et al. 2001, 2005), while Acp36DE may interact directly with sperm and/or other targets (Bloch Qazi and Wolfner 2003; Adams and Wolfner 2007). These observations are consistent with the hypothesis that females are already molecularly “poised” to respond to mating and to the Acps that they receive from their mates (Heifetz and Wolfner 2004; McGraw et al. 2004; Mack et al. 2006).
Two Acps trigger substantial effects on the female's transcriptome:
In contrast to ovulin and Acp36DE, Acp29AB and Acp62F appear to elicit a large number of gene expression changes in the mated female even after correcting for a conservative 5% FDR. Interestingly, although both of these Acps appear to contribute substantially to the upregulation of genes involved in egg production and in muscularity (see below), analyses of Acp29AB and Acp62F null males have not detected an ovulation or egg-laying phenotype, although these Acps may contribute to sperm storage and/or competition phenotypes (Mueller 2005; A. Wong, S. N. Albright, J. D. Giebel, K. Ravi Ram, S. Ji, A. C. Fiumera and M. F. Wolfner, unpublished results). It is possible that these Acps may exert effects at a later time points after mating than we assayed here.
Ovulin (Acp26Aa):
In the P ≤ 0.001 subset of genes whose expression was modulated in females by the receipt of ovulin in the seminal fluid, only 2 of 40 mating-regulated genes had twofold or greater increases in transcript levels (Chorion protein 15 and Jonah 25Bii; see Table 5) and no genes exhibited twofold or greater decreases in transcript abundance. We also examined this subset of 40 genes for gene classes that were represented more than would be expected by chance. Three (7.5%) of the genes encode predicted NADH dehydrogenases [CG3446, CG9140, and lethal (2) 35Di; EASE score = 0.01] although these genes have only slightly reduced transcript levels in females by receipt of ovulin. One gene of particular interest, CG13083, had slightly (less than twofold) elevated transcript levels when females received ovulin from their mates. Although its function is unknown, CG13083's gene product was previously shown in yeast two-hybrid analyses to interact with ovulin (Wong et al. 2006), although this has not been verified biochemically.
TABLE 5.
Genes exhibiting twofold or greater changes in transcript abundance when females received an Acp from their mate relative to females who did not receive an Acp from their mate at the test-wise significance cutoff of P < 0001
| Gene | Fold change | P-value |
|---|---|---|
| Genes regulated by ovulin | ||
| Chorion protein 15 | 2.19 | <0.001 |
| Jonah 25Bii | 1.31 | <0.001 |
| Genes regulated by Acp36DE | ||
| CG30497 | 1.03 | 0.001 |
| Genes regulated by Acp29AB | ||
| CG7953 | 1.43 | <0.001 |
| CG8693 | 1.34 | 0.001 |
| CG8997 | 1.30 | <0.001 |
| CG14368 | 1.03 | <0.001 |
| CG16775 | 1.02 | <0.001 |
| CG18180 | 1.28 | 0.001 |
| defective chorion 1 | 1.07 | <0.001 |
| la costa | 1.00 | <0.001 |
| pale | 1.05 | <0.001 |
| Lysozyme E | 1.44 | 0.001 |
| Neuropeptide-like precursor 3 | 2.30 | 0.001 |
| Senescence marker protein-30 | 1.03 | <0.001 |
| Vitelline membrane 32E | 1.17 | <0.001 |
| Genes regulated by Acp62F | ||
| CG12374 | 1.10 | 0.001 |
| CG16758 | 1.02 | <0.001 |
| Chorion protein 15 | 2.42 | <0.001 |
| Chorion protein 19 | 1.53 | 0.001 |
| Neuropeptide-like precursor 3 | 1.36 | 0.001 |
Only genes with twofold or greater changes in expression levels are shown. Fold change is given in log2.
The biological and physiological effects of ovulin on the mated female have been well characterized. Ovulin stimulates the release of oocytes from the ovary by increasing the ovulation rate, and its effects are most evident at ∼1.5 hr after the end of copulation (Heifetz et al. 2000). Shortly after entering the female reproductive tract it is proteolytically processed at three cleavage sites, and two processing products localize to the base of the ovary (Monsma and Wolfner 1988; Monsma et al. 1990; Park and Wolfner 1995) while full-length ovulin enters the hemolymph (Monsma et al. 1990; Herndon and Wolfner 1995; Heifetz et al. 2000). Ovulin can be detected in the mated female for ∼6 hr after its transfer (Monsma et al. 1990). Ovulin is thought to induce ovulation by interacting with neuromuscular targets near the lateral oviduct or by acting on neuroendocrine targets (Heifetz and Wolfner 2004; Heifetz et al. 2005). Apart from upregulation of transcript abundance of the eggshell protein Chorion protein 15 or ovulin's putative interactor, CG13083 (described above), other genes and gene classes whose transcript levels are affected by the receipt of ovulin are not obviously correlated with its known effects in the mated female. It will be of future value to determine if and how other ovulin-modulated genes contribute to its phenotype.
