Abstract
Using the compiled human genome sequence, we systematically cataloged all tandem repeats with periods between 20 and 2000 bp and defined two subsets whose consensus sequences were found at either single-locus tandem repeats (slTRs) or multilocus tandem repeats (mlTRs). Parameters compiled for these subsets provide insights into mechanisms underlying the creation and evolution of tandem repeats. Both subsets of tandem repeats are nonrandomly distributed in the genome, being found at higher frequency at many but not all chromosome ends and internal clusters of mlTRs were also observed. Despite the integral role of recombination in the biology of tandem repeats, recombination hotspots colocalized only with shorter microsatellites and not the longer repeats examined here. An increased frequency of slTRs was observed near imprinted genes, consistent with a functional role, while both slTRs and mlTRs were found more frequently near genes implicated in triplet expansion diseases, suggesting a general instability of these regions. Using our collated parameters, we identified 2230 slTRs as candidates for highly informative molecular markers.
TANDEMLY repeated sequences are common in higher eukaryotes, accounting for several percent of the human genome (Levy et al. 2007). While much of their functional nature remains enigmatic, tandem repeats have been implicated both in the regulation of gene expression (Nakamura et al. 1998) and human disease (Gatchel and Zoghbi 2005). The latter is epitomized by the triplet expansion diseases, which result from size inflation of both coding and noncoding microsatellite tracts due to replication slippage or unequal crossing over. An interesting behavior has been reported for a tandem repeat near the insulin gene where nontransmitted alleles have been proposed to influence the function of the transmitted alleles in predisposition to type I diabetes (Bennett et al. 1997). This observation is reminiscent of paramutation, a genetic phenomenon where alleles can heritably modify the expression of each other without demonstrable alteration of their underlying DNA sequence (Stam and Scheid 2005). The b1 locus in maize represents the best understood example of paramutation wherein seven tandem repeats of an 853-bp noncoding sequence located ∼100 kb upstream of the transcription start site mediate this phenomenon (Stam et al. 2002).
Most previous characterizations of human tandem repeats predated the availability of the complete genome sequence (Nakamura et al. 1987; Cox and Mirkin 1997) or focused on only a subset of chromosomes (Denoeud et al. 2003; Boby et al. 2005), hence our understanding of the nature and function of tandemly repeated sequences is based upon limited examples. The completion of the human genome sequence allows one to globally survey the tandem repeats in the genome and reexamine many ideas regarding their creation, distribution, maintenance, and function. Because microsatellites have already been extensively described (Li et al. 2002; Buschiazzo and Gemmell 2006), we undertook a comprehensive data-mining effort to characterize all of the tandem repeats in the human genome with periods ranging from 20 to 2000 bp. The lower bound was selected to support identification of their consensus sequences as unique or redundant entities in the genome and the higher bound was mandated by constraints intrinsic to Tandem Repeats Finder (TRF) (Benson 1999). Beginning with the output from this program, we defined two subsets of tandem repeats in the human genome on the basis of whether the sequence was located at a single site or repeated elsewhere in the genome, compiled descriptive characters for each subset, and examined the results in the context of current knowledge of tandem repeats.
MATERIALS AND METHODS
Identification of ALL TRs data set:
To warehouse the genomic data used in this project, a custom PostgreSQL database was developed, making use of the BLASTgres extension. All data were expressed relative to the NCBI 36.1 (March 2006) assembly of the human genome, and were, for the scope of this study, taken from three sources: Ensembl, the University of California Santa Cruz (UCSC) Genome Browser, and the Tandem Repeats Database (TRDB).
The project database was initially populated with all 947,696 tandem repeats identified by TRDB for the March 2006 human genome assembly. To eliminate any redundant entries from the data set, tandem repeats with the longest array length were first selected from among those with overlapping genome coordinates. Subsequently from this group, entries with the shortest period were identified and flagged as nonredundant; related entries with the same genome coordinates were discarded from further analyses. Within their coordinates, some tandem repeats contain internal tandem repeats with distinct period and copy values. We defined a tandem repeat as being a child of another if it was contained fully within the coordinate range of the parent repeat plus a flanking window of 5% of the parent's total length. Child tandem repeats were recursively identified and culled, leaving at each locus a single “root” tandem repeat, which was then also flagged as nonredundant. The flagged tandem repeats with periods between 20 and 2000 bp constitute the ALL TRs set.
Cluster analysis:
Cluster analysis was used to group tandem repeats with similar consensus sequences. Because tandem repeat copies are adjacent, the designation of the first position is entirely arbitrary (Benson 2005) and consensus sequences of two such repeats cannot be directly compared with confidence using single repeat sequences. Instead, we aligned single copies of the consensus sequence (both strands) of the tandem repeat from ALL TRs against two tandemly duplicated copies of all other consensus sequences with BLAST and looked for significant sequence alignments spanning at least 80% of the length of either the query or target, whichever was the longer of the two. The alignment was performed separately for two subsets of ALL TRs: those with periods between 20 and 39 bp and those with periods ≥40 bp. This allowed greater sensitivity for the shorter sequences while avoiding a flood of spurious alignments for the larger ones (E-values: 0.01 and 1e-6, respectively). To avoid corner cases where significant alignments of tandem repeats with different consensus sequence lengths might otherwise have been missed, the databases against which the queries were aligned included all flagged tandem repeats with periods up to 20% shorter or longer than the bounds of their respective query sets. Those tandem repeats for which no sufficiently long significant alignments were found were considered candidate slTRs and the remaining were labeled as mlTRs.
As a consensus sequence may be repeated in tandem at only one locus but may appear elsewhere in the genome as a single copy and we wished to identify candidate slTRs that were truly unique in the genome, the consensus sequences of the candidate slTRs were aligned with BLAST against the human genome sequence. Those for which there was found a significant alignment (≥80% sequence identity) spanning at least 80% of the query sequence were added to the mlTRs set; the remainder form the final set of slTRs. The TRDB identifications of both sets are available in supplemental File 1, and BED files suitable for use as custom tracks in the UCSC Genome Browser are available in supplemental File 2.
RepeatMasker analysis:
RepeatMasker was used to mask the repetitive elements from the ALL TRs set. Alignments were performed with WU-BLAST against version 12.03 of RepBase, using default parameters. Tandem repeats were considered significantly masked and thus placed into the repTR set when ≥80% of their consensus sequence was masked by RepeatMasker; all other remaining tandem repeats form the notRepTR set. TRDB identifications and BED files are also available for these sets in supplemental Files 1 and 2.
