Abstract
Aberrant expression of the steroidogenic acute regulatory (StAR) protein in human endometriotic stromal cells plays an important role in the development of endometriosis. Prostaglandin E2 (PGE2) is a potent inducer of StAR expression in these cells; however, the mechanisms responsible for the transcriptional regulation of StAR remain to be elucidated. Herein we report that PGE2-induced StAR expression is independent of the transcriptional suppressor DAX-1 but is regulated by the transcriptional activator cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein (CREB). A promoter activity assay revealed that the cis-element needed for the binding of the CCAAT/enhancer-binding protein (C/EBP) was critical for PGE2-induced StAR expression. Electrophoretic mobility shift assay demonstrated that this region of the StAR promoter was bound by C/EBPα, C/EBPβ, and CREB. Forced expression of either C/EBPα or C/EBPβ alone was sufficient to up-regulate StAR promoter activity whereas PGE2 was needed to induce StAR promoter activity in CREB-overexpressed cells. Results from a chromatin immunoprecipitation assay demonstrated that the binding of C/EBPβ to the StAR promoter was increased whereas CREB binding was unchanged after PGE2 treatment. Taken together, PGE2-induced StAR promoter activity appears to be regulated by CREB and C/EBPβ in a cooperative manner in ectopic human endometriotic stromal cells, providing a molecular framework for the etiology of endometriosis.
Endometriosis is a polygenic disease with a complex, multifactorial etiology that affects 8 to 10% of women in the reproductive age. Women with endometriosis suffer from symptoms such as dysmenorrhea, dyspareunia, and chronic pelvic pain.1 In addition, endometriosis is the primary cause of infertility in women with a prevalence rate of 20 to 50%.2 The high prevalence and severe outcomes associated with this disease have made endometriosis a major concern of public health in the modern society. Although the mechanism responsible for the initiation of this disease remains enigmatic, immune dysfunction3 and aberrant production of estrogen by ectopic endometriotic implants have been demonstrated to be critical factors in the maintenance of endometriosis.4,5,6,7
The biosynthesis of estrogen is controlled by two rate-limiting proteins namely steroidogenic acute regulatory protein (StAR) and aromatase. StAR plays a vital role in steroidogenesis by providing a continuous supply of cholesterol to the P450 side-chain cleavage enzyme. Previously, we reported that expression of StAR in endometriotic stromal cells is induced by PGE2 via EP2-coupled activation of protein kinase A (PKA) and is associated with cyclic adenosine 3′,5′-monophosphate (cAMP) responding element-binding protein (CREB) phosphorylation and CREB binding protein (CBP)-mediated histone acetylation.7,8 However, the molecular mechanism responsible for PGE2-induced StAR expression and their corresponding binding elements in StAR promoter were not characterized.
The StAR gene contains binding sites for transcriptional regulators such as steroidogenic factor-1 (SF-1), activator protein-1 (AP-1), GATA, SP1, CCAAT enhancer-binding protein (C/EBP), and dosage-sensitive sex reversal-adrenal hyperplasia congenital critical region on the X-chromosome gene-1 (DAX-1).9,10 Numerous studies have reported the regulation of StAR promoter through binding to these transcription factor-binding sites. For example, it has been shown that SF-1, SP1, GATA-4, C/EBPβ, and members of CREB family play a stimulatory role in basal and cAMP-mediated StAR promoter activity11,12,13,14,15 whereas DAX-1 and Ying Yang-1 (YY-1) inhibit StAR expression.16,17 However, other studies analyzing the activity of StAR promoter in SF-1-expressing cells did not support a role for SF-1 in a cAMP-inducible manner.13 Furthermore, Wang and colleagues18 demonstrated that DAX-1 inhibited the SF-1-mediated transactivation of the P450 aromatase promoter but did not repress the StAR promoter activity. These data suggested that acute regulation of StAR may be mediated via distinct transcription factors in different cells and certain regulators only exert their function in particular cell types under certain circumstance.