One caveat in this study was that ovulin null mutant males, in addition to not transferring ovulin, also did not transfer sperm to their mates due to a secondary mutation that arose in the mutant line. Thus, in addition to detecting female genes modulated by ovulin, we also expected to observe transcript differences in genes that were previously shown to be differentially expressed in females as a result of the receipt of sperm (McGraw et al. 2004). Oddly, of the 315 sperm-regulated genes that were detected by McGraw et al. (2004), only 2 were shown to be differentially expressed in females mated to the ovulin/sperm-less males in this study. Several potential explanations are suggested by the data. First, in both this study and the previous study (McGraw et al. 2004), most transcriptional differences were less than twofold in magnitude. As discussed above, these small differences may reflect subtle technical differences or complex interactions between genotypes and/or the environment. A second possibility is that sperm and seminal fluid proteins (such as ovulin) act synergistically to create differences in gene transcript levels in mated females. Future work will be necessary to tease apart these and other explanations.
Acp36DE:
Of the 17 genes (see supplemental material) whose expression was regulated in females by the receipt of Acp36DE in the seminal fluid at the P ≤ 0.001 test-wise threshold, 2 (12.5%) are involved in transcription corepressor activity (C-terminal binding protein and Hira, EASE score = 0.049), and only 1 gene (CG30497, whose function is unknown) had a greater than twofold increase in transcript levels (Table 5).
Acp36DE plays a role in facilitating the entry of sperm into the storage organs of the mated female and its effects are evident between 0.3 and 0.7 hr after the start of mating (Bloch Qazi and Wolfner 2003). Acp36DE undergoes proteolytic cleavage shortly after being transferred to the female (Bertram et al. 1996) and full-length Acp36DE, along with its cleavage products, localizes to sperm storage organs, the anterior mating plug, and at the base of the common oviduct (Bertram et al. 1996; Neubaum and Wolfner 1999; Bloch Qazi and Wolfner 2003) and can be detected in the female 3–4 hr after transfer (Bertram et al. 1996; Bloch Qazi and Wolfner 2003). That this Acp was observed to cause transcriptional changes in only 17 genes suggests that its role in sperm storage does not require modulation of the female's transcriptome. Several hypotheses to explain the action of Acp36DE have been proposed. For example, Acp36DE may act as a scaffold to guide sperm to storage or may corral sperm near the entrance of sperm storage organs or may affect muscle contraction in the female's reproductive tract that helps guide sperm into storage (see Bloch Qazi and Wolfner 2003). That Acp36E did not induce substantial transcriptome changes suggests that it interacts directly with sperm or with muscular targets in the female's reproductive tract.
Acp29AB:
One hundred and forty genes were regulated in females by receipt of Acp29AB in the seminal fluid at the test-wise threshold of P ≤ 0.001 (see supplemental material). All of these genes also fall into the statistically conservative set of genes that are robust to the Q-value cutoff. Expression levels of 13 of these genes were regulated twofold or more and all of these genes were expressed at higher levels when females received Acp29AB from their mates (Table 5). These genes include CG7953, CG8693, CG8997, CG14368, CG16775, CG18180, defective chorion 1, la costa, pale, Lysozyme E, Neuropeptide-like precursor 3, Senescence marker protein-30, and Vitelline membrane 32E. In the subset of genes regulated by Acp29AB, several gene classes are overrepresented, including four genes (2.86%) involved in vitelline membrane formation (Vitelline membrane 26Aa, Vitelline membrane 26Ab, Vitelline membrane 32E, and Vitelline membrane 34Ca; EASE score < 0.0001) and five genes with chymotrypsin activity (higher transcript levels: CG8329, CG17571, and CG18180; lower transcript levels: CG17234 and CG17239; EASE score = 0.05).
Although little is known about the localization or function of Acp29AB, two studies (Clark et al. 1995; Fiumera et al. 2005) and phenotypic characterization of a null mutant (A. Wong, S. N. Albright, J. D. Giebel, K. Ravi Ram, S. Ji, A. C. Fiumera and M. F. Wolfner, unpublished results) suggest that this Acp may play a role in sperm competition. Whether or not the genes whose transcript levels increase after receipt of Acp29AB contribute to sperm competition is unknown. Although Acp29AB has not been shown to contribute to elevated ovulation or egg-laying rates in the mated female, we find that it appears to contribute significantly to elevating transcript abundances of genes whose products compose the egg's vitelline membrane. It will be of future interest to determine Acp29AB's role in the processes of egg formation or movement of eggs within the female's reproductive tract.
