Abstract
The complex aspects linking the nucleolus and ribosome biogenesis to cancer are reviewed here. The available evidence indicates that the morphological and functional changes in the nucleolus, widely observed in cancer tissues, are a consequence of both the increased demand for ribosome biogenesis, which characterizes proliferating cells, and the changes in the mechanisms controlling cell proliferation. In fact, the loss or functional changes in the two major tumor suppressor proteins pRB and p53 cause an up-regulation of ribosome biogenesis in cancer tissues. In this context, the association in human carcinomas of nucleolar hypertrophy with bad prognoses is worthy of note. Further, an increasing amount of data coming from studies on both hepatitis virus-induced chronic liver diseases and a subset of rare inherited disorders, including X-linked dyskeratosis congenita, suggests an active role of the nucleolus in tumorigenesis. Both an up-regulation of ribosome production and changes in the ribosome structure might causally contribute to neoplastic transformation, by affecting the balance of protein translation, thus altering the synthesis of proteins that play an important role in the genesis of cancer.
The relationship between the nucleolus and cancer has been the subject of study for many years. Abnormalities in the nucleolar morphology of cancer cells attracted the attention of tumor pathologists as early as the late 19th century, when Pianese1 reported that hypertrophic and irregularly shaped nucleoli were characteristic of malignant cells. From that moment on, a series of studies have been performed to clarify whether these nucleolar changes were a consequence of the cancerous state or if, instead, they might represent a cause of neoplastic transformation.
The nucleolus is the organelle of the interphase cell nucleus where the biogenesis of ribosomes takes place.2,3 Nucleolar hypertrophy and increased ribosome biogenesis have been observed in all normal mammalian cells stimulated to proliferate as a result of the higher biosynthetic demand characterizing the cycling cells.4 Indeed, in proliferating cells the amount of cell constituents must be increased to ensure that daughter cells have the necessary complement for survival and normal functioning.5,6,7 This result is accomplished by increased synthesis of proteins that, in turn, is induced by an up-regulation of the rate of ribosome production. Cell proliferation appears to be closely coordinated with nucleolar function. In fact, there is evidence that the mechanisms controlling cell proliferation also regulate the rate of ribosome biogenesis. This relationship exists in cancer cells in which changes in either tumor suppressors or proto-oncogenes, which are responsible for uncontrolled cell proliferation, also up-regulate ribosome biogenesis, thus increasing the cell growth rate according to the modified cell cycle progression rate.8,9 Therefore, in this context, it is reasonable to consider nucleolar structural-functional changes in tumors as a mere consequence of both the proliferative activity of cancer cells and alterations of the mechanisms controlling cancer cell proliferation.
In recent years some data have been produced that also suggest an active role of ribosome biogenesis in tumorigenesis. Human nontumor lesions characterized by an up-regulation of nucleolar function were found to be associated with an increased risk of neoplastic transformation, and evidence shows that people with inherited diseases characterized by the production of abnormal ribosomes have a very high incidence of cancer. A mechanism of neoplastic transformation, supported by quantitative and qualitative defects in ribosome biogenesis, has been proposed. These defects might be responsible for alterations in the translation of specific mRNAs that induce altered expression of proteins involved in neoplastic transformation.10 Therefore, although on the one hand the nucleolar changes in cancer cells can be considered to be the consequence of neoplastic transformation, on the other hand, increasing evidence suggests that quantitative and qualitative changes in ribosomes may be involved in tumorigenesis.
The aim of the present work was to review these two different and complex aspects linking the nucleolus to cancer. For the functional aspects of the nucleolus that are only indirectly connected with its major function in ribosome biogenesis (such as viral replication, control of aging, cell cycle regulation, modifications of small nuclear ribonucleoproteins, sensing of cellular stress and telomerase maturation), but that may be of some interest for broadening the knowledge on the relationship between the nucleolus and cancer, we refer the reader to specific reviews.11,12,13,14,15
Nucleolar Changes as a Consequence of Neoplastic Transformation
Before discussing the relationship between neoplastic transformation and nucleolar changes, we will briefly describe the normal structural-functional organization of the nucleolus and how, in recent years, it has been possible to evaluate precisely the nucleolar changes in human tumors. The nucleolus can be easily detected by contrast-phase light microscopy in living cells, as well as by the higher concentration of RNA and proteins in the nucleolus than in the nucleoplasm (Figure 1a). In hematoxylin and eosin (H&E)-stained histological sections from routinely processed tissue samples, the nucleoli are intensely stained by eosin owing to their very high protein content (Figure 1b).
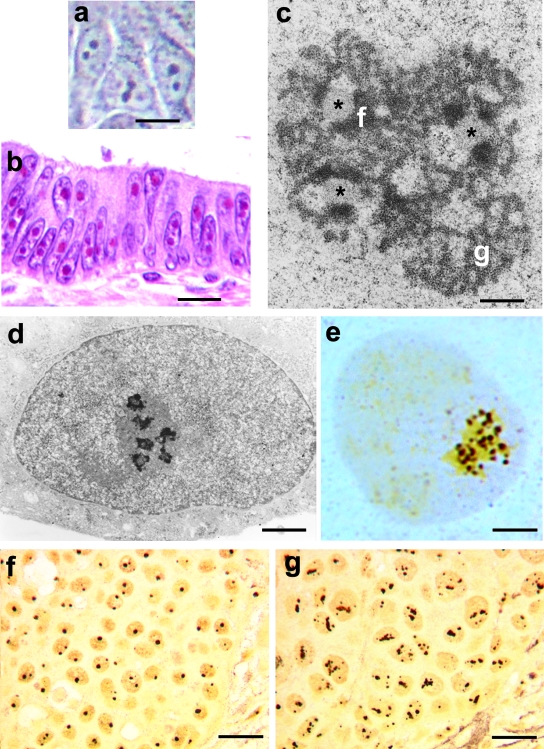
Figure 1.