Human genes and DNA samples analyzed in this study:
Genes were imported into the project database from version 42.36d of the Ensembl Homo sapiens database (August 2006 genebuild) (Curwen et al. 2004). Several subsets of genes were hand selected after an extensive literature review (i.e., imprinted genes, tumor suppressor genes, and genes related to trinucleotide-repeat expansion diseases); these are available in supplemental File 3.
Lymphoblastoid cell-line DNA samples from individuals selected to represent ethnic diversity were obtained from the Coriell Institute including: NA14935, NA14547, NA14772, NA14439, NA14529, NA04390, NA09820, NA03715, NA14537, NA14819, NA00726, NA00946, NA00037, and NA03469.
Analyses of recombination hotspots:
The set of recombination hotspots was exported from the snpRecombHotspotHapmap table in the UCSC Genome Browser hg17 database and converted to March 2006 coordinates with the LiftOver utility. To analyze the relationship between tandem repeats and recombination hotspots, we first partitioned the genome into a series of adjacent interhotspot regions delimited by consecutive recombination hotspots' midpoints. At the ends of chromosomes and next to centromeres, where no two consecutive hotspots are available, the nearest start or stop coordinates of the chromosome and centromere were used instead, respectively (centromere coordinates were exported from the gaps table in the UCSC Genome Browser hg18 database). Tandem repeats were then localized to an interhotspot region by their midpoints, and the distance from each tandem repeat's midpoint to its region's start coordinate was recorded. This distance was then normalized to fall between 0 and 1, with the start of the interhotspot region represented by 0, and the end by 1. This distribution of normalized distances was then plotted with the understanding that a distributional bias of tandem repeats relative to recombination hotspots would be evinced by a curve, as was observed with microsatellites, although not with mlTRs, slTRs, or the candidate tandem repeats for array length polymorphism (ALL TRs with copy numbers ≥5 and match percentages ≥95%).
To verify random distribution of tandem repeats relative to recombination hotspots, we performed a randomization experiment. For each tandem repeat, the number of recombination hotspots falling at least partially within a window formed by extending the tandem repeat's coordinates 100 kb in both directions were counted, defining a reference distribution for that set. One thousand tandem repeats were then sampled randomly from the applicable set and placed at random across the genome. The number of recombination hotspots within each randomly placed tandem's window was then counted, defining a new distribution, which was compared to the reference distribution using a chi-square test. After repeating this process 100 times, no randomly positioned set of tandem repeats was found to have a significantly different distribution from its reference, corroborating the initial result.
RESULTS
Classification of human tandem repeats:
The Tandem Repeats Database (TRDB, http://tandem.bu.edu/cgi-bin/trdb/trdb.exe) provides a compilation of all tandem repeats in genome sequences (Gelfand et al. 2007) with periods between 1 and 2000 bp in length and the output data from its analysis of the human genome sequence formed the starting point for this study. Our approach was to first identify two complementary subsets of tandem repeats: the single-locus tandem repeats (slTRs), whose consensus sequence (the sequence that is repeated to produce the tandem repeat) is found at only a single position in the human genome and the multilocus tandem repeats (mlTRs), whose consensus sequence is found at more than one site. Our motivation derived from an interest to better understand the sequence origin of tandem repeats as well as the expectation that slTRs would more easily serve to interrogate both genomic DNA and expressed RNA in a variety of molecular and genetic applications. We began with the 947,696 tandem repeats from TRDB, identified from the March 2006 assembly of the human genome. TRDB often describes a given tandem repeat at a single location with multiple entries, differing in “period” (the length of the consensus sequence that is repeated) and “copies” (the number of times the consensus sequence is found within this tandem repeat). We sought to eliminate this redundancy by first selecting the entries with the longest array length for each genome location and subsequently for the single subentry among them with the shortest period. This allowed us to recognize the entry with the smallest consensus sequence for each location, and by discarding its overlapping alternatives, we identified 788,596 independent tandem repeats. We then removed those tandem repeats with periods of <20 bp and the remaining set, n = 241,253 entries, is denoted as “ALL TRs.”
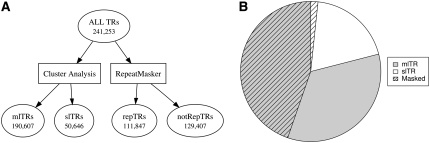
Progress in our approach to delineate this nonredundant set of tandem repeats into two subsets differing by the representation of their consensus sequences in the human genome is tracked in Figure 1A. To identify “unique” tandem repeats within this population, we first performed a cluster analysis that produced two distinct tandem repeat subsets: (1) those whose consensus sequence is found only once in TRDB and (2) those whose consensus sequence is related by sequence (≥80% identity) to other entries in TRDB. The resulting unique set was subsequently aligned against the entire human genome sequence using BLAST to determine whether any of these tandem repeat consensus sequences were found elsewhere in the genome (presumably as a single copy sequence rather than as tandem repeats). This final step produced the two exclusive subsets of tandem repeats, the slTRs (n = 50,646) whose consensus sequence is represented only once in the human genome sequence and the remaining mlTRs (n = 190,607), which represent tandem repeats that share sequence similarity with other genome regions.
Figure 1.—
Classification of human tandem repeats. (A) Steps taken to produce two paired subsets of tandemly repeated sequences in the human genome sequence. Numbers indicate the number of tandem repeats in each subset. The 241,653 entries in the ALL TRs set were chosen from the complete list of tandem repeats in TRDB by selecting only those with periods between 20 and 2000 bp and removing entries redundant by identical genome locations. slTRs or “single-locus tandem repeats” possess consensus sequences found only once in the human genome while the consensus sequences for mlTRs or “multilocus tandem repeats” are related to sequences elsewhere in the genome. repTRs are tandem repeats whose consensus sequence is “masked” by RepeatMasker due to similarity to known human repetitive elements; notRepTRs represent the remaining tandem repeats unmasked by RepeatMasker. (B) Pie chart showing the composition of the ALL TRs by comparing the two pairs of subsets, mlTRs and slTRs vs. repTRs and notRepTRs. The hatched overlay depicts the fraction of ALL TRs masked by RepeatMasker.