Acp62F:
There were 172 genes showing altered transcript levels due to receipt of Acp62F in the seminal fluid at the test-wise significance threshold of P < 0.001 and, like the genes affected by Acp29AB, all of these genes are also included in the Q-value set. Five genes had twofold or higher transcript levels when females receive Acp62F (Table 5; CG12374, CG16758, Chorion protein 15, Chorion protein 19, and Neuropeptide-like precursor 3). Thirteen genes (7.56%) involved in RNA binding are overrepresented in this group (higher transcript levels: Elav; lower transcript levels: CG6937, CG7006, piwi, nanos, U2af38, Nucleolar KKE/D repeat protein, modulo, Negative elongation factor B homolog, Probable ribonuclease HI large subunit, cysteinyl-trna synthetase, Signal recognition particle 19 kD protein; EASE score = 0.04). Six genes (3.49%) involved in eggshell formation (higher transcript levels: Chorion protein 15, Chorion protein 19, and Vitelline membrane 32E; lower transcript levels: cdc42, Lissencephaly-1, and rhino; EASE score < 0.006) and 4 genes (2.33%) involved in immune/defense response (higher transcript levels: Attacin-A, Attacin-B, Drosocin, and Immune induced molecule 3; EASE score < 0.02) are also overrepresented in this group of genes modulated by Acp62F.
Like ovulin and Acp36DE, Acp62F is detectable only in the female for a few hours after mating (Lung and Wolfner 1999). Although the function of Acp62F is still unknown, it has been suggested to play a role in the decreased life span observed in mated females since it is toxic upon ectopic expression in preadult and adult flies (Lung et al. 2002) and may play a role in mediating sperm competition (Mueller et al. 2008). Thus, it is intriguing that this Acp also strongly contributes to upregulation of antimicrobial peptides in the mated female. In addition, localization of this Acp to the sperm storage organs suggests a possible role in sperm storage and/or maintenance (Lung et al. 2002); however, it is unclear how genes modulated by this Acp in females might contribute to these processes nor is it immediately apparent why this Acp contributes to increased transcript abundance of genes encoding eggshell proteins.
Acps may act antagonistically or additively in altering female gene expression changes:
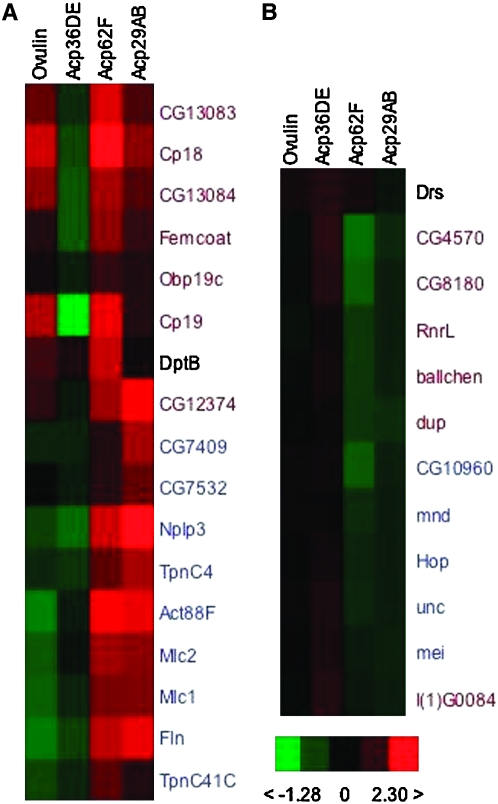
Of the subset of D. melanogaster genes represented on these arrays, expression levels of 29 genes were modulated by all four of the Acps that we tested at the nominal P-value threshold of 5% (Figure 1), although at the more conservative P < 0.001 cutoff, only 2 genes are represented. Of these 29 genes, only transcript levels of Diptericin B were increased by all four Acps.
Figure 1.—
Clustering of genes whose transcript levels were regulated by all four Acps tested. (A and B) Two distinct clustering nodes. Names of genes regulated in one direction by ovulin and Acp36DE and in the opposite direction by Acp29AB and Acp62F are in blue type. Names of genes regulated in an inconsistent direction by Acp36DE are in red type. Red, increased expression; green, decreased expression relative to normalized average expression for that gene among all arrays; black, no change in expression. (Bottom right) Fold change on the log2 scale.