a: Human breast cancer cells cultured in vitro visualized using the contrast phase microscope. The nucleoli appear as highly refractive bodies. b: Histological section of human colon carcinoma stained with H&E. The nucleoli are intensely stained by eosin. c: Electron microscope visualization of the nucleolus of a TG cell (established cell line from a human fallopian tube cancer) after uranium and lead staining. Three fibrillar centers are present (asterisks), with the closely associated dense fibrillar component (f). The granular component (g) is organized in cord-like structures. d: TG cell, after specific AgNOR protein staining, at the electron microscopy level. Only the fibrillar centers and the associated dense fibrillar component are stained. e: Same preparation as in d, but visualized with light microscopy. The silver-stained structures appeared as well-defined, darkly stained dots clustered in the nucleolus. f and g: Histological sections of two human breast carcinomas, stained for the AgNOR proteins. The nucleoli are specifically stained. Note the small size on the nucleoli in the case reported in f, which was characterized by a good prognosis, and the larger size of nucleoli in the case reported in g, characterized by a fatal outcome. Scale bars: 12 μm (a); 18 μm (b); 0.3 μm (c); 1.25 μm (d); 3.25 μm (e); 20 μm (f, g).
At the electron microscope level, mammalian cell nucleoli constantly exhibit three major components: fibrillar centers, which appear as roundish structures of varying size, with a very low electron opacity; the dense fibrillar component, which frequently constitutes a rim intimately associated with the fibrillar centers, composed of densely packed fibrils; and the granular component, composed of granules that surround the fibrillar components (Figure 1c).2,3 Transcriptionally active ribosomal genes are located in the fibrillar centers and the intimately associated dense fibrillar component. Therefore, the fibrillar centers plus the associated dense fibrillar component can be considered to represent the structural-functional units of the nucleolus producing rRNA molecules that migrate to the granular component where they undergo maturation for ribosome subunit constitution.16,17 Some of the proteins located in the fibrillar components, and necessary for rRNA transcription and processing [nucleolin, nucleophosmin, the upstream binding factor (UBF), the largest RNA polymerase I (RPI) subunit], are selectively stained by the same silver-staining method used to visualize the nucleolar organizer regions (NORs) on metaphase chromosomes (Figure 1d); therefore, these proteins are called AgNOR proteins, and the silver-stained fibrillar components are called interphase AgNORs.18,19 The silver staining procedure for the AgNOR proteins makes it possible to visualize—even at the light microscope level—the interphase NORs, the structural-functional units of the nucleolus, as well-defined black dots within the nucleolar body (Figure 1e); these can therefore be quantified very precisely by morphometric analysis.20 Because the number and the area occupied by interphase AgNORs within the nucleolus are directly related to both the whole nucleolar size21 and its transcriptional activity,22,23 the evaluation of the distribution of the interphase AgNORs appears to be a very simple method for quantifying nucleolar changes and also for obtaining precise information on the ribosome biogenesis activity of the cell at the light microscope level. This staining procedure has been applied to routinely processed histological tumor samples24 and is a very useful tool for studying the nucleolar changes in tumor pathology.25,26
The nucleolar changes of practically all human cancers have been evaluated and shown to be highly variable and independent of the histogenesis of the tumors as well as within the same tumor sample.26 Interestingly, even though nucleolar hypertrophy and functional up-regulation of the nucleolus are generally considered to be a characteristic of cancer cells, these studies demonstrate that this was not always true: the nucleolar size and functional activity is sometimes lower than those of the corresponding normal cells. In fact, the nucleolar changes in tumors are closely related to the number of proliferating cells within the cancer tissue and the rapidity of proliferation of the cycling cells, kinetics parameters that are highly variable in human tumors and sometimes of lower value than in the corresponding proliferating normal tissues.25,26 There is now evidence that the up-regulation of the nucleolar function occurring in proliferating cells is attributable to the products of the same proto-oncogene and tumor suppressor genes that control cell proliferation.
Changes of proto-oncogenes and tumor suppressor genes occur very frequently in a variety of human cancers that are responsible not only for the loss of the normal control mechanisms of cell proliferation and cell cycle progression,27 but also for an enhanced ribosome biogenesis.8,9 c-Myc, which is necessary and sufficient for cell-cycle entry,28 may be overexpressed in a variety of human hematological malignancies and solid tumors29 and directly enhances RNA polymerase I transcription activity by binding to specific consensus elements of rDNA and recruiting the selectivity factor 1 (SL1) to the rDNA promoter.30,31 In fact, SL1 is necessary for rDNA transcription by recruiting RNA polymerase I, in a complex together with the UBF, to the rRNA gene promoter.32 Furthermore, the Myc oncoprotein directly controls the transcription of several nucleolar proteins necessary for ribosome biogenesis.33 Cyclin D and E, which control the normal cell cycle progression and which may be overexpressed or altered in a number of human tumors,34 also induce the phosphorylation of UBF by cyclin-dependent kinase 4 (Cdk4)-cyclin D1- and Cdk2-cyclin E mechanism, thus enhancing the transcription of ribosome genes.35 However, among the genetic changes leading to neoplastic transformation that are of special importance for both their highly frequent occurrence and their effects on ribosome biogenesis, those involving the retinoblastoma tumor suppressor protein (pRB) and p53 pathways are undoubtedly the most relevant.
