Abstract
A putative advantage of allopolyploidy is the possibility of differential selection of duplicated (homeologous) genes originating from two different progenitor genomes. In this note we explore this hypothesis using a high throughput, SNP-specific microarray technology applied to seed trichomes (cotton) harvested from three developmental time points in wild and modern accessions of two independently domesticated cotton species, Gossypium hirsutum and G. barbadense. We show that homeolog expression ratios are dynamic both developmentally and over the several-thousand-year period encompassed by domestication and crop improvement, and that domestication increased the modulation of homeologous gene expression. In both species, D-genome expression was preferentially enhanced under human selection pressure, but for nonoverlapping sets of genes for the two independent domestication events. Our data suggest that human selection may have operated on different components of the fiber developmental genetic program in G. hirsutum and G. barbadense, leading to convergent rather than parallel genetic alterations and resulting morphology.
GOSSYPIUM seed trichomes, colloquially termed “cotton fiber,” compose the foundation of the world's most important textile fiber and also represent one of the most distinct cell types in the plant kingdom (Kim and Triplett 2001). The long, strong, and fine fibers of modern cotton cultivars were generated through a long history of natural and human-mediated selection (Brubaker et al. 1999; Wendel and Cronn 2003). Wild diploid species usually have short (mostly <5 mm), coarse, and tightly adherent trichomes that would not be recognized visually as “cotton.” Longer, ultimately spinable fibers evolved in the ancestors of the modern A-genome diploids (Applequist et al. 2001; Hovav et al. 2008b), and a second level of morphological innovation was precipitated by polyploid formation. The latter entailed transoceanic dispersal of an A-genome species to the New World, hybridization between the immigrant A-genome species and a native D-genome species (Endrizzi et al. 1985; Wendel 1995), and genome doubling. This event is thought to have occurred ∼1–2 MYA (Wendel and Cronn 2003) and led to the evolution of the five wild allotetraploid species that occur in seasonally arid regions of the American tropics and subtropics (Fryxell 1979).
Superimposed on these natural events were later innovations caused by human domestication. Remarkably, four wild Gossypium species were independently domesticated by aboriginal domesticators ≥∼5000 years ago and transformed into fiber and oilseed plants (Wendel 1995; Dillehay et al. 2007). Two of these (Gossypium arboreum L. and G. herbaceum L.) are A-genome diploids from the Old World, whereas the other two, G. hirsutum L. (the source of “Upland” cotton) and G. barbadense L. (the source of “Pima” or “Egyptian” cotton), are AD-genome allotetraploids. Both allotetraploid species have large indigenous ranges (wild ranges, expanded by thousands of years of human-mediated, pre-Colombian geographic diffusion), with G. hirsutum predominantly distributed in Mesoamerica and the Caribbean and G. barbadense having a more southerly distribution in South America and the Caribbean (Brubaker and Wendel 1994; Brubaker et al. 1999). Previous studies that included RFLP data in conjunction with morphological and anthropological information suggested the Yucatan peninsula as the primary site of earliest G. hirsutum domestication (Brubaker and Wendel 1994). Agronomically advanced forms developed in southern Mexico and Guatemala appear to have been derived from these wild Yucatan peninsular forms, creating a secondary center of diversity. In a parallel way, archeological (Dillehay et al. 2007) and molecular (Percy and Wendel 1990; Westengen et al. 2005) evidence indicate that G. barbadense was first domesticated on the western slopes of the northern Peruvian Andes, with subsequent diffusion pathways into regions east of the Andes and the Caribbean. In post-Colombian times, both G. hirsutum and G. barbadense were further dispersed globally, and cultivars derived from these two allotetraploid species now dominate world cotton commerce.
Domestication and breeding of both allotetraploid cottons has resulted in yield and quality levels superior to those achieved by parallel development of the A-genome diploid cottons. Evidence suggests that the ultimate agricultural dominance of allotetraploid cultivated species is directly related to the biological reunion of the two diverged diploid genomes (A and D), which by virtue of genomewide gene duplication permitted the evolution of novel and agronomically important fiber traits (Jiang et al. 1998; Udall and Wendel 2006; Rong et al. 2007). The merger of two differentiated genomes in a common nucleus is accompanied by myriad genomic alterations and gene expression changes (Wendel 2000; Liu and Wendel 2003; Osborn 2004; Adams and Wendel 2005; Chen 2007; Chen et al. 2007; Gaeta et al. 2007) and is thought to provide the raw material for the origin of morphological novelty, adaptation, and speciation. The duplicated genes (termed homeologs) are subject to a dynamic tension between mutational decay and fixation by selective forces, including natural and human selection (Force et al. 1999; Lynch and Conery 2000). In this regard, alterations in mRNA levels derived from artificial selection of one of the two homeologs of each gene may contribute to the total phenotype and thus be thought of as genes “recruited” into a pathway or process subsequent to polyploid formation.