Examination of the remaining 28 genes reveals two unique patterns. First, RNA levels of 14 of these genes are modulated in one direction by ovulin and Acp36DE and in the opposite direction by Acp29AB and Acp62F. For example, 9 of these 14 genes (CG7409, CG7532, Actin88F, flightin, Myosin alkali light chain 1, Myosin light chain 2, Neuropeptide-like precursor 3, Troponin C at 41C, and TpnC4) had lower transcript levels when females received ovulin and Acp36DE, but higher transcript levels when females received Acp29AB and Acp62F (Figure 1A). Interestingly, 6 (Actin88F, flightin, Myosin alkali light chain 1, Myosin light chain 2, Troponin C at 41C, and TpnC4) of these 9 genes are involved in muscle contraction, although the role of these genes in relation to the female's reproductive physiology is unknown. Acps, as a whole, are known to induce morphological changes in the female's reproductive that may occur as a result of muscle contraction/relaxation (Adams and Wolfner 2007) and it is possible that the overall downregulation of 5 of these genes observed at 1–3 hr post-mating may contribute to these changes. These conformational changes may perhaps facilitate sperm movement toward the sperm storage organs. In contrast to the genes just described, 5 of these 14 genes (CG10960, Hsp70/Hsp90 organizing protein homolog, mei-P26, minidisks, and unc-119) had higher transcript levels when females received ovulin and Acp36DE, but lower transcript levels when females received Acp29AB and Acp62F (Figure 1B). The second striking feature of these 29 genes whose RNA levels were found to be modulated by all four Acps is that, for 13 of these genes, Acp36DE drives expression in the opposite direction than do the other three Acps. For example, 6 of these 13 genes (CG4570, CG8180, enhancer of yellow 3, ballchen, double parked, and Ribonucleoside diphosphate reductase large subunit) had higher transcript levels when females received Acp36DE, but lower transcript levels when females received the other three Acps. The remaining 7 genes in the class (CG12374, CG13083, CG13084, Chorion protein 18, Chorion protein 19, femcoat, and odorant-binding protein 19c) had lower transcript levels when females received Acp36DE but higher transcript levels when females received the other three Acps (Figure 1). Three genes that play a role in egg formation are represented in this set of genes. Drosomycin was unique in that its transcript levels were lower when females received Acp29AB, but higher when females received the other three Acps.
Our observations that the individual Acps that we examined in this study can modulate expression of genes in different directions suggests two possible modes of Acp action. First, since we examined gene expression changes in whole female flies, it is possible that Acps may have different effects on transcript levels in different tissues. For example, it is plausible that one Acp causes increased transcript abundance of a gene in the reproductive tract while another Acp causes a decrease in transcript abundance of the same gene in a different tissue. This would be especially true if genes whose transcript levels are affected by Acps exhibit large levels of pleiotropy, like many other genes involved in behavior (see Baker et al. 2001). Consistent with these ideas, the four Acps that we examined target multiple tissues, each with a unique pattern (Lung and Wolfner 1999; Heifetz et al. 2000; Ravi Ram et al. 2005).
Another possible explanation is that some Acps act antagonistically in altering the female's gene expression profiles. This explanation is intriguing from an evolutionary perspective. Compared to nonreproductive proteins, an unusually large number of Acps show evidence of rapid evolution at the sequence level (Swanson et al. 2001), including ovulin, Acp36DE, and Acp29AB (Aguadé et al. 1992; Tsaur and Wu 1997; Aguadé 1998, 1999; Begun et al. 2000; Swanson et al. 2001; Kern et al. 2004). Sexual conflict models are among those that have been proposed to explain the rapid evolution of Acp genes (Swanson and Vacquier 2002b). Under these scenarios, it is often assumed that high rates of evolutionary change in Acps will be compensated for by congruent changes in female Acp receptors (Swanson and Vacquier 2002a,b). Our results raise the possibility that sexually antagonistic coevolution might also occur at the regulatory level. For instance, if a mutation in an Acp causes increased transcription of a female gene that benefits the male, yet comes at a cost to the female, females may adapt by evolving a mechanism to abate this effect by downregulating the gene. Other Acps may then be free to manipulate expression of that gene through other mechanisms or pathways. Future studies to dissect the mating-related functions of potential antagonistically regulated female genes will be necessary to test this idea.