During the cell cycle pRB controls passage through the G1/S phase restriction point by interacting with the family of transcription regulators called E2Fs,36 which regulate the expression of those genes whose products are necessary for S phase progression.37 In its active hypophosphorylated form, pRB is bound to E2Fs and prevents them from activating E2F target genes whereas in the hyperphosphorylated form, pRB no longer binds to E2Fs, which are let free to activate the target genes. pRB undergoes a progressive phosphorylation through the cell cycle phases. Phosphorylation of pRB is triggered in the early G1 phase by cyclin D-cyclin-Cdk-4 and -6 complexes and is completed, at the end of the G1 phase, by cyclin E-Cdk-2 complexes. The activities of the Cdks are in turn constrained by the Cdk inhibitors: Cdk-4 and Cdk-6 are inhibited mainly by p16Ink4a whereas the Cdk-2 is negatively regulated by p21Cip1 and p27.38 There is evidence that pRB, in addition to controlling the transition from G1 to S phase, also modulates ribosome biogenesis, thus directly linking cell growth to cell proliferation. Active, nonphosphorylated pRB inhibits rRNA synthesis by binding to UBF.39,40,41,42,43 Therefore, in cycling cells the progressive phosphorylation of pRB from early G1 phase to G2 phase44 induces a progressive increase of the rRNA transcription rate, along with a progressive enlargement of the nucleolar size, from the G1 to the G2 phase. In human cancers, genetic changes involving the pathway leading to pRB inactivation, such as RB1 mutation or deletion, INK4a mutation, deletion or gene silencing and cyclin D1 or Cdk4 overexpression, are observed rather frequently.36 These changes, causing either pRB loss or more frequently pRB hyperphosphorylation, cancel or strongly reduce the negative control over rRNA transcription according the mechanisms described above.
TP53 mutations, resulting in p53 inactivation and nuclear accumulation, are also very frequently found in human tumors.45,46 p53, in addition to its role in response to a variety of cellular stresses leading to either apoptosis or induction of the Cdk2 inhibitor p21Cip1, inhibition of cyclin E/Cdk2, and pRB-dependent cell cycle arrest,47 directly influences ribosome biogenesis. Accumulation of the wild-type p53 inhibits RNA Pol I transcription by binding to the selectivity factor SL1, thus hindering the formation of the UBF-SL1 complex necessary for RNA polymerase I recruitment to the rRNA gene promoter.48 Mutated, inactive p53 can no longer exert its negative control over both cell proliferation and rRNA transcription. The activity of p53 may also be influenced by changes in the expression of p14Arf, the tumor suppressor protein encoded by the Arf gene located in the Ink4a-Arf locus.49 The Arf gene is induced by a series of stress signals such as hyperproliferative signals emanating from oncogenic Ras and overexpressed Myc and may be mutated or silenced in tumor cells.50,51 p14Arf helps the stabilization of p53 by binding to Hdm2, the factor responsible for p53 degradation.52 p14Arf, in addition to the activation of the p53 pathway, hinders the ribosome biogenesis by inhibiting UBF recruitment on the transcription complex53 and by lengthening rRNA processing.54 Therefore the loss of p14Arf expression may be responsible for the enhancement of ribosome biogenesis both directly and through the action on p53 stabilization.
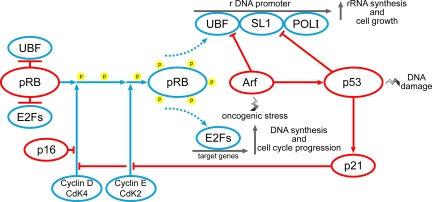
These data, taken together, demonstrate that the up-regulation of ribosome biogenesis and the consequent nucleolar changes in cancer cells are the consequence of the functional changes of those oncogene and tumor suppressor proteins that control cell proliferation, with the high variability of the severity of these changes explaining the high variability of the nucleolar changes in cancer. Figure 2 shows a simplified diagram of the complex relationship between the mechanisms controlling cell proliferation and ribosome biogenesis, in which the importance of pRB and p53 is stressed.
Figure 2.
Simplified diagram of the control mechanisms of cell proliferation and ribosome biogenesis by pRB and p53. Inactive pRB is linked to UBF and E2Fs. When the cell enters the cell cycle, phosphorylation of pRB by cyclin-dependent kinases 4 and 2 occurs, freeing UBF and the E2Fs. UBF, together with SL1, binds to the rDNA promoter, thus allowing rDNA transcription by RNA polymerase I (Pol I). The E2Fs bind to their target genes, thus allowing DNA duplication and cell cycle progression. DNA damage stabilizes p53, which blocks ribosome biogenesis both by directly binding to SL1 and by hindering pRB phosphorylation through the inhibitory action of p21 on Cdk4 and 2. Cdk4 may be also constrained by p16Ink4a. Ribosome biogenesis may also be inhibited by Arf, which binds to SL1 and stabilizes p53.
This representation of the relationship between factors regulating cell proliferation and ribosome biogenesis in cancer cells has also been validated by the observation that in human tumors the degree of nucleolar hypertrophy is directly related to the severity of the pRB and p53 pathway alterations.55 It is worth recalling that these findings have practical relevance in tumor pathology constituting the rational basis for the use of the nucleolus parameter as an indicator of the clinical outcome of cancer disease. Disruption of the pRB and p53 pathway induces genetic instability in cancer cells that affects a series of processes leading to an increased biological aggressiveness of cancer and, from the clinical point of view, to a bad prognosis of the cancer disease.56 Because pRB and p53 inactivation are also responsible for the up-regulation of ribosome biogenesis, the presence of very enlarged nucleoli in cancer tissues is frequently associated with a poor clinical outcome,57 being that nucleolar hypertrophy is a valuable prognostic parameter in tumor pathology.7,20,27 Breast histological samples from a cancer without changes of pRB and p53 and with small nucleoli (Figure 1f) and from a cancer with RB1 loss and mutated p53 and with hypertrophic nucleoli (Figure 1g) are shown, characterizing good and poor prognosis, respectively.