In a recent study (Hovav et al. 2008a) we used a novel high-resolution microarray methodology (Udall et al. 2006) to demonstrate the dynamics of homeologous gene expression for ∼1500 gene pairs during development of the allotetraploid cotton fiber. We showed that it is common for homeologs to contribute unequally to the transcriptome during fiber development and that there exists an overall bias toward D-genome transcription. In this note we expand our previous study emphasizing evolution under domestication, in parallel, and across a developmental time course. By using the same SNP-specific microarray design as employed earlier (Hovav et al. 2008a), we report on duplicate gene expression patterns accompanying the transition from wild to domesticated cotton in both G. hirsutum and G. barbadense. Our effort was to evaluate: (i) if domestication was accompanied by enhanced genome-specific bias toward either the A- or D-genome, (ii) if parallel domestication led to similar responses in the two species, and (iii) if polyploidy and domestication was accompanied by novel gene “recruitment.”
Changes in duplicate gene expression during fiber development:
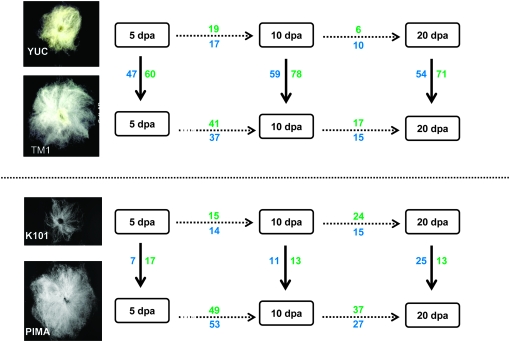
A SNP-specific microarray platform was used to estimate homeolog expression ratios for 1484 gene pairs at three time points in wild and domesticated accessions of both G. hirsutum and G. barbadense (Figure 1). Comparisons among time points and accessions permitted an analysis of significant changes [false discovery rate (FDR) < 0.05] in homeolog expression ratios. As shown, this analysis was conducted for both the several-thousand-year temporal transitions from wild to domesticated and the several-day temporal transitions corresponding to fiber developmental stages. Overall, the numbers of genes that changed homeolog expression ratios between forms and between developmental time points (within forms) were relatively low, with percentages ranging from 2.0 to 9.2 among comparisons (out of total genes detected). This result is consistent with our former study (Hovav et al. 2008a), which showed that most bias in homeolog expression resulted from genome merger and polyploid formation as opposed to human domestication. Yet, as shown in Figure 1, homeolog-expression ratios are not static, but are dynamic on both temporal scales explored. Notably, domestication generally appears to have resulted in a larger number of changes in expression ratios during development. In G. hirsutum, for example, the number of expression ratio changes in the first [5–10 days postanthesis (DPA)] and the second (10–20 DPA) developmental transitions were 36 and 16 for the wild accession, respectively, whereas the corresponding numbers for the domesticated form were twice as high, i.e., 78 and 32. Similarly, in G. barbadense, the number of expression ratio changes in the first and the second developmental transitions were 29 and 39 for K101, respectively, whereas the corresponding numbers for Pima S-7 were 3.5 and 1.6 times as large, respectively, i.e., 102 and 64. When examined from the standpoint of overall change from 5 to 20 DPA, 93 and 122 genes changed expression ratios in wild and domesticated G. hirsutum, respectively, whereas 98 and 168 genes, respectively, did so in wild and domesticated G. barbadense. With a χ2-value <0.05 for all of the above comparisons (H0:50%), these data indicate that in both species, thousands of years of human selection under domestication have altered the fiber transcriptome in such a manner that homeolog ratios are more dynamic overall and more unstable developmentally.