Comparison of these data with previous reports of genes regulated by Acps or mating in D. melanogaster females:
In our previous Affymetrix microarray experiment (McGraw et al. 2004), we identified ∼1783 genes whose expression levels in females were altered by sperm, by Acps as a whole, or by other non-sperm/non-Acp components of mating. We examined the sets of genes that were assayed in both experiments and compared the genes whose expression levels were observed to change in this experiment (at the test-wise threshold of P ≤ 0.001) to results from our previous experiment. These results are summarized in Table 6. Surprisingly, only a small portion of the genes that we identified to be regulated by ovulin, Acp36DE, Acp29AB, or Acp62F in this study were also shown to be regulated by Acps as a group in our previous screen (but note that the previous screen examined the presence/absence of all Acps, rather than targeting them one at a time).
TABLE 6.
Comparison of Acp-regulated genes to genes previously shown to be regulated by Acps, sperm, or non-sperm/non-Acp components of mating
| Regulated by | Total overlap (%) | Consistent (%) |
|---|---|---|
| Ovulin | ||
| Acps | 1/86 (1.16) | 1/1 (100) |
| Sperm | 2/315 (0.63) | 2/2 (100) |
| Non-sperm/non-Acp | 8/368 (2.17) | 5/8 (62.5) |
| Total | 11/769 (1.43) | 8/11 (72.73) |
| Acp36DE | ||
| Acps | 0/31 | — |
| Sperm | 0/71 | — |
| Non-sperm/non-Acp | 0/314 | — |
| Total | 0/416 | — |
| Acp29AB | ||
| Acps | 1/140 (0.71) | 0/1 |
| Sperm | 14/311 (4.50) | 2/14 (16.67) |
| Non-sperm/non-Acp | 13/401 (3.24) | 4/13 (36.36) |
| Total | 18/853 (3.29) | 6/18 (33.33) |
| Acp62F | ||
| Acps | 2/123 (1.63) | 2/2 (100) |
| Sperm | 8/366 (2.19) | 2/8 (25) |
| Non-sperm/non-Acp | 10/562 (1.78) | 5/10 (50) |
| Total | 20/612 (3.27) | 8/20 (40) |
For each Acp assayed, the total number of genes found to be significantly differentially regulated in this study is given over the total number of genes that were significantly differentially regulated in McGraw et al. (2004). Only genes that were represented on both microarray platforms were considered. The percentage of this total is given in parentheses. “Consistent” indicates the total number of genes that exhibited consistency in the directionality of changes (e.g., increased or decreased transcript levels) in both studies over the total number of genes significantly differentially expressed in both studies. The percentage of this total is also given in parentheses.
The role of Acps in upregulating genes involved in the female's immune response:
Our previous microarray screen demonstrated that, as a group, Acps upregulate a large number of genes involved in the female's immune response (McGraw et al. 2004). In particular, that study showed that of the 12 immune response genes whose transcript levels increased in females after receiving Acps from their mates, 5 showed twofold or greater differences in transcript levels. This magnitude of change in transcript levels was dramatic in comparison to all of the other genes and gene classes identified in that screen. That Acps contribute to upregulating the female's antimicrobial peptide transcript levels was also demonstrated in a study of the Acp, SP (Peng et al. 2005b). Compared to females who did not receive SP from their mates, females who received SP have higher levels of Metchnikowin transcripts and RNAs encoding other antimicrobial peptides regulated by both the Toll and Imd immune response pathways (Tzou et al. 2002). Our study extends previous data (Lawniczak and Begun 2004; McGraw et al. 2004; Mueller 2005; Peng et al. 2005b) suggesting that the female's immune responsive gene network is elevated after mating (but see Fedorka et al. 2007). Here, we show that this response is not the consequence of the receipt of a single Acp. Instead, this study, along with a study by Peng et al. (2005b), suggests that several Acps might act additively in generating this effect. Although some of the Acp-regulated immune response genes were not represented on the arrays in this study (e.g., Attacin B), of the ones that were, we find that all four Acps that we analyzed contribute to the upregulation of at least one of these genes (Table 7).
TABLE 7.
Differentially regulated immune response genes
| Mtk | Dro | Drs | DptB | AttA | CecA2 | Im1 | |
|---|---|---|---|---|---|---|---|
| Ovulin | 0.80* | 0.53 | 0.26 | 0.54 | 0.46 | 0.53* | — |
| Acp36DE | — | — | — | 0.33 | — | — | — |
| Acp29AB | 0.23 | — | −0.20 | 0.19 | — | — | — |
| Acp62F | 0.98 | 0.98 | 0.27 | 1.31 | 0.69 | 0.21 | 0.89 |
Fold change is given in log2 where P < 0.05. *P ≤ 0.001.