Nucleolar Functional Up-Regulation as a Risk Factor in Developing Cancer
With regard to the relationship between the nucleolus and cancer, the results reported in the previous section indicate that the nucleolar changes in cancer cells can be considered to simply represent an adaptation of the nucleolus to the acquired new functional characteristics of the transformed cells. However, there is also evidence of an active role of the nucleolus in cancer. Several human nontumor tissue lesions are characterized by an increased risk of neoplastic transformation. Among these, chronic liver disease, caused by viral hepatitis, alcohol abuse, and inborn metabolic errors, is one of the most important and widespread conditions associated with cancer development. Eighty percent of all hepatocellular carcinomas (HCCs) worldwide occur when chronic liver disease has reached the cirrhotic stage.58 The mechanisms underlying the development of HCC in a chronically diseased liver are currently being investigated, with special emphasis on the role of the genetic alterations occurring during the hepatocarcinogenesis process. These studies have not identified a common profile of genetic changes leading to a common pathway of neoplastic transformation. In fact, gene expression analysis shows that each HCC is characterized by its own unique profile of genetic changes.59
Interestingly, there is evidence that, as far as chronic liver diseases from viral infections are concerned, an up-regulation of hepatocyte ribosome biogenesis is constantly associated with later onset of HCC.60,61 Both hepatitis B (HBV) and hepatitis C (HCV) viruses, which are responsible for the development of chronic liver disease, have been shown to up-regulate the activity of the RNA polymerase I and III.62,63 The HBV X protein, required for viral replication and correlated with the hepatocarcinogenesis of individuals chronically infected with HBV, modulates RNA polymerase I-dependent rRNA transcription by enhancing the rRNA promoter activity,62 and activates specific RNA polymerase III-dependent promoters.63 The HCV core protein, implicated in the control of cell growth and proliferation,64,65 enhances the recruitment of UBF and RNA polymerase I to the rRNA promoter and cooperates in the activation of RNA polymerase III-dependent transcription.66 According to these findings, it has been suggested that the increased ribosome production induced by these viral proteins may play an important role during tumorigenesis by sustaining an enhanced rate of cell proliferation. A series of studies performed to define whether nucleolar changes might be related to neoplastic transformation in chronic liver disease from viral infection demonstrated that the presence of abnormally enlarged hepatocyte nucleoli represent a very strong risk factor for developing HCC, mainly in the HBV-related cirrhotic livers.61 In other words, only those chronic liver lesions in which the viral infection induced hypertrophy, and consequently a functional up-regulation of nucleoli, would be susceptible to cancer development.
The observation that the up-regulation of ribosome biogenesis represents a risk factor for developing HCC does not permit the deduction of any cause-effect relationship. The HBV and HCV proteins that stimulate the RNA polymerase I and III activity are transcriptional activators that also modulate the expression of genes involved in the control of the cell cycle machinery.67,68 Therefore, nucleolar changes may only be the consequence of the activation of the cell cycle-related genes. However, it is worth noting that the nucleolar hypertrophy, which precedes the onset of cancer, appears to be independent of cell proliferation: in fact, most hepatocytes characterized by the presence of enlarged nucleoli constitute dysplastic foci in which no proliferative activity was found.69 Thus, it is likely that functional up-regulation of the nucleolus may either be necessary for, or at least facilitate, progression toward neoplastic transformation of chronically diseased liver cells. Additionally, as will be discussed in the next section, an increased production of ribosomes, not coordinated with cell division, may also alter the translational mechanisms, causing quantitative variations of the synthesis of specific proteins that control cell cycle progression, thus favoring cell proliferation.
Alterations of Ribosome Biogenesis as a Cause of Neoplastic Transformation
At the present time, one major question is indeed whether a perturbation in the normal process of ribosome production can induce, or contribute to inducing, neoplastic transformation by itself. The ultimate products of ribosome biogenesis, the ribosomes, act as the central players of the translation of mRNAs into proteins. Deregulation of mRNA translation has already been demonstrated to trigger neoplastic transformation. This is observed, for instance, when the process of translation initiation is modified by the overexpression of the eukaryotic initiation factor 4E (eIF4E), a rate-limiting protein in the initiation process.70 An increased initiation process rate leads to selective changes in mRNA translation resulting in increased levels of a panel of proteins (such as proto-oncogenes, anti-apoptotic factors, growth factors, and matrix remodeling proteins), whose functions support neoplastic transformation.71 In vitro overexpression of eIF4E, in cooperation with other pro-oncogenic proteins such as v-myc and E1A, is able to induce in primary cells the morphological changes and the capacity to grow in soft agar that are characteristic of neoplastic transformation.70 In vivo, transgenic mice expressing higher levels of eIF4E display a marked increase in tumorigenesis.72 On the other hand, a similar mechanism can also be hypothesized in the case in which a defect in ribosome biogenesis may generate ribosomes with an intrinsically altered translation capability, thus affecting levels of the proteins playing an important role in neoplastic transformation and progression. The whole ribosome is constituted by almost 7000 RNA nucleotides in length (∼200 of which are modified), assembled with at least 82 different proteins. Therefore, a variety of different possible modifications can alter such a complex structure at different levels.