We also used RepeatMasker (Jurka et al. 2005) as another means to identify members of the ALL TRs population that were not unique in the genome by virtue of their derivation from previously described repetitive elements. This analysis again produced two subsets, one that is “masked” and consists of sequences derived from known repetitive elements (here termed “repTRs”), and a second set that is unmasked and includes the tandem repeats that are not related to any known repetitive element (here termed “notRepTRs”). We expected the former to overlap substantially with the mlTR class and the latter with the slTR class. While the repTRs were constituted mostly of mlTRs (Figure 1B), the remaining 43.4% (n = 82,788) of mlTRs were unrelated to a known repetitive element. Examination of the slTR and notRepTR populations indicates that 92% of the slTRs (n = 46,618) overlapped with the notRepTR population. The remaining 4028 slTRs found in the masked set were most often ascribed to “simple” and “low complexity” classifications by RepeatMasker, but detected only once in the human genome sequence by BLAST analysis.
The application of RepeatMasker proved less effective in identifying single-locus tandem repeats, but it enabled a determination of the repetitive element composition of the ALL TRs subset and provided some instructive insights into their origins (Table 1). The representation of sequences similar to short interspersed repetitive elements (SINE) and long terminal repeat (LTR) repetitive elements within ALL TRs parallels their general representation within the human genome sequence, together accounting for ∼48% of all repetitive elements. In contrast, long interspersed repetitive elements (LINEs) are underrepresented compared to their general proportion of the genome. Given that the length of full-size LINEs ranges from ∼6000 to 7000 bp in length (Babushok and Kazazian 2007) and the limitation of periods to no more than 2000 bp by the algorithm used in Tandem Repeats Finder, duplication of intact LINEs in the genome sequence are unlikely to be recognized by TRF and would account for this apparent discrepancy. The classes of simple, low complexity, and satellite repeats all appear to be “overrepresented” in our tandem repeat data relative to their occurrence in the genome sequence, but the innate tandemly repeated nature of these classes insures that virtually all of these genome sequences are also included in this tandem repeats category. Given the correspondences noted here as well as the relative concurrence of the repTR and notRepTR subsets with the general frequency of repetitive and single-copy sequences in the human genome, our conclusion is that the process that creates tandem repeats operates on a largely nonselective basis with regard to target genomic sequences.
TABLE 1.
Repetitive element composition of ALL TRs
| Repeat type | No. | Occurrence/ data set (%) | Occurrence/ genome (%) |
|---|---|---|---|
| SINE | 53,475 | 36.4 | 35.3 |
| LTR | 18,366 | 12.5 | 13.0 |
| LINE | 6,850 | 4.7 | 27.8 |
| Simple | 39,013 | 26.5 | 8.4 |
| Low complexity | 15,927 | 10.8 | 7.3 |
| Satellite | 5,029 | 3.4 | 0.2 |
| Other | 7,287 | 5.0 | 0.1 |
| DNA | 1,150 | 0.8 | 7.8 |
| RNA | 12 | 0.0 | 0.2 |
| Unknown | 2 | 0.0 | 0.0 |
No. represents the quantity of tandem repeats in the ALL TRs set that were identified by RepeatMasker as having significant sequence identity with known repetitive elements. Occurrence/data set presents these data as the percentage of the sum of all such entries. Occurrence/genome reports the percentage of bases in the human genome sequence masked by each type of repetitive element.
Characterization of human slTRs and mlTRs:
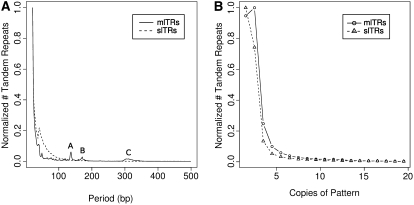
TRDB compiles several parameters to characterize tandem repeats, such as their period, copies, and “match percentage” (the percentage of sequence identity shared across those copies). We compared the slTR and mlTR subsets for each of these characters to identify general similarities and distinctions among these two groups. As shown in Figure 2A both populations are dominated by tandem repeats with short periods (mean for slTRs is 48 bp and for mlTRs is 67 bp). There are three regions where mlTRs break step from an otherwise smooth curve—A (130–145 bp), B (168–175 bp), and C (300–400 bp)—and these regions predominantly feature tandem repeats derived from Alu repetitive elements (A = 88.5%, B = 86.2%, and C = 55.7%), which are the most abundant repetitive element in primate genomes and often exhibit a size length of ∼300 bp (Batzeer and Deininger 2002). A small discontinuity in the slTR distribution at ∼40 bp is an anomaly created by different criteria used for BLAST analyses on either side of this value (see materials and methods) and does not reflect any actual biological significance. Both subsets also primarily contain repeats with three or fewer copies (87.43% of slTRs and 84.19% of mlTRs) as shown in Figure 2B. Evaluation of this parameter could be compromised when the period of tandem repeats approaches and exceeds sequencing read length and match percentage nears 100% such that larger arrays of tandem repeats could artificially collapse to two to three copies during the genome assembly process. However as large periods represent such a small proportion of our dataset, genome assembly errors are unlikely to significantly confound this analysis. Our conclusion from these results is that both subsets of tandem repeats in the human genome primarily contain simple duplications strongly biased to shorter tracts, as further exemplified by the >500,000 repeats with periods <20 bp that were removed in creating the ALL TRs class.
Figure 2.—
Period length and copy distribution for mlTRs and slTRs. (A) The relative frequency of periods (i.e., the length of each tandemly repeating unit) from 20 to 500 bp is shown for human mlTRs and slTRs. Both curves have been normalized such that a value of “1” on the y-axis corresponds to the greatest number of tandem repeats with any given period for that class. The “peaks” in the mlTR distribution at ∼140 bp (A), 170 bp (B), and 300 bp (C) derive from lengths associated with particularly abundant repetitive element families such as Alu elements and satellite DNA. (B) The frequency of copies (i.e., the number of tandem duplications within a single repeated consensus sequence) ranging from 1.8 to 20 is shown for human slTRs and mlTRs. Both curves have been normalized with respect to each other such that a value of “1” on the y-axis corresponds to the greatest number of tandem repeats with any given copy number in that class.