Figure 1.—
Changes in duplicate gene ratios during fiber development and between wild and cultivated cotton. Analyses were performed on fibers isolated from ovules from three stages of development [5, 10, and 20 days postanthesis (DPA)], representing early and mid elongation and the transition to secondary cell-wall synthesis. Three biological replicated blocks of four allotetraploid cotton accessions were included, corresponding to wild and modern domesticated versions of the two important cotton species. For G. hirsutum (top), we included the domesticated genetic standard stock TM1 (TM1) and the wild shrub from the coastal Yucatan Peninsula, G. hirsutum var. yucatanense (accession TX2094; YUC). For G. barbadense (bottom), we included the elite cultivar Pima S-7 (PIMA) and a primitive Bolivian form, G. barbadense accession K101 (K101). A SNP-specific microarray (Udall et al. 2006) was used, including 11,350 probe sets designed to target individual homeologs (A or D). Messenger RNAs were isolated, amplified, and hybridized to the microarrays as described (Flagel et al. 2008; Hovav et al. 2008a; Hovav et al. 2008c). In addition, RNAs derived from two diploid species, G. raimondii (D5) and G. arboreum (A2) were hybridized to the same microarray platform and used for diagnostic probes selection. Raw data values for each microarray were natural-log transformed, median centered, and scale normalized across all arrays prior to performing a mixed linear model (LM), as described (Hovav et al. 2008a). The LM was used to find diagnostic, homeolog-specific probe sets by identifying those probe sets for which the expression level for a given A-genome probe was significantly greater (FDR ≤ 0.05) than the corresponding D-genome probe when hybridized with A-genome RNA, and vice versa when hybridized with D-genome RNA. Only probes that met these conditions for all three time points were considered as diagnostic and were used further. Of the 22,798 probes representing 2028 contigs, 5078 probes representing 1484 contigs were analyzed further. For each contig targeted by multiple probes, a Tukey biweight correction was calculated. The log (A-probe)–log (D-probe) expression values [log(A/D)] were calculated for each gene, representing the relative contribution of each homeolog to the allotetraploid transcriptome. To analyze gene bias changes between accessions or during development, a linear model that included accession, time point, and the interaction was used. The difference of the log(A/D) ratio between species or time points was calculated for each of 1484 gene pairs. The 1484 P-values from each comparison were converted to q-values using the method of Storey and Tibshirani (2003). These q-values were used to identify the number of differentially biased genes for a given comparison when controlling the false discovery rate (FDR) at various levels. Shown in the figure are changes in homeolog ratios among developmental stages and between wild and cultivated forms. Solid arrows indicate the wild-to-domesticated transitions, whereas developmental transitions are denoted by dashed arrows. Significant changes in homeolog ratios are divided into those increasing toward A-genome bias (blue) and those increasing in D-genome bias (green).
Interestingly, the foregoing conclusion does not appear to be random with respect to direction of homeolog bias. Instead, comparisons of wild and domesticated forms (solid arrows in Figure 1) in both G. hirsutum and G. barbadense show that there are more changes from A-biased to D-biased (green numbers) than the reverse (blue numbers), except for the single comparison at 20 DPA in G. barbadense. From the standpoint of morphology, this might be considered an unexpected result, given that A-genome species have relatively long, spinable fibers whereas D-genome species have shorter, tightly adherent seed trichomes. Hence one might postulate, a priori, that any increase in biased expression accompanying domestication would favor the A genome. Particularly relevant, in light of the D-genome preference demonstrated here, are results of numerous QTL studies, a recent meta-analysis of which demonstrates that a majority of QTL for important fiber traits are located on chromosomes derived from the D-genome parent (Jiang et al. 1998; Rong et al. 2007). Our data may thus comprise transcript-level evidence of this possibility of recruitment of D-genome homeologs following polyploid formation, manifested as novel or enhanced expression levels and thereby potentially contributing to superior commercial cotton, or by extension to natural processes, evolutionary innovation. This speculation awaits functional confirmation, but expression analyses may provide useful clues in this regard.
Parallel domestication generated different biased genes and biological processes:
Relevant to the possibility of novel gene recruitment being facilitated by polyploidy, we partitioned the expression ratio changes into categories representing either increased A-homeolog or increased D-homeolog bias. Biological processes corresponding to these two categories, accompanying domestication of both G. hirsutum and G. barbadense and at three fiber developmental stages (5, 10, and 20 DPA), are shown in Table 1. This tabulation shows that within each of the two species, different biological processes are represented by genes that increased A- or D-homeolog expression following domestication.
TABLE 1.