Such an intriguing hypothesis can be used to explain what occurs in a rare inherited syndrome called X-linked dyskeratosis congenita (DC), attributable to a mutation in the DKC1 gene. DC is characterized by skin, mucosal, and bone marrow failure associated with an increased risk of developing tumors.73 The DKC1 product, dyskerin, is a nucleolar protein that has at least two different major functions. First, dyskerin is a component of small nucleolar ribonucleoproteic particles (snoRNPs) involved in rRNA processing. In particular, dyskerin is necessary for the site-specific conversion of uridine residues in rRNA to pseudouridine.74 Secondly, dyskerin binds to the telomerase RNA component stabilizing the telomerase enzymatic complex.75 Therefore, the alterations of dyskerin result in defects of ribosome biogenesis, such as the reduction of rRNA pseudouridylation and the slowing down of ribosome rRNA processing rate76 as well as in defective telomerase function attributable to the degradation of the telomerase RNA component, causing the impairment of the enzymatic activity of the telomerase complex and the progressive telomere shortening of proliferating cells.75,76,77,78
Recent reports have sought to address the specific function of dyskerin that, when lost, contributes to tumorigenesis. Loss of the telomerase function may lead to an increased susceptibility to cancer, as observed in mice lacking telomerase activity because of the deletion of the telomerase RNA component. In this case, telomere attrition—which in mice generates functional consequences only after being present in the hTR knockout mice for five to six generations—leads to a limited increase in the incidence of tumors in late-generation mice, probably because of chromosomal instability.79 On the other hand, an increased incidence of tumors has been observed in a hypomorphic mouse in which the DKC1 gene is targeted, and dyskerin expression is reduced to 30% as compared to control littermates.76 In this mouse model that, because the DKC1 knockout is embryonic lethal,80 is the only one available so far for in vivo studies, the reduced expression of dyskerin results in a reduction of both telomerase activity and rRNA pseudouridylation. Interestingly, in the DKC1 hypomorphic mice the increase in tumor incidence is also observed in the early generations, when telomeres are still very long. It has been therefore concluded that the defective pseudouridylation of rRNA might itself play a role in promoting tumor genesis. Moreover, in cells from both DC patients and the DKC1 hypomorphic mice, a selective defect has been demonstrated in the translation of a group of mRNAs containing internal ribosome entry site elements.81 Internal ribosome entry site nucleotide sequences, originally described in viral mRNA, are also present in some eukaryotic mRNA molecules. Their presence in a mRNA makes it possible to accomplish its translation independently of other elements generally required for translation initiation, including the mRNA 5′ cap and the availability of translation initiation factors. The internal ribosome entry site translation defects in DC cells, if associated with a reduction of 5′ cap-dependent translation initiation, result in the relative impairment of the translation of some mRNAs, including those encoding the tumor suppressor p27, as well as antiapoptotic factors such as Bcl-xL and XIAP.81 The defective translation of tumor suppressors might then contribute to the neoplastic phenotype, at least in tumors arising in DC patients. It would be interesting to know whether such a ribosome defect might support malignant transformation in tumors arising in the general population also. In fact, it has been demonstrated that dyskerin expression is extremely variable in human tumors of various histological origins, such as breast, lung, and colon carcinomas, and B-cell lymphomas. Moreover, in breast carcinomas, dyskerin expression levels are actually closely related with its functions. In particular, a group of tumors characterized by very low dyskerin expression also exhibited a defective rRNA pseudouridylation.82 These results, together with the data obtained studying DC patients and the DC mouse model, are compatible with a scenario in which intrinsic ribosome alterations because of defective dyskerin function may contribute to neoplastic transformation by an altered translation-mediated mechanism both in DC and in a subset of human tumors.
In addition to DC, there is evidence that an altered ribosome biogenesis is associated with increased tumor susceptibility in other inherited disorders. These diseases include Diamond-Blackfan anemia, cartilage hair hypoplasia, and Shwachman-Diamond syndrome. Interestingly, all these disorders are characterized by some grade of bone marrow failure as in DC. Although the molecular pathogenesis of these diseases is less characterized than in DC, it is known that the increase in cancer susceptibility is associated with defects in ribosome biogenesis that may occur at different levels. For instance, in 25% of the familial and sporadic cases of Diamond-Blackfan anemia, the ribosome defect was attributable to mutations in RPS19, the gene coding for protein 19 in ribosomal small subunits.83 It is not clear what the specific consequences of these mutations on mRNA translation are; however S19 ribosomal protein is necessary for proper 18S rRNA processing and 40S subunit maturation in yeast.84 In another 2% of cases of Diamond-Blackfan anemia, the ribosome defect was attributable to mutations of the RPS24 gene coding for small ribosomal subunit protein 24.85 In cartilage hair hypoplasia the noncoding RNA component of the ribonucleoprotein complex RNase MRP, called RMRP, is mutated. This noncoding RNA has been shown to have several different functions, including mitochondrial DNA replication, cell cycle progression at the end of mitosis, and ribosomal RNA processing.86,87 The mutations described in cartilage hair hypoplasia primarily affect the processing of 5.8S rRNA, suggesting a ribosome involvement in the pathogenesis of this disorder also.88 A defect in rRNA processing is also behind Shwachman-Diamond syndrome. The function of the product of the SBDS gene, which is mutated in this syndrome,89 has been characterized only recently. Phylogenetic studies show that through evolution it shares a high similarity with a group of proteins involved in RNA metabolism, translation, and ribosome-associated function.90 Moreover, recent studies have demonstrated that the SBDS protein is localized in the nucleolus and is essential for a key step in 60S maturation and for the translational activation of ribosomes such as the shuttling of the ribosome subunit anti-associating factor eIF6.91
Interestingly, there is at least one additional inherited disorder in which ribosome biogenesis may be altered. Indeed, mutations of the Tcof1 gene have been reported in Treacher-Collins syndrome.92 The Tcof1 gene product, named treacle, is a nucleolar protein linking transcription of rRNA to its posttranscriptional site-specific methylation.93,94 In this disorder, mainly characterized by craniofacial deformities, no increase in tumor susceptibility and no bone marrow failure have been observed, indicating that these two types of consequences are not necessarily shared by every perturbation of ribosome biogenesis. Table 1 summarizes the main features of inherited disorders in which ribosome biogenesis is altered.