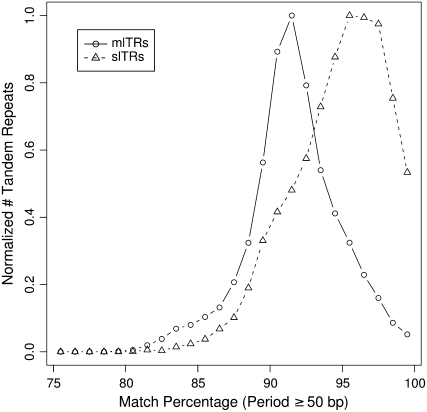
One of the most distinctive parameters between the two subsets was their distributions for match percentage (Figure 3), with the majority of slTRs exhibiting a higher degree of internal similarity (∼95%) across their repeated sequences relative to mlTRs (∼90%). This difference might be due to either a difference in age of these repeats or different decay rates. It has been shown that tandem repeats derived from Alu elements can diverge in their consensus sequence due to exchange events with related partners elsewhere in the genome (Jurka and Gentles 2005). Our mlTR subset by definition possesses related but not necessarily identical partners throughout the genome that they can recombine with, while slTRs are by definition unique in the genome. Consequently these two subsets might be expected to possess inherently different decay rates that could account for some of this difference in divergence.
Figure 3.—
Match-percentage distribution of mlTRs and slTRs. Match percentage (i.e., the percentage of bases shared across each copy of the tandem repeat's consensus sequence) is shown for human mlTRs and slTRs with periods from 50 to 2000 bp. Tandem repeats with periods <50 bp were omitted, as their inclusion exerts a negative bias on higher match percentages for the trivial reason that match percentages from 96 to 99% are impossible for tandem repeats with periods of 20 bp.
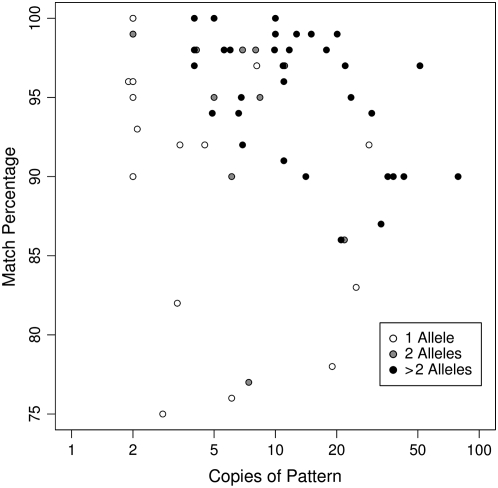
Higher match percentage is one component used by many models to predict genetic polymorphism for tandem repeats (Denoeud et al. 2003; Näslund et al. 2005; Legendre et al. 2007) and this attribute of slTRs along with their unique representation in the genome suggests that this subset is likely to contain many useful molecular markers. We evaluated sixty-two slTRs for array-length polymorphism in a panel of lymphoblastoid cell DNAs obtained from 14 individuals selected for ethnic diversity. It is evident in Figure 4 that genetically variable tandem repeats can be predicted effectively from the compiled human genome sequence using just two characters found in TRDB, values of five or more copies, and a match percentage of ≥95%, consistent with other models (Stephan and Cho 1994; Näslund et al. 2005; Legendre et al. 2007). Given these stringent criteria and our analyses of slTRs in the completed human genome sequence, we predict 2230 slTRs that could serve as highly informative genetic markers (supplemental File 1). Relaxation of the match percentage to 90% would produce even more candidates but include additional monomorphic loci as well. While the increased emphasis upon SNPs in current genetic analyses is understandable given their tractability for very high throughput, the multiallelic nature of tandem repeats and their high information content offer strong advantages for certain applications.
Figure 4.—
Genetic variation of array length for selected slTRs. Array length for a set of 62 slTRs was determined in a panel of genomic DNA from lymphoblastoid cell lines for 14 individuals, selected for ethnic diversity. PCR was performed with primers flanking the entire repeat followed by amplicon separation via agarose gel electrophoresis. The number of alleles for each tandem repeat detected in this panel is indicated by the darkness of the circle. The number of copies for each tandem repeat is plotted on the x-axis (log scale) against its corresponding match percentage on the y-axis.
Chromosomal distribution of tandem repeats:
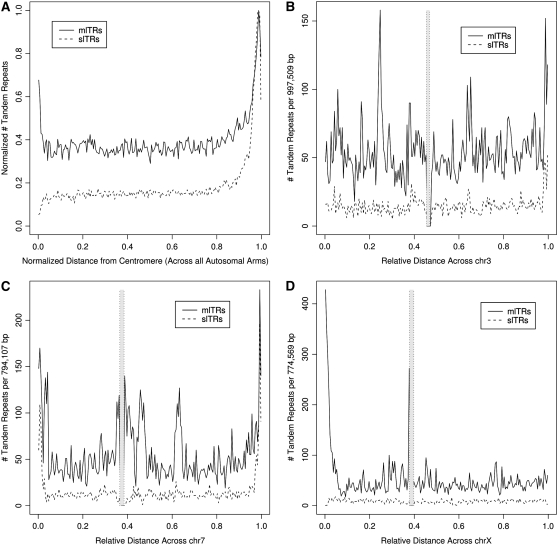
Previous analyses of the chromosomal positions of tandem repeats have suggested that they are not randomly distributed, with tandem repeats preferentially located near chromosome ends (Royle et al. 1988, Amarger et al. 1998). Using the entire genome sequence and summarizing across all of the chromosome arms, we do find clusters of tandem repeats located at the chromosome ends (Figure 5A), a tendency even more pronounced for slTRs. There is also an increased frequency of mlTRs near the centromere, an observation attributable to the highly repetitive satellite repeats that are preferentially located in pericentric regions. However, closer inspection of frequency-distribution plots of both slTRs and mlTRs across individual chromosomes reveals exceptions to this generalized consensus. Of the metacentric autosomes, chromosome (chr) 3 (Figure 5B) along with chrs 5, 9, 18, and 20, all exhibit an obvious concentration of tandem repeats at only one end, whereas others such as chrs 2, 7, 8, 10, 16, and 17 (Figure 5C and supplemental Figure 1) demonstrate the generalized pattern of concentrations at both chromosome ends. The acrocentric chromosomes (chrs 13, 14, 15, 21, and 22) all exhibit a single concentration of both slTR and mlTR tandem repeats opposite their centromere. Interestingly, clusters of both mlTRs and slTRs are either both present at a chromosome end or both absent, suggesting that the same general forces operate on the creation and maintenance of both subsets but that those forces are not consistent across all chromosome ends. While the preferential location of hypervariable minisatellites at the ends of human chromosomes was noted early on (Royle et al. 1988), it is by no means a generalized observation across all species. The mouse, rat, and pig genomes do not exhibit this distribution pattern near chromosome ends and likewise lack the hypermutable minisatellites that are evident in humans despite extensive searches (Bois 2003).