Biological processes corresponding to changes in expression ratios accompanying domestication of G. hirsutum and G. barbadense at three fiber developmental stages (5, 10, and 20 DPA)
| G. hirsutum comparison | N | D-biased processes under domestication | GO | N | A-biased processes under domestication | GO |
|---|---|---|---|---|---|---|
| 5 DPA | 60 | Nuclear pore | GO:0005643 | 47 | Methylenetetrahydrofolate reductase (NADPH) activity | GO:0004489 |
| Glycerol metabolic process | GO:0006071 | Group transfer coenzyme metabolic process | GO:0006752 | |||
| Unlocalized protein complex | GO:0005941 | Ceramidase activity | GO:0017040 | |||
| Nucleoside metabolic process | GO:0009116 | Malate transport | GO:0015743 | |||
| Signal transduction | GO:0007165 | Pathogenesis | GO:0009405 | |||
| Protein targeting | GO:0006605 | rRegulation of translational fidelity | GO:0006450 | |||
| Kinase activity | GO:0016301 | Coenzyme biosynthetic process | GO:0009108 | |||
| Phosphoethanolamine N-methyltransferase activity | GO:0000234 | Golgi apparatus part | GO:0044431 | |||
| Hydrolase activity | GO:0016823 | Plastid | GO:0009536 | |||
| Trypsin activity | GO:0004295 | |||||
| UTP:glucose-1-phosphate uridylyltransferase activity | GO:0003983 | |||||
| Pyrimidine dimer repair | GO:0006290 | |||||
| Aldehyde dehydrogenase (NAD) activity | GO:0004029 | |||||
| 3-oxo-5-α-steroid 4-dehydrogenase activity | GO:0003865 | |||||
| Amidase activity | GO:0004040 | |||||
| 10 DPA | 78 | Response to ethylene stimulus | GO:0009723 | 59 | Methylenetetrahydrofolate reductase (NADPH) activity | GO:0004489 |
| Sugar binding | GO:0005529 | Group transfer coenzyme metabolic process | GO:0006752 | |||
| Cell communication | GO:0007154 | Cell cycle | GO:0007049 | |||
| Kinase activity | GO:0016301 | Lipid binding | GO:0008289 | |||
| Nucleoside metabolic process | GO:0009116 | ATP metabolic process | GO:0046034 | |||
| Aromatic compound metabolic process | GO:0006725 | Regulation of translational fidelity | GO:0006450 | |||
| UDP-sugar pyrophosphorylase activity | GO:0051748 | DNA ligase activity | GO:0003909 | |||
| amidase activity | GO:0004040 | Malate transport | GO:0015743 | |||
| Chlorophyll binding | GO:0016168 | Fructokinase activity | GO:0008865 | |||
| Selenium binding | GO:0008430 | |||||
| Pectin metabolic process | GO:0045488 | |||||
| Cellular morphogenesis during vegetative growth | GO:0000903 | |||||
| ESCRT III complex | GO:0000815 | |||||
| Cytosolic large ribosomal subunit (sensu Bacteria) | GO:0009282 | |||||
| 3-Hydroxyacyl-CoA dehydratase activity | GO:0018812 | |||||
| Negative regulation of programmed cell death | GO:0043069 | |||||
| Aminomethyltransferase activity | GO:0004047 | |||||
| Aldehyde dehydrogenase (NAD) activity | GO:0004029 | |||||
| Pyrimidine dimer repair | GO:0006290 | |||||
| 20 DPA | 71 | Unlocalized protein complex | GO:0005941 | 54 | Methylenetetrahydrofolate reductase (NADPH) activity | GO:0004489 |
| Transferase activity, transferring phosphorus-containing groups | GO:0016772 | Cation-transporting ATPase activity | GO:0019829 | |||
| Nucleobase, nucleoside, nucleotide kinase activity | GO:0019205 | Cell division | GO:0051301 | |||
| Kinase activity | GO:0016301 | Methionine metabolic process | GO:0006555 | |||
| Glycerol-3-phosphate dehydrogenase complex | GO:0009331 | DNA ligase activity | GO:0003909 | |||
| Two-component signal transduction system (phosphorelay) | GO:0000160 | Xanthophyll metabolic process | GO:0016122 | |||
| Electron transport | GO:0006118 | 5S rRNA binding | GO:0008097 | |||
| Magnesium ion binding | GO:0000287 | Regulation of translational fidelity | GO:0006450 | |||
| Phosphoric ester hydrolase activity | GO:0042578 | Dipeptidase activity | GO:0016805 | |||
| Oxidoreductase activity, acting on the CH-CH group of donors | GO:0016627 | β-Carotene hydroxylase activity | GO:0042411 | |||
| Pectin biosynthetic process | GO:0045489 | |||||
| Amidase activity | GO:0004040 | |||||
| Cellular morphogenesis during vegetative growth | GO:0000903 | |||||
| Negative regulation of apoptosis | GO:0043066 | |||||
| Pyrimidine dimer repair | GO:0006290 | |||||
| Aldehyde dehydrogenase (NAD) activity | GO:0004029 | |||||
| Chlorophyll binding | GO:0016168 | |||||
| G. barbadense comparison | N | D-biased biological processes under domestication | GO | N | A-biased biological processes under domestication | GO |
| 5 DPA | 17 | GTPase activity | GO:0003924 | 7 | Protein phosphatase type 2A activity | GO:0000158 |
| ATP-binding cassette (ABC) transporter complex | GO:0043190 | FK506 binding | GO:0005528 | |||
| Protein polymerization | GO:0051258 | |||||
| Microtubule-based movement | GO:0007018 | |||||
| Positive regulation of protein kinase activity | GO:0045860 | |||||
| Plastid membrane | GO:0042170 | |||||