The significance of post-mating upregulation of immune response genes by Acps is still unknown. Upregulation of these genes may help the female, the female's reproductive tract, or sperm to combat pathogens introduced via copulatory wounding (Kamimura 2007) or via the ejaculate or to prevent future infections (reviewed in Lawniczak et al. 2007). Alternatively, upregulation of antimicrobial peptide genes may occur if the female perceives Acps as non-self molecules.
Conclusions:
To gain a clearer perspective of the genetics underlying the female's post-mating response, we further dissected the transcript-level changes that occur in mated female D. melanogaster. By examining the female's transcriptome at several time points post-mating, we provide additional evidence that females are molecularly poised to respond to mating. During development, females appear to have already synthesized most of the transcripts needed to initially respond to mating. We show that in whole female flies, between 1 and 3 hr post-mating, 32.1% of assayed genes exhibit alterations in transcript abundance compared to virgin females. At later time points following mating, the number of genes with differences in transcript levels is much smaller, although a larger proportion of those genes exhibit twofold or greater changes in transcript abundance and their regulation coincides with the known timing of increased egg production. Comparing the whole-body transcriptome changes that we identified here to the transcriptome changes reported in a previous study that examined only lower reproductive tracts suggests that many mating-induced transcriptional changes are not limited to the reproductive tract but also occur in other tissues of the fly.
We have also demonstrated that four individual seminal fluid proteins contribute to the transcriptome changes between 1 and 3 hr after mating, the time at which the first behavioral and physiological effects are observed in the mated female. We found that while ovulin and Acp36DE caused few to no changes soon after mating, Acp29AB and Acp62F caused numerous transcriptome changes. Each of the Acps that we examined also contributed to the upregulation of antimicrobial peptides in the female; thus the effects of Acps appear to be additive in this respect. However, in other cases, the Acps that we examined appear to have antagonistic effects with one another: some genes are downregulated by one Acp, yet upregulated by another. This apparent fluidity of regulatory control may help explain how male–female interactions can drive the unusually rapid evolution of reproductive genes. The experiments reported here consider only the consequences of either the presence or the absence of targeted Acps on transcript abundance in a single inbred laboratory strain of D. melanogaster. In natural populations, females' transcript-level responses may vary if males possess different allelic forms of a given Acp, further fueling the opportunity for rapid evolution. Modulation of gene regulation in response to male–female interactions results in an intriguing confounding of cause and effect in the evolution of organismal physiology and behavior at the gene regulatory level.
Acknowledgments
We thank C. Aquadro, K. Ravi Ram, L. Sirot, A. Wong, and two anonymous reviewers for helpful comments on the manuscript. We thank the National Institutes of Health (NIH) Neuroscience Microarray Consortium and Duke University Microarray Facility for assistance with expression profiling. Microarrays were obtained from the Drosophila Genomics Resource Center. This work was supported by NIH grant HD38921 to M.F.W., National Science Foundation grant DEB-0108965 to A.G.C., and a traineeship to L.A.M. from NIH training grant T32-GM07617.
References
- Adams, E. M., and M. F. Wolfner, 2007. Seminal proteins but not sperm induce morphological changes in the Drosophila melanogaster female reproductive tract during sperm storage. J. Insect Physiol. 53 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., N. Miyashita and C. Langley, 1992. Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics 132 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigaki, T., I. Fleischmann, P. S. Chen and E. Kubli, 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7 557–563. [DOI] [PubMed] [Google Scholar]
- Amitin, E. G., and S. Pitnick, 2006. Influence of developmental environment on male- and female-mediated sperm precedence in Drosophila melanogaster. J. Evol. Biol. 20 381–391. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., and T. F. C. Mackay, 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 5 838–849. [DOI] [PubMed] [Google Scholar]
- Baker, B. S., B. J. Taylor and J. C. Hall, 2001. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell 105 13–24. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, M. J., D. M. Neubaum and M. F. Wolfner, 1996. Localization of the Drosophila accessory gland protein Acp36DE in the mated female suggests a role in sperm storage. Insect Biochem. Mol. 26 971–980. [DOI] [PubMed] [Google Scholar]
- Bloch Qazi, M. C., and M. F. Wolfner, 2003. An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. J. Exp. Biol. 206 3521–3528. [DOI] [PubMed] [Google Scholar]
- Carvalho, G. B., P. Kapahi, D. J. Anderson and S. Benzer, 2006. Allocrine modulation of feeding behavior by the sex peptide of Drosophila. Curr. Biol. 16 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., 2001. Seminal fluid-mediated fitness traits in Drosophila. Heredity 87 511–521. [DOI] [PubMed] [Google Scholar]
- Chapman, T., and S. J. Davies, 2004. Functions and analysis of the seminal fluid proteins of male Drosophila melanogaster fruit flies. Peptides 25 1477–1490. [DOI] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. S., E. Stumm-Zollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54 291–298. [DOI] [PubMed] [Google Scholar]
- Chu, T. M., B. Weir and R. Wolfinger, 2002. A systematic statistical linear modeling approach to oligonucleotide array experiments. Math. Biosci. 176 35–51. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorka, K. M., J. E. Linder, W. Winterhalter and D. Promislow, 2007. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc. R. Soc. Lond. Ser. B 274 1211–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G., and R. Wolfinger, 2004. Gene expression profiling using mixed models, pp. 251–278 in Genetic Analysis of Complex Traits Using SAS, edited by A. M. Saxton. SAS Users Press, Cary, NC.