Figure 5.—
Distribution of tandem repeats across human autosomes. The summed distribution of tandem repeats along all chromosomes is shown schematically in A. The x-axis represents the normalized lengths of each arm with the centromere set to “0” and the telomere to “1.” The y-axis reports the normalized number of both subsets of tandem repeats in 0.5% intervals across the compiled chromosome arms. Plots for individual chromosomes are also presented for chromosome 3 (B), chromosome 7 (C), and the X chromosome (D). Here the physical length for each chromosome has been normalized and is distributed between “0” and “1” on the x-axis to facilitate comparisons between chromosomes. The numbers of slTRs and mlTRs in 0.5% intervals across the chromosome were summed and plotted on the y-axis. The two parallel and vertical dotted lines indicate the approximate centromeric region of each chromosome. All other chromosomes are shown in supplemental Figure 1.
A second observation, evident in the inspection of individual chromosome distributions but not in the generalized pattern shown in Figure 5A, is that mlTRs can also be found in concentrations internal to some chromosome arms. Chromosome 7 (Figure 5C) illustrates an obvious concentration of mlTRs on its long arm; inspection of the sequences comprising this region did not reveal any particular repetitive sequence as dominant. Similar clusters can be seen on the short arms of chromosomes 3 and 6 as well as the long arm of 11 (supplemental Figure 1). Interestingly, conspicuous internal concentrations of slTRs were not seen on any of the chromosomes.
It was suspected that the distribution and abundance of tandem repeats on sex chromosomes differed from autosomes, with few resident minisatellites reported previously except for one cluster in the pseudoautosomal region involved in X–Y pairing (Cooke et al. 1985). The distribution of tandem repeats on the sex chromosomes (Figure 5D and supplemental Figure 1) is distinct from the autosomes, with a general lack of slTRs and no end bias in evidence. mlTRs are also much rarer on these chromosomes but do account for the concentration of tandem repeats at the X–Y pseudoautosomal region.
Tandem repeat distribution relative to genes:
As tandem repeats are found at higher density near the ends of many chromosomes, regions that also generally possess greater gene density, we examined the relative locations of tandem repeats and genes with respect to each other. Looking at the content of tandem repeats within genes (using the Ensembl gene model), 41.7% of genes include one or more mlTRs within their transcribed regions, 25.0% were shown to include at least one slTR, and 21.4% of genes include both a slTR and an mlTR. Looking from a different perspective, 44.5% of slTRs and 36.5% of mlTRs were identified as intragenic and the remainders are found outside of transcribed gene boundaries. These values are similar to the fraction of the genome accounted for by genes of 31.1% (total genome = 3,652,768,762 and bounded by gene transcription = 1,136,952,915), suggesting that the mechanisms that create and maintain tandem repeats are randomly sampling sequences, not only by type (unique vs. repetitive) but also with regard to their location near genes.
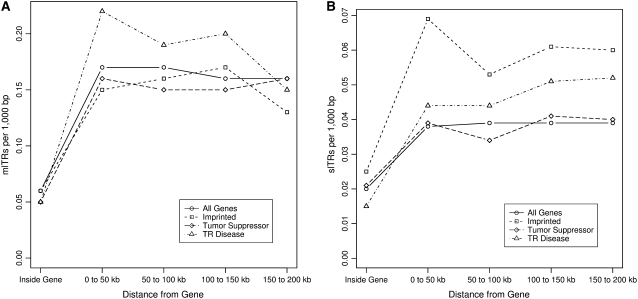
Given their purported roles in modulating gene function, such as variable number tandem repeats (VNTRs) associated with INS, SLC6A3, SLC6A4, and IGF2 (Bennett et al. 1997; Paquette et al. 1998; Bradley et al. 2005; Kelada et al. 2005) and with so many tandem repeats in close proximity to genes, there are likely to be many more genes whose expression is influenced by tandem repeats. Hence we next examined the proximity of our two tandem repeat subsets with respect to different gene groupings: all (n = 31,105), imprinted (n = 45), tandem repeat disease genes (those shown to cause disease by triplet expansions; n = 24), and tumor suppressor (n = 182), with each set listed in supplemental Table 1. As shown in Figure 6, A and B, there are fewer tandem repeats of both subsets within genes than in the regions immediately flanking their transcribed segments. Given that these repeats with their longer periods would be very disruptive if located within coding segments, their occurrence within genes must necessarily be constrained to intronic and untranslated regions. Looking at mlTRs (Figure 6A), we find relatively little difference in their distribution nearby tumor suppressor and imprinted genes with both groups indistinguishable from the inclusive all gene group. In contrast, mlTRs are located more densely external to the tandem repeat disease genes. Even though these diseases are caused by aberrant expansion of a single triplet microsatellite within each gene—tandem repeats that as a group are excluded from our data sets due to their small period—this observation of increased frequency of nearby larger tandem repeats suggests that the environment around these genes is more conducive to general instability. Searching for other genes with higher densities of surrounding mlTRs may identify other unstable regions not yet associated with any specific disease syndrome.
Figure 6.—
Density of mlTRs and slTRs around genes. We defined four groups of human genes: all, imprinted, tumor suppressor, and tandem repeat disease, each delineated by a different symbol. (A) The density of mlTRs inside and around genes of each group is illustrated; (B) data for the slTRs subset. “Inside Gene” represents the internal gene from start to stop of transcription and given that each gene length will vary greatly, we normalized these values to 50 kb to facilitate comparison with those intervals external to the gene. The next region denotes the first 50 kb interval flanking either side of the gene. Similarly the other three entries on the x-axis reflect additional 50 kb intervals extending outward from the gene in both directions for a cumulative distance 200 kb in each direction from the gene's transcriptional boundaries.
The proximity of slTRs to genes (Figure 6B) resembles the general distribution for mlTRs around genes and again the class of tandem repeat disease genes appears to be surrounded by more of this subset of tandem repeats as compared to both the all and tumor suppressor gene groups. We also observe a greater frequency of slTRs around imprinted genes, most strikingly in the nearest 50-kb intervals flanking these genes. To date, at least 23 tandem repeats have been shown to be positioned within or near differentially methylated regions (DMRs) of imprinted genes (Walter et al. 2006), and these have been postulated to serve as a hallmark for these genes (Neumann et al. 1995). This class of parent-of-origin, monoallelically expressed genes also contains a greater representation of tandem repeats within their CpG islands (Hutter et al. 2006) and a higher proportion of repetitive elements, such as LINEs and SINEs, although another study concluded that SINEs are excluded from imprinted regions of the genome (Greally 2002). Our analyses suggest that the general environment around imprinted genes, looking as far out as 150–200 kb, is associated with a higher density of slTRs but not tandem repeats derived from repetitive sequences. This distinction might reflect a functional aspect of these tandem repeats, perhaps facilitating communication between unlinked loci as has been shown for CTCF (CCCTC-binding factor) interactions with imprinting control regions on different chromosomes (Ling et al. 2006). Tandem repeats derived from repetitive sequences might compromise the specificity required for this function and could conceivably be selected against.