| 10 DPA | 13 | Peptidyl-proline 4-dioxygenase activity | GO:0031545 | 11 | Detection of cytokinin stimulus | GO:0009722 |
| NAD biosynthetic process | GO:0009435 | Bis(5′-adenosyl)-triphosphatase activity | GO:0047710 | |||
| Phosphoglucomutase activity | GO:0004614 | Regulation of actin filament length | GO:0030832 | |||
| Pollen germination | GO:0009846 | FK506 binding | GO:0005528 | |||
| Regulation of protein kinase activity | GO:0045859 | Membrane-bound vesicle | GO:0031988 | |||
| G-protein signaling, coupled to IP3 second messenger | GO:0007200 | Peptidyl-prolyl cis-trans isomerase activity | GO:0003755 | |||
| Protein kinase activity | GO:0004672 | |||||
| 20 DPA | 21 | Pollen germination | GO:0009846 | 25 | Cystathionine γ-synthase activity | GO:0003962 |
| Fatty acid metabolic process | GO:0006631 | Vesicle docking during exocytosis | GO:0006904 | |||
| Peptidyl-amino acid modification | GO:0018193 | ATP citrate synthase activity | GO:0003878 | |||
| Nucleotide diphosphatase activity | GO:0004551 | Antioxidant activity | GO:0016209 | |||
| Oxidoreductase activity | GO:0016705 | Actin filament organization | GO:0007015 | |||
| Hydrolase activity, acting on ester bonds | GO:0016788 | Response to cytokinin stimulus | GO:0009735 | |||
| G-protein signaling, coupled to IP3 second messenger | GO:0007200 | |||||
| Nucleoside-triphosphatase activity | GO:0017111 |
Data are partitioned into processes corresponding to genes biased toward either the A-genome or D-genome homeolog. Blast2GO (http://www.blast2go.de/) was used to identify biochemical pathways involved in a given comparison and to calculate the statistical significance of each pathway. Blast2GO includes the Gossip package (Bluthgen et al. 2005) for statistical assessment of annotation differences between two sets of sequences, using Fisher's exact test for each GO term. P-values (P < 0.05) were used for the assessment of significant metabolic pathways. A complete list of genes is at (http://www.eeob.iastate.edu/faculty/WendelJ/ranhovav.htm).
A notable feature of the gene lists for G. hirsutum and G. barbadense is that the set of genes and processes are nonoverlapping for the two independent domestication events. Remarkably, not even a single gene was shared in the gene lists compiled for the two species at any time point. Recalling that allotetraploid cotton was domesticated twice at geographically isolated locations, each thousands of years ago (Wendel 1995), our data suggest that this parallel selection and resulting morphology entailed human selection for different underlying biological processes. Phrased alternatively, human selection may have operated on different components of the fiber developmental genetic program in G. hirsutum and G. barbadense, leading to convergent rather than parallel genetic alterations and resulting morphology. This possibility is bolstered by ongoing studies in our laboratory using conventional (not homeolog-specific) microarrays, which reveal similar nonoverlapping sets of genes (our unpublished data). One caveat to these speculations is that because both of the wild forms used may not be fully representative of the progenitor lineage of modern, domesticated G. hirsutum and G. barbadense, respectively, differences in gene expression between the wild and domesticated forms may reflect factors in addition to domestication, such as infraspecific polymorphism and random, neutral change.
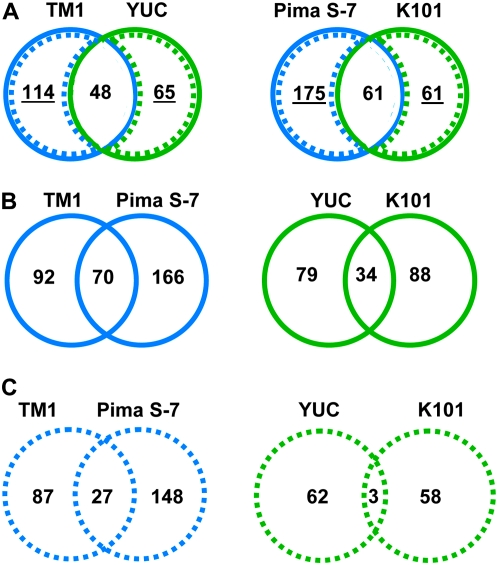
In contrast to the comparisons described above between wild and domesticated forms (vertical comparisons in Figure 1), many A- or D-biased genes were actually shared between forms and even between species during fiber development (horizontal comparisons in Figure 1). This is reflected in Figure 2, which shows the numbers of genes that changed homeolog expression ratios between any two time points among the three stages of fiber development studied. As shown, many genes that changed expression ratios were shared between wild and domesticated forms in both G. hirsutum and G. barbadense (Figure 2A), with 21.1 and 20.5% of shared genes between TM1/YUC and PIMA/K101, respectively, for the two species. In addition, many of the genes exhibiting homeolog expression ratio changes were shared across species as well (Figure 2B), with 21.3 and 16.9% for the domesticated (TM1/PIMA) and wild (YUC/K101) comparisons, respectively. Yet, the number of these changes that were shared between domestication events is much lower. Considering only the group of genes that changed bias in domesticated forms (blue dashed lines in Figure 2A), 10.3% were shared between TM1 and PIMA (Figure 2C, left). A similar comparison for wild forms (green dashed lines in Figure 2A) shows an even lower level (2.4%) of shared genes (Figure 2C, right).