- Heifetz, Y., and M. F. Wolfner, 2004. Mating, seminal fluid components, and sperm cause changes in vesicle release in the Drosophila female reproductive tract. Proc. Natl. Acad. Sci. USA 101 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz, Y., O. Lung, E. A. Frongillo and M. F. Wolfner, 2000. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr. Biol. 10 99–102. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., J. Yu and M. F. Wolfner, 2001. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 234 416–424. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., L. N. Vandenberg, H. I. Cohn and M. F. Wolfner, 2005. Two cleavage products of the Drosophila accessory gland protein ovulin can independently induce ovulation. Proc. Natl. Acad. Sci. USA 102 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon, L. A., and M. F. Wolfner, 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg-laying in females for one day following mating. Proc. Natl. Acad. Sci. USA 92 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack, D. A., G. Dennis, B. T. Sherman, H. C. Lane and R. A. Lempicki, 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70. [DOI] [PMC free article] [PubMed]
- Hsieh, W. P., T. M. Chu, R. D. Wolfinger and G. Gibson, 2003. Mixed-model reanalysis of primate data suggests tissue and species biases in oligonucleotide-based gene expression profiles. Genetics 165 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel et al., 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29 389–395. [DOI] [PubMed] [Google Scholar]
- Johnston, R., B. Wang, R. Nuttall, M. Doctolero, P. Edwards et al., 2004. FlyGEM, a full transcriptome array platform for the Drosophila community. Genome Biol. 5 R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura, Y., 2007. Twin intromittent organs of Drosophila for traumatic insemination. Biol. Lett. 3 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2004. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics 167 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli, E., 1996. The Drosophila sex-peptide: a peptide pheromone involved in reproduction. Adv. Dev. Biochem. 4 99–128. [Google Scholar]
- Kubli, E., 2003. Sex-peptides: seminal peptides of the Drosophila male. Cell. Mol. Life Sci. 60 1689–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak, M. K., and D. J. Begun, 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47 900–910. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K., A. I. Barnes, J. R. Linklater, J. M. Boone, S. Wigby et al., 2007. Mating and immunity in invertebrates. Trends Ecol. Evol. 22 48–55. [DOI] [PubMed] [Google Scholar]
- Liu, H., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O., and M. F. Wolfner, 1999. Drosophila seminal fluid proteins enter the circulatory system through the walls of the posterior vagina. Insect Biochem. Mol. Biol. 29 1043–1052. [DOI] [PubMed] [Google Scholar]
- Lung, O., U. Tram, C. Finnerty, M. Eipper-Mains, J. M. Kalb et al., 2002. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics 160 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, P. D., A. Kapelnikov, Y. Heifetz and M. Bender, 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald, A. P., and M. Kambysellis, 1980. Oogenesis, pp. 141–224 in The Genetics and Biology of Drosophila, edited by M. Ashburner and T. Wright. Academic Press, New York.
- Manning, A., 1967. The control of sexual receptivity in female Drosophila. Anim. Behav. 15 239–250. [DOI] [PubMed] [Google Scholar]
- McGraw, L. A., G. Gibson, A. G. Clark and M. F. Wolfner, 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14 1509–1514. [DOI] [PubMed] [Google Scholar]
- McGraw, L. A., A. C. Fiumera, M. Ramakrishnan, S. Madhavarapu, A. G. Clark et al., 2007. Larval rearing environment affects several post-copulatory traits in Drosophila melanogaster. Biol. Lett. 3 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma, S. A., and M. F. Wolfner, 1988. Structure and expression of a Drosophila male accessory gland gene whose product resembles a peptide pheromone precursor. Genes Dev. 2 1063–1073. [DOI] [PubMed] [Google Scholar]
- Monsma, S. A., H. A. Harada and M. F. Wolfner, 1990. Synthesis of two Drosophila male accessory gland proteins and their fate after transfer to the female during mating. Dev. Biol. 142 465–475. [DOI] [PubMed] [Google Scholar]
- Moshitzky, P., I. Fleischmann, N. Chaimov, P. Saudan, S. Klauser et al., 1996. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch. Insect Biochem. Physiol. 32 363–374. [DOI] [PubMed] [Google Scholar]
- Mueller, J. L., 2005. Evolutionary, structural and functional analysis of Drosophila melanogaster seminal fluid proteins. Ph.D. Thesis, Cornell University, Ithaca, NY.