Comparison of tandem repeats and recombination hotspots:
The rate of recombination is generally repressed near centromeres and increased near the telomeres, results that were substantially confirmed once the draft human genome sequence was compiled and analyzed (International Human Genome Sequencing Consortium 2001). Since homologous recombination is central to the creation and degradation of tandem repeats (Bois and Jeffreys 1999), it has been suggested that the concentrations of tandem repeats at chromosome ends might reflect these locally higher rates of recombination. Colocalization of a human hypermutable minisatellite with a nearby recombination “hotspot” has been reported (Jeffreys et al. 1998; Jeffreys and Neumann 2005), but it is unclear how prevalent this relationship is. Therefore, we tested for associations between the locations of our subsets of tandem repeats and recombination hotspots in the human genome by two strategies.
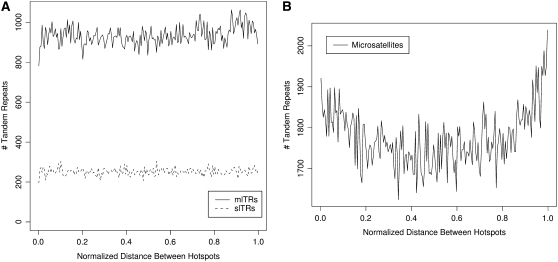
We first compared actual distances between nearest members of both tandem repeats and recombination hotspots (UCSC Genome Browser hg17 database). The analysis presented in Figure 7A normalizes every interval between adjacent hotspots to “1” and then depicts the locations of any tandem repeats within that interval as a fractional value along this distance. While there are minor fluctuations along the distributions for slTRs and mlTRs, the entire distribution of each subset is relatively invariant between hotspots, suggesting that hotspots do not exert a strong impact upon creation of tandem repeats with periods between 20 and 2000 bp and conversely that these tandem repeats do not significantly influence recombination. In contrast a similar analysis of shorter repeats (Figure 7B), with periods ranging from 1 to 10 bp, reveals a nonrandom bias of residency closer to hotspots, suggesting either that the creation of microsatellites is influenced by recombination hotspots or conversely that microsatellites themselves contribute to the existence of recombination hotspots. Given that microsatellites possess higher copies of their repeat consensus sequence than the average larger tandem repeat (with copy values closer to “2–3”), we speculated that a nonrandom association might be more evident near tandem repeats with both higher copy values and match percentage (i.e., VNTRs or hypervariable repeats); however, reexamination of this subgroup of larger tandem repeats still failed to detect any nonrandom association with recombinational hotspots (data not shown).
Figure 7.—
Density of recombination hotspots around human tandem repeats. The length between each pair of adjacent recombination hotspots was normalized to “1.0” and plotted on the x-axis, and the relative positions of tandem repeats along this distance are summarized on the y-axis. (A) The relative positions of slTRs and mlTRs between all adjacent pairs of recombinational hotspots in the human genome are shown; (B) analogous data for tandem repeats with comparatively shorter periods of 1–10 bp.
In contrast to evaluating the length intervals between tandem repeats and recombination hotspots, perhaps a more relevant parameter to demonstrating a functional interaction might be revealed in the density of recombination hotspots around these larger tandem repeats. Three discrete groups of 1000 tandem repeats each were created by random selection from the three subsets of mlTRs, slTRs, and highly polymorphic slTRs. Density of recombination hotspots was measured in a 200 kb window flanking each of these tandem repeats and then 100 random repetitions were performed by relocating these tandem repeats randomly around the genome. It was found that actual densities of recombination hotspots relative to mlTRs and slTRs, as well as high-copy high-similarity tandem repeats, were all statistically indistinguishable from the randomized set (P ≥ 0.23), corroborating our initial result that did not detect any colocalization of slTRs and mlTRs with recombination hotspots. These observations suggest that either the association between tandem repeats and greater frequency of recombination at chromosomal ends (Jeffreys et al. 1998; Jeffreys and Neumann 2005) is coincidental or that these two different approaches are incapable of detecting such a functional interaction.
DISCUSSION
Tandem repeats come in many flavors, thus understanding their creation and evolution can be confounded by whether one examines microsatellites, minisatellites, VNTRs, or hypervariable repeats, whether germinal or somatic variation is investigated, and which organism serves as the basis for the analyses. Much of our current understanding is also based upon the examination of a limited number of repeats, often not selected at random but for the characteristic of polymorphism. Using the results of the whole-genome survey described herein, we can reexamine current ideas for the creation and variation of tandem repeats and how they inform us as to the evolution of the genome. Of course any such survey is a “snapshot” in evolution of the current genome being interrogated and must reflect the cumulative effect of many different processes influencing creation, mutation, maintenance, degradation, and loss; yet a survey based upon a completed genome sequence can provide much more comprehensive data than earlier investigations.
One question for tandem repeat evolution is in ascertaining the relative influences of its consensus sequence vs. the surrounding genome environment. Earlier studies commented upon the higher GC content of minisatellites (Nakamura et al. 1987; Vergnaud and Denoeud 2000); however, by most measures we are unable to detect any distinguishing characteristics of the consensus sequences themselves. The average GC content of the ALL TRs group, which includes all nonredundant tandem repeats with periods between 20 and 2000 bp, is ∼40% and not substantially different from the average value for the entire genome of ∼38%. Recognizing that these earlier studies focused upon hypervariable minisatellites, we examined the GC content of just the polymorphic candidate subset of slTRs as a more direct comparison and obtained a value of ∼56%. Hence, while the GC content is not obviously contributory toward the generation of a tandem repeat, higher GC content may play a role in the more extensive amplification to five or more copies. We also find that the frequency of unique and most repetitive sequences found in tandem repeats mirrors their frequency in the genome, suggesting that the creation and maintenance of tandem repeats is relatively nonspecific to this nature of the consensus sequence as well. Any other aspect of their consensus sequence that might contribute to the development of tandem repeats is not obvious in our data and suggests further exploration of their genome environment might be more insightful.