Table 1.
Human Inherited Disorders Attributable to Defects of Ribosomal Biogenesis and Cancer Susceptibility
| Disease | Gene(s) | Function(s) | Bone marrow failure | Cancer susceptibility |
|---|---|---|---|---|
| X-linked dyskeratosis congenita | DKC1 | rRNA and snRNA pseudouridylation, telomerase stabilization | Yes | Yes: lymphomas, squamous carcinomas |
| Diamond-Blackfan anemia | RPS19 (25%), RPS26 (2%), others? | 28S rRNA processing, 40S subunit maturation | Yes | Yes: leukemia, osteosarcoma |
| Cartilage-hair hypoplasia | RMRP | pre-rRNA cleavage | Yes | Non-Hodgkin’s lymphoma, basal cell carcinoma |
| Shwachman-Diamond syndrome | SBDS, others | rRNA processing—ribosome maturation? Nucleolar localization | Yes | Leukemia |
| Treacher-Collins syndrome | Tcfo1 | rRNA transcription and maturation | No | No |
Alterations of ribosome biogenesis and cancer susceptibility in inherited disorders.
In addition to these important observations in human pathology, in vivo experimental data from animal models also support the hypothesis that alterations in ribosome biogenesis may promote neoplastic transformation. In drosophila mutations, reducing the expression of the gene coding for ribosomal protein S6 can cause aberrant cell growth and neoplastic transformation of the hematopoietic system.95,96 In zebrafish many ribosomal protein genes behave as tumor suppressors. Indeed, heterozygous mutations in 11 different ribosomal protein genes lead to elevated cancer susceptibility.97 In mice, in addition to the DKC1 hypomorphic mouse model described above, another example is represented by the effect of the targeting of NPM, the gene coding for the nucleolar protein nucleophosmin. Nucleophosmin is a multifunctional protein involved in rRNA processing and export, the response to stress stimuli, and the control of cellular genomic integrity and ploidy.98 Although NPM knockout is embryonic lethal, the reduction of nucleophosmin expression (such as in the NPM wild-type/knockout heterozygous mice) is associated with accelerated tumorigenesis.98 However, in this case, because of the variety of nucleophosmin functions, its role in ribosome biogenesis cannot be identified as the factor responsible for the tumor susceptibility.
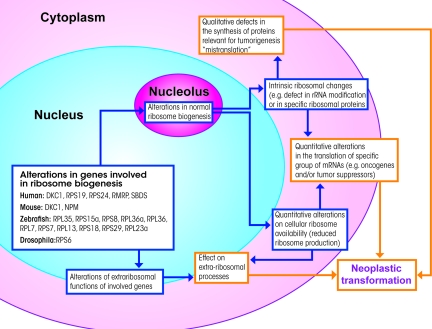
Most of the examples reported here are perfectly in line with the hypothesis of a mechanism of neoplastic transformation supported by defects in ribosome biogenesis. As hypothesized, this can be explained by ribosome intrinsic defects leading to alterations in the translation of specific mRNAs that are important for tumorigenesis. However, alternative possible explanations also exist. First of all, alterations in ribosome biogenesis may produce quantitative changes in cellular ribosome availability that could influence the translation of genes involved in neoplastic transformation. Second, most of the proteins involved, including ribosomal proteins,99 have extra-ribosomal functions. These other functions may support tumorigenesis independently of ribosome function and mRNA translation. Last, the disruption of a normally well-coordinated process such as ribosome biogenesis may generate by-products (eg, unassembled ribosomal proteins) that may affect other cellular functions that are relevant in tumor genesis (eg, checkpoints).100 Figure 3 assembles all these proposed hypotheses. In any case, it is clear that several, independent alterations of ribosome biogenesis are causally related to an increase in tumor development. This implies a series of relevant consequences, not only for the related human disorders but also for the understanding of basic biological processes.
Figure 3.
Schematic representation of the different cellular events through which altered ribosome biogenesis might contribute to neoplastic transformation. Alterations in the genes involved in ribosome biogenesis may generate intrinsic altered ribosomes (eg, hypopseudouridylated ribosomes in X-linked dyskeratosis congenita) characterized by an altered capacity for translating mRNAs coding for proteins associated with tumorigenesis. Alternatively, altered ribosome biogenesis may be associated with quantitative changes in cellular ribosome availability, which could influence translation of the genes involved in neoplastic transformation as well. It should also be considered that most of the proteins involved in ribosome biogenesis have extra-ribosomal functions that may support tumorigenesis independently of ribosome function and mRNA translation.
Conclusions
The relationship between the nucleolus and cancer appears to be much more complex than that known just a few years ago. The morphological and functional changes of the nucleolus, widely observed in cancer tissues, are the consequence of the increased metabolic necessities that characterize proliferating cells. Moreover, the most frequent gene alterations occurring in human tumors are responsible for up-regulation of the rRNA transcriptional activity. In this context, which is still currently true, we can reasonably speak of an adaptation of the nucleolus in the transformed cells. On the other hand, there is increasing evidence, coming from the studies of certain human diseases, that also suggests an active role of ribosome biogenesis in tumorigenesis. In chronic liver diseases, especially in those from HBV infection, the presence of hepatocyte nucleolar hypertrophy is closely associated with the later development of HCC. In this case an up-regulated ribosome biogenesis rate might be responsible for changes in the balance of the translational processes, with an increased quantity of ribosomes making it easier to translate those mRNAs (whose product might be involved in tumorigenesis) but that were not, or to a lesser degree, translated in normal conditions. In addition to quantitative changes of ribosomes, qualitative ribosome modifications have also been found to be associated with an increased frequency of cancer development, as in the case the inherited disorder DC. Ribosomes with an intrinsically altered translation capability might affect the balance of protein translation by favoring the synthesis of these proteins, playing an important role in neoplastic transformation.