Figure 2.—
Comparisons of changes in homeolog expression bias during fiber development in wild and cultivated G. hirsutum (wild, Yuc; domesticated, TM1) and G. barbadense (wild, K101; domesticated, Pima S-7). To analyze changes in homeolog bias during development, a linear model that included only one effect (time point, with three levels 5, 10, and 20) and an error was used. The 1484 P-values from each comparison were converted to q-values using the method of Storey and Tibshirani (2003). These q-values were used to identify the number of differentially biased genes for a given comparison when controlling the false discovery rate (FDR) at various levels. For each accession shown, genes exhibiting changes in homeolog bias were concatenated into a single list. (A) Numbers of shared and nonshared changes in homeolog expression bias between wild and domesticated forms of G. hirsutum (left) and G. barbadense (right). (B) Number of shared and nonshared changes between two different domesticated (left) and wild (right) species. (C) Number of shared and nonshared changes using only those genes that changed homeolog bias during domestication (left) or in the wild (right).
Genome-specific gene recruitment during cotton domestication:
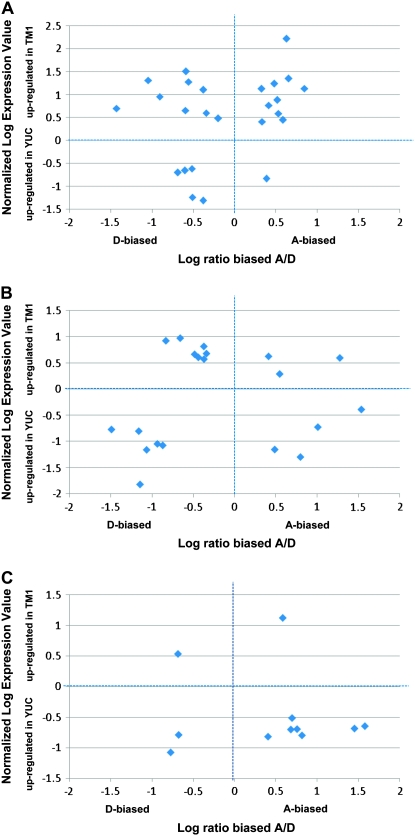
As discussed elsewhere (Udall and Wendel 2006) and noted above, one advantage of human mediated selection in an allotetraploid crop species is the ability to utilize or recruit genes from two different progenitor genomes. To explore this possibility in our system, we inspected the nature and expression levels of genes that were highly differentially biased toward A or D genomes as a result of human domestication. Although genes expressed at very low levels can have profound phenotypic effects, it may be that for many processes of relevance, overall level of expression is important. Accordingly, we measured overall gene expression for genes of interest by using a set of seven generic (non-SNP-specific) probes that were also spotted on the microarray for each of the 1484 duplicated genes. The relationship between homeolog-specific expression ratios and overall gene expression (generic probes) is summarized in Figure 3 using only genes that changed in both bias and expression (FDR < 0.05) during G. hirsutum domestication at the three developmental stages. Genes that increased in degree of D bias and are upregulated overall (i.e., as a gene pair) in the domesticated accession may be considered to be “D-recruited genes,” while those exhibiting enhanced A bias under domestication while being upregulated may be considered A recruited. At least half of the significantly biased and differentially expressed genes were upregulated in domesticated G. hirsutum relative to the wild form at 5 and at 10 DPA (Figure 3, A and B). The number of A-genome biased and D-genome biased genes is approximately equal. By 20 DPA (Figure 3C), most of the biased genes were actually downregulated in the domesticated form. Because there were fewer recruitments at the 20-DPA stage than at earlier stages, perhaps the total change in expression for this set of genes is less important than in the earlier fiber developmental stages.
Figure 3.—
Gene recruitment during G. hirsutum domestication. In addition to homeolog-specific probes, the microarray platform contains “generic” probes targeting both homeologs. Thus, this microarray platform interrogates both homeolog-specific and total (duplicate) gene expression. Significant differences in gene expression during domestication were determinate as previously described (Hovav et al. 2008a). Shown are the relationships between homeolog-specific expression ratios, using only genes that changed both in bias (x-axis) and in total expression (y-axis) during domestication, using q-values of FDR < 0.05 as the threshold. A, B, and C are for 5, 10, and 20 DPA, respectively. Genes upregulated in the domesticated form (top) can be considered as candidates for D genome (left) or A genome (right) recruited during G. hirsutum domestication. The list of genes and their putative biological roles are shown in supplemental Table 1.