Current models for the creation and evolution of minisatellites in the germ line often describe two discrete steps, an initial duplication event followed by expansion and contraction that can lead to multiple alleles and even complete loss of the tandem repeat. Haber and Louis (1998) observed one influence of the surrounding genome environment in the presence of short 5- to 10-bp repeat sequences flanking several yeast and human tandem repeats and proposed that replication slippage or unequal crossing over centered upon these flanking sequences is responsible for initial duplication. Richard and Dujon (2006) subsequently extended this work by analyzing the entire genome of yeast and confirmed the general existence of short repeats flanking most tandemly repeated sequences. Several mechanisms for amplifying this initial duplication and creating heritable variants in copy number have been suggested, including slippage during replication, mitotic recombination, or meiotic recombination, but early studies of minisatellites indicated that crossovers were not usually involved and that most of the heritable changes were consistent with gene conversion, probably initiated as repair of double-stranded breaks (Buard and Vergnaud 1994; Jeffreys et al. 1994; Paques and Haber 1999; reviewed in Richard and Paques 2000).
By our own analyses, repeat periods shorter than 100 bp are far more common in the human genome, which is even more striking if one includes tandem repeats with periods from 1 to 19 bp as well. Tandem repeats with low copies, just 2–3 repeats, also predominate in the human genome. While both parameters are subject to contrasting processes of creation and degradation, one parsimonious explanation for our results is that an initial duplication event is far more frequent than subsequent amplification events and that it also occurs more frequently with smaller periods. As the initial duplication could derive from replication slippage, we note that this process represents an intramolecular event that is thermodynamically simpler than a subsequent amplification step based upon recombinational exchanges between different sequences and hence could be more frequent in its occurrence. Replication slippage is also more compatible with shorter segments (Jeffreys et al. 1988). Additional selective pressures may also be working to degrade larger- and higher-copy tandem repeats relative to shorter- and low-copy repeats.
The most distinctive characteristic between slTRs and mlTRs, match percentage, could be explained by differences in age or functional conservation of the two subsets. An additional explanation we favor derives from the observation that divergence of minisatellites can occur via gene conversion with related but not identical sequences including Alu elements (Jurka and Gentles 2005). Because the mlTR subset is defined by the existence of related, but often not identical, sequences elsewhere in the genome, intergenic conversion cannot only lead to new alleles differing for copy values but also likely degrade the sequence between repeats. On the other hand slTRs are by definition unique in the genome, they have no partners with which to recombine other than their homolog on the other paired chromosome. This exchange between alleles at the same locus can produce new alleles of both fewer and more repeats but with higher fidelity. Hence an exchange mechanism common to both subsets could not only account for expansion of initial duplication events but also for differences in match percentage of the two subsets. Utilizing this intrinsically higher match percentage of slTRs, combined with high-copy number of some members and their unique genomic locations, we could efficiently predict tandem repeats representing the best candidates for highly polymorphic molecular markers.
While our data are generally consistent with models for the creation and evolution of tandem repeats, the forces that trigger this process to create such strongly nonrandom distributions in the genome are less obvious. Initial generalizations regarding the predilections of tandem repeats for the ends of human chromosomes were generally confirmed in our data, but the absence of foci on some chromosome arms is notable. Similarly the internal concentrations of mlTRs for some chromosomal regions but the absence of any comparable slTR concentrations remains unexplained. The intimate role for recombination in the generation of these tandem repeats and its higher frequency near chromosome ends suggest that it could account for the nonrandom distribution of tandem repeats as noted by reports of colocalization (Jeffreys et al. 1998; Jeffreys and Neumann 2005); however, we were unable to confirm a general colocalization of tandem repeats and recombination hotspots in the human genome, which is consistent with other observations. Recombination is more frequent near chromosome ends in many organisms, yet mouse chromosomes do not exhibit a similarly higher frequency of tandem repeats near their ends (Bois and Jeffreys 1999). If higher recombination frequency is critical to creation of concentrations of tandem repeats, then the concentrations of pericentric satellite repeats are problematic to reconcile with their locations in regions with greatly suppressed recombination. Tandem repeats are strongly conserved between humans and chimpanzee but there is very little conservation of recombination hotspots between them (Winckler 2005). Finally an analysis in yeast also failed to find any colocalization of tandem repeats near recombination hotspots (Richard and Dujon 2006). Despite the preponderance of studies citing a primary role of recombination in the development of the larger tandem repeats examined in this study, we are unable to implicate hotspots as driving the general distribution of tandem repeats, perhaps reflecting subtler aspects of the recombination mechanisms that must contribute to their derivation.
Considering the potential impacts of tandem repeats upon gene function, it is interesting to note their nonrandom distribution around two classes of genes. That slTRs but not mlTRs are more abundant around imprinted genes might signal a functional contribution to this phenomenon in contrast to the greater frequency of both subsets around genes implicated in triplet expansion diseases, this latter case potentially reflecting a general instability of these genomic regions. Acknowledging reported instances of tandem repeats that influence nearby gene expression, we have looked for other instances of polymorphic tandem repeats found within 2 kb of the boundaries of gene transcription. We find >300 examples in our compilation, many representing genes implicated in human disease, and in a few that we have evaluated, and the results are consistent with the influence of the tandem repeat upon that gene's expression (data not shown here). Use of this catalog and its characterization of the entire genome clearly provides for a more systematic discovery process that facilitates identification of genes whose function is affected by nearby tandem repeats and may add to our understanding of the molecular basis of both genome function and human disease.
Acknowledgments
We especially thank Gary Benson of Boston University for his contributions to this study and help with the Tandem Repeats Database and Bruce Walsh of the University of Arizona for assistance with statistical analysis. This research was funded through a National Institutes of Health award 5DP10D575-2 to V.L.C.