- Mueller, J. L., D. R. Ripoll, C. F. Aquadro and M. F. Wolfner, 2004. Comparative structural modeling and inference of conserved protein classes in Drosophila seminal fluid. Proc. Natl. Acad. Sci. USA 101 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., J. L. Page and M. F. Wolfner, 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics 175 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., J. Linklater, K. Ravi Ram, T. Chapman and M. F. Wolfner, 2008. Targeted gene deletion and phenotypic analysis of the Drosophila melanogaster seminal fluid protease inhibitor Acp62F. Genetics 178 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum, D. M., and M. F. Wolfner, 1999. Drosophila females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., 1991. Drosophila chorion genes: cracking the eggshell's secrets. BioEssays 13 97–105. [DOI] [PubMed] [Google Scholar]
- Park, M., and M. F. Wolfner, 1995. Male and female cooperate in the processing of a Drosophila melanogaster seminal fluid protein. Dev. Biol. 171 694–702. [DOI] [PubMed] [Google Scholar]
- Peng, J., S. Chen, S. Büsser, H. Liu, T. Honegger et al., 2005. a Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr. Biol. 15 207–213. [DOI] [PubMed] [Google Scholar]
- Peng, J., P. Zipperlen and E. Kubli, 2005. b Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15 1690–1694. [DOI] [PubMed] [Google Scholar]
- Poiani, A., 2006. Complexity of seminal fluid: a review. Behav. Ecol. Sociobiol. 60 289–310. [Google Scholar]
- Ravi Ram, K., and M. F. Wolfner, 2007. a Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47 427–445. [DOI] [PubMed] [Google Scholar]
- Ravi Ram, K., and M. F. Wolfner, 2007. b Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 3: e238. [DOI] [PMC free article] [PubMed]
- Ravi Ram, K., S. Ji and M. F. Wolfner, 2005. Fates and targets of male accessory gland proteins in mated female Drosophila melanogaster. Insect Biochem. Mol. Biol. 35 1059–1071. [DOI] [PubMed] [Google Scholar]
- Soller, M., M. Bownes and E. Kubli, 1997. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur. J. Biochem. 243 732–738. [DOI] [PubMed] [Google Scholar]
- Soller, M., M. Bownes and E. Kubli, 1999. Control of oocyte maturation in sexually mature Drosophila females. Dev. Biol. 208 337–351. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., 2003. Sex peptide and the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 9643–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. a Reproductive protein evolution. Annu. Rev. Ecol. Syst. 33 161–179. [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. b The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., Z. Yang, M. F. Wolfner and C. F. Aquadro, 2001. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 98 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., and C.-I. Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14 544–549. [DOI] [PubMed] [Google Scholar]
- Tzou, P., E. De Gregorio and B. Lemaitre, 2002. How Drosophila combats microbial infection: a model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 5 102–110. [DOI] [PubMed] [Google Scholar]
- Wigby, S., and T. Chapman, 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15 316–321. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88 85–93. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., S. Applebaum and Y. Heifetz, 2005. Insect gonadal glands and their gene products, pp. 179–212 in Comprehensive Insect Physiology, Pharmacology and Molecular Biology, edited by L. Gilbert, K. Iatrou and S. Gill. Elsevier, Amsterdam/New York.
- Wong, A., and M. F. Wolfner, 2006. Sexual behavior: a seminal peptide stimulates appetites. Curr. Biol. 16 R256–R257. [DOI] [PubMed] [Google Scholar]
- Wong, A., S. A. Albright and M. F. Wolfner, 2006. Evidence for structural constraint on ovulin, a rapidly evolving Drosophila melanogaster seminal protein. Proc. Natl. Acad. Sci. USA 103 18644–18649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L., and M. Noll, 2000. Drosophila female sexual behavior induced by sterile males showing copulation complementation. Proc. Natl. Acad. Sci. USA 97 3272–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]