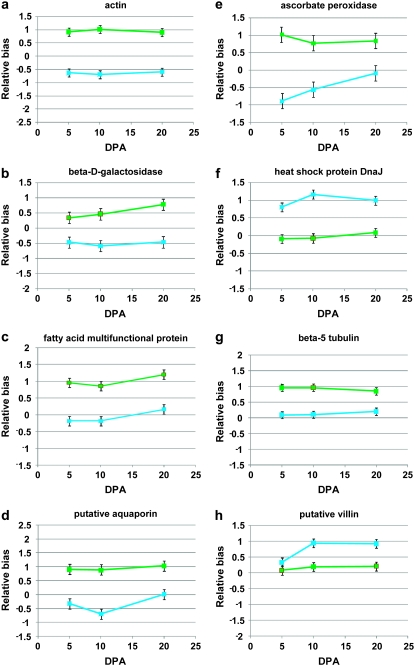
To the extent that total overexpression for any given gene pair in the domesticated form is affected by a change in homeolog bias, candidates for genes recruited under domestication might possibly be inferred from this type of analysis. An example of the recruitment expression pattern of a number of fiber-related genes that exhibit homeolog bias during fiber development is presented in Figure 4. Some of these genes show changes in total expression as well (Generic probes; Figure 4, A–C). The actin, β-5-tubulin, and villin genes (the first is changed in G. hirsutum and the other two in G. barbadense), for example, are part of the microtubule and cytoskeleton formation system, a process important to plant cell elongation in general and cotton fiber elongation in particular (Whittaker and Triplett 1999; Taliercio and Boykin 2007; Xiang et al. 2007; Hovav et al. 2008c). The β-d-galactosidase gene, important for primary cell-wall synthesis, has been described as predominantly expressed in fiber cells and tightly associated with fiber elongation (Zhao et al. 2001; Hovav et al. 2008c). Another interesting group is the one containing ascorbate peroxidase and the heat-shock protein Dnaj. These were previously shown to be stress-related genes, assisting in the regulation of reactive oxygen species levels and maintaining cell elongation under high redox-related conditions associated with fiber elongation (Hong-Bin et al. 2007; Hovav et al. 2008b).
Figure 4.—
Biased homeolog expression patterns for fiber-related genes during fiber development (DPA, days postanthesis). Shown are differences in homeolog contributions to the transcriptome between domesticated (blue) and wild (green) forms, where positive and negative relative bias indicate preferential accumulation of A-genome and D-genome homeologs, respectively. A–F are for G. hirsutum, whereas G and H are for G. barbadense. A–C show genes that were upregulated as a duplicate gene pair in the domesticated form and also changed bias (cf. Figure 3).
Concluding remarks:
This study has used a high throughput SNP-specific technology for detecting changes in homeolog expression in domesticated and wild tetraploid cotton plants. We show that two independent cotton domestication events were characterized by different degrees of genome-specific bias. In both events, however, the D genome was preferentially expressed, and the changes in homeolog expression bias during fiber development were higher in the cultivated form. Examples for several genes related to fiber development are provided. Given that the total number of genes in the cotton genome may be ∼50,000 (Rabinowicz et al. 2005), the 1484 gene pairs tested here include perhaps 5% of the total genic diversity. This suggests that for every 5 genes suggested as candidates for novel gene recruitment, another 95 exist that were not explored. Even though our platform permitted discrimination among homeologs for a relatively small proportion of the total genic content of the cotton genome, we used a large number designed as an unbiased, random sample. This study suggests that temporal partitioning of duplicate gene expression may, in aggregate, contribute significantly to processes important in fiber development and evolution under domestication. By extension, the data point to a hitherto underexplored dimension to the adaptive significance of polyploidy, namely, the coordination and regulation of newly combined transcriptional networks that lead to physiological and/or morphological innovation under human selection.
References
- Adams, K. L., and J. F. Wendel, 2005. Novel patterns of gene expression in polyploid plants. Trends Genet. 21 539–543. [DOI] [PubMed] [Google Scholar]
- Applequist, W. L., R. Cronn and J. F. Wendel, 2001. Comparative development of fiber in wild and cultivated cotton. Evol. Dev. 3 3–17. [DOI] [PubMed] [Google Scholar]
- Bluthgen, N., K. Brand, B. Cajavec, M. Swat, H. Herzel et al., 2005. Biological profiling of gene groups utilizing gene ontology. Genome Inform. Ser. Workshop Genome Inform. 16 106–115. [PubMed] [Google Scholar]
- Brubaker, C. L., F. M. Bourland and J. F. Wendel, 1999. The origin and domestication of cotton, pp. 3–31 in Cotton: Origin, History, Technology and Production, edited by C. W. Smith and J. T. Cothren. John Wiley & Sons, New York.