References
- Amarger, V., D. Gauguier, M. Yerle, F. Apiou, P. Pinton et al., 1998. Analysis of distribution in the human, pig, and rat genomes points toward a general subtelomeric origin of minisatellite structures. Genomics 52 62–71. [DOI] [PubMed] [Google Scholar]
- Babushok, D. V., and H. H. Kazazian, 2007. Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28 527–539. [DOI] [PubMed] [Google Scholar]
- Batzeer, M. A., and P. L. Deininger, 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3 370–380. [DOI] [PubMed] [Google Scholar]
- Bennett, S. T., A. J. Wilson, L. Esposito, N. Bouzekri, D. E. Undlien et al., 1997. Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. Nat. Genet. 17 350–352. [DOI] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G., 2005. Tandem cyclic alignment. Discrete Appl. Math. 146 124–133. [Google Scholar]
- Boby, T., A. M. Patch and S. J. Aves, 2005. TRbase: a database relating tandem repeats to disease genes for the human genome. Bioinformatics 21 811–816. [DOI] [PubMed] [Google Scholar]
- Bois, P. R. J., 2003. Hypermutable minisatellites, a human affair. Genomics 81 349–355. [DOI] [PubMed] [Google Scholar]
- Bois, P., and A. J. Jeffreys, 1999. Minisatellite instability and germline mutation. Cell. Mol. Life Sci. 55 1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, S. L., K. Dodelzon, H. K. Sandhu and R. A. Philibert, 2005. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am. J. Med. Genet. B Neuropsychiatr. Genet. 136B 58–61. [DOI] [PubMed] [Google Scholar]
- Buschiazzo, E., and N. J. Gemmell, 2006. The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays 28 1040–1050. [DOI] [PubMed] [Google Scholar]
- Buard, J., and G. Vergnaud, 1994. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J. 13 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, H. J., W. R. A. Brown and G. A. Rappold, 1985. Hypervariable telomeric sequences from the human sex chromosomes are pseudoautosomal. Nature 317 687–692. [DOI] [PubMed] [Google Scholar]
- Cox, R., and S. M. Mirkin, 1997. Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. USA 94 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen, V., E. Eyras, T. D. Andrews, L. Clarke, E. Mongin et al., 2004. The Ensembl automatic gene annotation system. Genome Res. 14 942–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud, F., G. Vergnaud and G. Benson, 2003. Predicting human minisatellite polymorphism. Genome Res. 5 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchel, J. R., and H. Y. Zoghbi, 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6 743–755. [DOI] [PubMed] [Google Scholar]
- Gelfand, Y., A. Rodriguez and G. Benson, 2007. TRDB—the Tandem Repeats Database. Nucleic Acids Res. 35 D80–D87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally, J. M., 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. USA 99 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., and E. J. Louis, 1998. Microsatellite origins in yeast and humans. Genomics 48 132–135. [DOI] [PubMed] [Google Scholar]
- Hutter, B., V. Helms and M. Paulsen, 2006. Tandem repeats in the CpG islands of imprinted genes. Genomics 88 323–332. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., K. Tamaki, A. MacLeod, D. G. Monckton, D. L. Neil et al., 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6 136–145. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., and R. Neumann, 2005. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 14 2277–2287. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., N. J. Royle, V. Wilson and Z. Wong, 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in humans. Nature 332 278–281. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., J. Murray and R. Neumann, 1998. High-resolution mapping of crossovers in human sperm defined a minisatellite-associated recombination hotspot. Mol. Cell 2 267–273. [DOI] [PubMed] [Google Scholar]
- Jurka, J., and A. J. Gentles, 2005. Origin and diversification of minisatellites derived from human Alu sequences. Gene 365 21–26. [DOI] [PubMed] [Google Scholar]
- Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany et al., 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110 462–467. [DOI] [PubMed] [Google Scholar]
- Kelada, S. N., P. Costa-Mallen, H. Checkoway, C. S. Carlson, T. S. Weller et al., 2005. Dopamine transporter (SLC6A3) 5′ region haplotypes significantly affect transcriptional activity in vitro but are not associated with Parkinson's disease. Pharmacogenet. Genomics 15 659–668. [DOI] [PubMed] [Google Scholar]
- Legendre, M., N. Pochet, T. Pak and K. J. Verstrepen, 2007. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 17 1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., G. Sutton, P. C. Ng, L. Feuk, A. L. Halpern et al., 2007. The diploid genome sequence of an individual human. PLoS Biol. 5 2113–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., A. B. Korol, T. Fahima, A. Beiles and E. Nevo, 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11 2453–2465. [DOI] [PubMed] [Google Scholar]
- Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen et al., 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312 269–272. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., M. Leppert, P. O'Connell, R. Wolff, T. Holm et al., 1987. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235 1616–1622. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., K. Koyama and M. Matsushima, 1998. VNTR (variable number tandem repeat) sequences as transcriptional, translational, or functional regulators. J. Hum. Genet. 43 149–152. [DOI] [PubMed] [Google Scholar]
- Näslund, K., P. Saetre, J. von Salomé, T. F. Bergström, N. Jareborg et al., 2005. Genome-wide prediction of human VNTRs. Genomics 85 24–35. [DOI] [PubMed] [Google Scholar]
- Neumann, B., P. Kubicka and D. P Barlow, 1995. Characteristics of imprinted genes. Nat. Genet. 9 12–13. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette, J., N. Giannoukakis, C. Polychronakos, P. Vafiadis and C. Deal, 1998. The INS 5′ variable number of tandem repeats is associated with IGF2 expression in humans. J. Biol. Chem. 273 14158–14164. [DOI] [PubMed] [Google Scholar]
- Richard, G.-F., and B. Dujon, 2006. Molecular evolution of minisatellites in hemiaxcomycetous yeasts. Mol. Biol. Evol. 23 189–202. [DOI] [PubMed] [Google Scholar]
- Richard, G.-F., and F. Paques, 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle, N. J., R. E. Clarkson, Z. Wong and A. J. Jeffreys, 1988. Clustering of hypervariable minisatellites in the proterminal regions of human autosomes. Genomics 3 352–360. [DOI] [PubMed] [Google Scholar]
- Stam, M., and O. M. Scheid, 2005. Paramutation: an encounter leaving a lasting impression. Trends Plant Sci. 10 283–290. [DOI] [PubMed] [Google Scholar]
- Stam, M., C. Belele, W. Ramakrishna, J. E. Dorweiler, J. L. Bennetzen et al., 2002. The regulatory regions required for B' paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., and S. Cho, 1994. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics 136 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnaud, G., and F. Denoeud, 2000. Minisatellites: mutability and genome architecture. Genome Res. 10 899–907. [DOI] [PubMed] [Google Scholar]
- Walter, J., B. Hutter, T. Khare and M. Paulsen, 2006. Repetitive elements in imprinted genes. Cytogenet. Genome Res. 113 109–115. [DOI] [PubMed] [Google Scholar]
- Winckler, W., 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308 107–111. [DOI] [PubMed] [Google Scholar]