The study of the quantitative and qualitative changes of ribosome biogenesis as possible causes of cancer represents a new, exiting research topic both for gaining more insight into the mechanisms involved in the transformation from a normal to a neoplastic cell and for trying new therapeutic strategies for cancer disease treatment. However, the enthusiasm for this topic is counterbalanced by the difficulties in obtaining clear-cut models to use for experimental studies. Indeed, the modification of a fundamental process such as ribosome biogenesis almost inevitably leads to a variety of effects. This makes it extremely difficult to draw definitive conclusions. Hopefully, new technological approaches and the definition of more selective experimental models will make it possible in the near future to characterize precisely the mechanisms linking ribosome biogenesis changes to cancer. Given the available evidence discussed earlier, it seems appropriate to apply these approaches to experimental models reproducing human pathological conditions in which a link between changes in ribosome biogenesis and neoplastic transformation has been observed, such as in chronic liver diseases and in inherited disorders with mutations in ribosome biogenesis genes. This could lead to the identification of the translational targets of the ribosomal changes associated with cancer susceptibility, providing more detailed knowledge of the molecular mechanisms underlying the contribution of ribosome biogenesis to neoplastic transformation. This knowledge could be also useful in studies aiming to clarify whether these defects might support malignant transformation in subsets of human tumors of different origins, thus representing one mechanism of tumorigenesis.
Acknowledgments
We thank the anonymous reviewers whose suggestions allowed the quality of the manuscript to be greatly improved.
Footnotes
Address reprint requests to Prof. Massimo Derenzini, Alma Mater Studiorum–Università di Bologna, Dipartimento di Patologia Sperimentale, Via San Giacomo 14, 40126 Bologna, Italy. E-mail: massimo.derenzini@unibo.it.
Supported by the Pallotti’s Legacy for Cancer Research, the Bologna University (funds for Ricerca Fondamentale Orientata), and the Italian Ministry of University and Research (funds for Programmi di Ricerca di Rilevante Interesse Nazionale 2006).
References
- Pianese G. Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und experimentelle Untersuchungen. Beitr Pathol Anat Allgem Pathol. 1896;142:1–193. [Google Scholar]
- Busch H, Smetana K. New York: Academic Press,; The Nucleolus. 1970 [Google Scholar]
- Hadjiolov AA. The nucleolus and ribosome biogenesis. Vienna: Springer Verlag,; 1985:pp 1–267. [Google Scholar]
- Derenzini M, Ploton D. Interphase nucleolar organizer regions in cancer cells. Int Rev Exp Pathol. 1991;32:149–192. doi: 10.1016/b978-0-12-364932-4.50008-3. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nat Cell Biol. 2000;2:E71–E72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty T. The flexible evolutionary anchorage-dependent Pardee’s restriction point of mammalian cells: how its deregulation may lead to cancer. Biochim Biophys Acta. 2006;1765:38–66. doi: 10.1016/j.bbcan.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Sulić S, Panic L, Dikic I, Volarevic S. Deregulation of cell growth and malignant transformation. Croat Med J. 2005;46:622–638. [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Marciniak RA, Guarente L. Telomeres, the nucleolus and aging. Curr Opin Cell Biol. 1998;10:332–338. doi: 10.1016/s0955-0674(98)80008-2. [DOI] [PubMed] [Google Scholar]
- Olson MO, Hingorani K, Szebeni A. Conventional and nonconventional roles of the nucleolus. Int Rev Cytol. 2002;219:199–266. doi: 10.1016/S0074-7696(02)19014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA. The nucleolus—a gateway to viral infection? Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB, Jr, Weber JD. Nucleolar adaptation in human cancer. Cancer Invest. 2005;23:599–608. doi: 10.1080/07357900500283085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- Ploton D, O'Donohue M-F, Cheutin T, Beorchia A, Kaplan H, Thiry M. Three-dimensional organization of rDNA and transcription. Olson MOJ, editor. New York: Kluwer/Plenum,; The Nucleolus. 2004:pp 154–169. [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Structural organization of the nucleolus in mammalian cells. Methods Achiev Exp Pathol. 1986;12:26–62. [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez-Verdun D. The AgNOR proteins: qualitative and quantitative changes during the cell cycle. Micron. 2000;31:121–126. doi: 10.1016/s0968-4328(99)00068-2. [DOI] [PubMed] [Google Scholar]
- Trerè D. AgNOR staining and quantification. Micron. 2000;31:127–131. doi: 10.1016/s0968-4328(99)00069-4. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Farabegoli F, Trere D. Relationship between interphase AgNOR distribution and nucleolar size in cancer cells. Histochem J. 1992;24:951–956. doi: 10.1007/BF01046500. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Trerè D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Ploton D, Menager M, Jeannesson P, Himber G, Pigeon F, Adnet JJ. Improvement in the staining and in the visualization of the argyrophilic proteins of the nucleolar organizer region at the optical level. Histochem J. 1986;18:5–14. doi: 10.1007/BF01676192. [DOI] [PubMed] [Google Scholar]
- Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, O'Donohue M-F, Ploton D. Interphase nucleolar organizer regions in tumor pathology. Crocker J, Murray PG, editors. London: Wiley,; Molecular Pathology in Cellular Pathology. 2003:pp 138–152. [Google Scholar]
- Sherr CJ. The Pezcoller Lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Schlosser I, Hölzel M, Mürnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dross R, Browning PJ, Pelling JC. Do truncated cyclins contribute to aberrant cyclin expression in cancer? Cell Cycle. 2006;5:472–477. doi: 10.4161/cc.5.5.2516. [DOI] [PubMed] [Google Scholar]
- Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific Cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- White RJ, Trouche D, Martin K, Jackson SP, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene. 2000;19:4988–4999. doi: 10.1038/sj.onc.1203875. [DOI] [PubMed] [Google Scholar]
- Ciarmatori S, Scott PH, Sutcliffe JE, McLees A, Alzuherri HM, Dannenberg JH, te Riele H, Grummt I, Voit R, White RJ. Overlapping functions of the ppRB family in the regulation of rRNA synthesis. Mol Cell Biol. 2001;21:5806–5814. doi: 10.1128/MCB.21.17.5806-5814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donjerkovic D, Scott DW. Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 2000;10:1–16. doi: 10.1038/sj.cr.7290031. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Weber DJ, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activate p53. Nat Cell Biol. 1999;1:20–25. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treré D, Ceccarelli C, Montanaro L, Tosti E, Derenzini M. Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J Histochem Cytochem. 2004;52:1601–1607. doi: 10.1369/jhc.4A6454.2004. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C. Mutations of cell cycle regulators. Biological and clinical implications for human neoplasia. Am J Pathol. 1995;147:545–560. [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Ceccarelli C, Santini D, Taffurelli M, Trere D. The prognostic value of the AgNOR parameter in human breast cancer depends on the pRb and p53 status. J Clin Pathol. 2004;57:755–761. doi: 10.1136/jcp.2003.015917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht L, Desmet V, Roskams T. Preneoplastic lesions in human hepatocarcinogenesis. Liver Int. 2005;25:16–17. doi: 10.1111/j.1478-3231.2005.01016.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, Van De Rijn M, Botstein D, Brown PO. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Oliveri F, David E, Colombatto P, Bonino F, Brunetto MR. Is high AgNOR quantity in hepatocytes associated with increased risk of hepatocellular carcinoma in chronic liver disease? J Clin Pathol. 1993;46:727–729. doi: 10.1136/jcp.46.8.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treré D, Borzio M, Morabito A, Borzio F, Roncalli M, Derenzini M. Nucleolar hypertrophy correlates with hepatocellular carcinoma development in cirrhosis due to HBV infection. Hepatology. 2003;37:72–78. doi: 10.1053/jhep.2003.50039. [DOI] [PubMed] [Google Scholar]
- Wang HD, Trivedi A, Johnson DL. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol Cell Biol. 1998;18:7086–7094. doi: 10.1128/mcb.18.12.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufiero B, Schneider RJ. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- Kawamura H, Govindarajan S, Aswad F, Machida K, Lai MM, Sung VM, Dennert G. HCV core expression in hepatocytes protects against autoimmune liver injury and promotes liver regeneration in mice. Hepatology. 2006;44:936–944. doi: 10.1002/hep.21360. [DOI] [PubMed] [Google Scholar]
- Kao CF, Chen SY, Lee YH. Activation of RNA polymerase I transcription by hepatitis C virus core protein. J Biomed Sci. 2004;11:72–94. doi: 10.1007/BF02256551. [DOI] [PubMed] [Google Scholar]
- Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34(Suppl 1):S75–S78. doi: 10.1016/s1386-6532(05)80014-9. [DOI] [PubMed] [Google Scholar]
- Koike K. Molecular basis of hepatitis C virus-associated hepatocarcinogenesis: lessons from animal model studies. Clin Gastroenterol Hepatol. 2005;3:S132–S135. doi: 10.1016/s1542-3565(05)00700-7. [DOI] [PubMed] [Google Scholar]
- Lee RG, Tsamandas AC, Demetris AJ. Large cell change (liver cell dysplasia) and hepatocellular carcinoma in cirrhosis: matched case-control study, pathological analysis, and pathogenetic hypothesis. Hepatology. 1997;26:1415–1422. doi: 10.1002/hep.510260607. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Pandolfi PP. Initiation of mRNA translation in oncogenesis: the role of eIF4E. Cell Cycle. 2004;3:1387–1389. doi: 10.4161/cc.3.11.1251. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Query CC, Meyer UT. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol Cell Biol. 2002;22:8457–8466. doi: 10.1128/MCB.22.24.8457-8466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Wong JM, Kollins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Tazzari PL, Derenzini M. Enhanced telomere shortening in transformed lymphoblasts from patients with X linked dyskeratosis. J Clin Pathol. 2003;56:583–586. doi: 10.1136/jcp.56.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, Bessler M, Mason PJ. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene. 2002;21:7740–7744. doi: 10.1038/sj.onc.1205969. [DOI] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, Taffurelli M, Calienni M, Teruya-Feldstein J, Trerè D, Pandolfi PP, Derenzini M. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, Gleizes PE, Elliss SR. Specific role for yeast homologs of the Diamond Blackfan anemia associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, Shaw C, Zabel B, Lee B. Consequences of mutations in the non coding RMRP RNA in cartilage-hair hypoplasia. Hum Mol Genet. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- Boocock GR, Marit MR, Rommens JM. Phylogeny, sequence conservation, and functional complementation of the SBDS protein family. Genomics. 2006;87:758–771. doi: 10.1016/j.ygeno.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- Dixon J, Edwards SJ, Gladwin AJ, Dixon MJ, Loftus SK, Bonner CA, Koprivnikar K, Wasmuth JJ, The Treacher Collins Syndrome Collaborative Group Positional cloning of a gene involved in the pathogenesis of Treacher Collins Syndrome. Nature Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet. 2005;14:2035–2043. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- Stewart MJ, Denell R. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol. 1993;13:2524–2535. doi: 10.1128/mcb.13.4.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- Chen FW, Ioannu IA. Ribosomal proteins in cell proliferation and apoptosis. Int Rev Immunol. 1999;18:429–448. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]