- Brubaker, C. L., and J. F. Wendel, 1994. Reevaluating the origin of domesticated cotton (Gossypium-hirsutum Malvaceae) using nuclear restriction-fragment-length-polymorphisms (Rflps). Am. J. Bot. 81 1309–1326. [Google Scholar]
- Chen, Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant. Biol. 58 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., M. Ha and D. Soltis, 2007. Polyploidy: genome obesity and its consequences. New Phytol. 174 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillehay, T. D., J. Rossen, T. C. Andres and D. E. Williams, 2007. Preceramic adoption of peanut, squash, and cotton in northern Peru. Science 316 1890–1893. [DOI] [PubMed] [Google Scholar]
- Endrizzi, J. E., E. L. Turcotte and R. J. Kohel, 1985. Genetics, cytology, and evolution of Gossypium. Adv. Genet. 23 271–375. [Google Scholar]
- Flagel, L., J. A. Udall, D. Nettleton and J. F. Wendel, 2008. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryxell, P. A., 1979. The Natural History of the Cotton Tribe. Texas A&M University Press, College Station, TX.
- Gaeta, R. T., J. C. Pires, F. Iniguez-Luy, E. Leon and T. C. Osborn, 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong-Bin, L., Y.-M. Qin, P. Yu, S. Wen-Qiang, M. Wen-Qian et al., 2007. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 175 462–471. [DOI] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, B. Chaudhary, L. Flagel, R. Rapp et al., 2008. a Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. USA 105 6191–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, B. Chaudhary, E. Hovav, L. Flagel et al., 2008. b The evolution of spinable cotton fiber entailed natural selection for prolonged development and a novel metabolism. PLoS Genet. 4 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav, R., J. A. Udall, E. Hovav, R. A. Rapp, L. Flagel et al., 2008. c A majority of genes are expressed in the single-celled seed trichome of cotton. Planta 227 319–329. [DOI] [PubMed] [Google Scholar]
- Jiang, C. X., R. J. Wright, K. M. El-Zik and A. H. Paterson, 1998. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 95 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. J., and B. A. Triplett, 2001. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Liu, B., and J. F. Wendel, 2003. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29 365–379. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Osborn, T. C., 2004. The contribution of polyploidy to variation in Brassica species. Plant Physiol. 121 531–536. [Google Scholar]
- Percy, R. G., and J. F. Wendel, 1990. Allozyme evidence for the origin and diversification of Gossypium-barbadense L. Theor. Appl. Genet. 79 529–542. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P. D., R. Citek, M. A. Budiman, A. Nunberg, J. A. Bedell et al., 2005. Differential methylation of genes and repeats in land plants. Genome Res. 15 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, J., E. A. Feltus, V. N. Waghmare, G. J. Pierce, P. W. Chee et al., 2007. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 176 2577–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. SAM thresholding and false discovery rates for detecting differential gene expression in DNA microarrays, pp. 272–290 in The Analysis of Gene Expression Data: Methods and Software, edited by G. Parmigiani, E. S. Garrett, R. A. Irizarry and S. L. Zeger. Springer, New York.
- Taliercio, E. W., and D. Boykin, 2007. Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 7 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J. A., and J. F. Wendel, 2006. Polyploidy and crop improvement. Crop Sci. 46 S3–S14. [Google Scholar]
- Udall, J. A., J. M. Swanson, D. Nettleton, R. J. Percifield and J. F. Wendel, 2006. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 173 1823–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., 1995. Cotton, pp. 358–366 in Evolution of Crop Plants, edited by N. Simmonds and J. Smartt. Longman, London.
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Wendel, J. F., and R. C. Cronn, 2003. Polyploidy and the evolutionary history of cotton. Adv. Agron. 78 139–186. [Google Scholar]
- Westengen, O., Z. Huaman and M. Heun, 2005. Genetic diversity and geographic pattern in early South American cotton domestication. Theor. Appl. Genet. 110 392–402. [DOI] [PubMed] [Google Scholar]
- Whittaker, D. J., and B. A. Triplett, 1999. Gene-specific changes in alpha-tubulin transcript accumulation in developing cotton fibers. Plant Physiol. 121 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y., X. Huang, T. Wang, Y. Zhang, Q. W. Liu et al., 2007. ACTIN BINDING PROTEIN29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19 1930–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, G. R., J. Y. Liu and X. M. Du, 2001. Molecular cloning and characterization of cotton cDNAs expressed in developing fiber cells. Biosci. Biotechnol. Biochem. 65 2789–2793. [DOI] [PubMed] [Google Scholar]