An intriguing phenomenon, which remains unresolved, is that in many cases, transcriptional regulation of StAR expression is cAMP/PKA-dependent whereas there is no consensus CRE identified in StAR promoter. Manna and colleagues12 identified three putative half-sites for 5′-canonical CRE sequences (TGAC) in mouse StAR promoter, which could be bound by CREB/CREM to stimulate StAR promoter activity in a mouse Leydig tumor cell line, MA-10. Although similar but less perfect CRE half sites are predicted in human StAR promoter, whether CREB and/or its family members can bind to these half sites and exert a stimulatory effect remains as an open question. Alternatively, cAMP-induced StAR promoter activity may be mediated by C/EBPs via direct or indirect mechanisms. It has been shown that C/EBPβ can be phosphorylated by PKA in its bZIP domain, which can in turn affect its DNA-binding activity.19 However, other members of C/EBP family, C/EBPα for example, lack a PKA phosphorylation site but still exert cAMP responsiveness. Thus, it is reasonable to hypothesize that there may be a co-factor that is able to interact with C/EBP when itself being phosphorylated by PKA. For instance, PKA phosphorylates GATA-4, which then dimerizes with C/EBPβ and cooperatively induced StAR promoter activity in MA-10 cells and transiently transfected CV-1 cells.13,20
In the current study, we characterized the involvement of transcription factors in the regulation of StAR promoter activity by PGE2. Considering the overexpression of cyclooxygenase-2 (COX-2) gene leading to elevation of PGE2 in peritoneal fluid of women with endometriosis21,22 and the critical role of estrogen in the development of endometriosis, results from this study may provide a molecular framework that helps explain the etiology of this disease. Herein we demonstrate that CREB and C/EBPβ binds to the same CCAAT cis-element of StAR promoter and the binding is required for activation of StAR transcription in PGE2-treated human endometriotic stromal cells.
Materials and Methods
Materials
Prostaglandin E2, butaprost (EP2 agonist), sulprostone (EP3 agonist), PGE1 alcohol (EP4 agonist), and anti-PGE2 antibody were purchased from Cayman Biochemical Co. (Ann Arbor, MI). Anti-StAR antiserum was a generous gift from Dr. Jerome F. Strauss III (University of Pennsylvania, Philadelphia, PA). Anti-CREB, anti-phosphoCREB (ser133P), and anti-CBP antibodies were from Upstate Inc. (Waltham, MA). Anti-CREB monoclonal antibody (sc-271), anti C/EBPα, and anti-C/EBPβ antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). TaqDNA polymerase, fetal bovine serum, Dulbecco’s modified Eagle’s medium/F-12, antibiotics, and 1-kb DNA ladders were from Life Technologies, Inc. (Gaithersburg, MD). Western blot chemiluminescence reagents were from NEN Life Science Products (Boston, MA). The cytomegalovirus-driven renilla reporter system and pGL3-basic vector and S133A CREB plasmid were purchased from Promega Corp. (Madison, WI). The expression plasmid C/EBPα and C/EBPβ were provided by Dr. Peter Johnson (National Cancer Institute–Frederick, Frederick, MD).
Stromal Cell Preparation and Culture
Ovarian endometrioma (n = 25) from patients with endometriosis and normal endometria (n = 10) from patients of reproductive age undergoing hysterectomy for leiomyoma or ovarian pathology were collected to process for stromal cell culture as previously described.7,8,22,23,24 All endometriosis samples were graded according to revised American Society of Reproductive Medicine25 and were histologically confirmed. All ovarian endometrioma samples were derived from advanced endometriosis (moderate and severe phases) with detailed information reported previously.7,8,22,23,24 All patients were of reproductive age with normal menstrual cycles. Human ethics approval was obtained from the Clinical Research Ethics Committee at the National Cheng Kung University Medical Center and informed consent was obtained from each patient.
Endometriotic stromal cells were dissociated, purified, and validated as previously described.7,8,24 Stromal cells were cultured in Dulbecco’s modified Eagle’s medium/F12 supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), streptomycin (100 U/ml), and fungizone (50 μg/ml) in a humidified atmosphere with 5% CO2 at 37°C. When cells reached 70% confluence, they were placed in serum-free, phenol red-free Dulbecco’s modified Eagle’s medium/F-12 medium for 36 hours and subjected to various treatment regimens.
Promoter Constructs
A previously published sequence of the 5′-flanking region of the human StAR gene was used to clone most of the StAR promoter constructs by a polymerase chain reaction (PCR)-based approach. To generate the −1297/+19 DNA fragment, primers (5′-TGTCTTTTCAAGCTTTCTGCAC-3′ and 5′-AGGGTGGTTCTTCGTCCTTC-3′) were included in the PCR reaction using human genomic DNA as template. The PCR proceeded with preheating at 95°C for 2 minutes to denature genomic DNA and then 35 cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 1.5 minutes at 72°C, followed by 10 minutes at 72°C. The PCR product was digested with HindIII to release a fragment of 1316 bp (−1297/+19), which was then ligated into the pGL3-basic vector (E1751, Promega). The resulting construct was referred to as −1.3kStAR. Constructs harboring deleted promoters or mutated transcription factor binding sites were generated from the −1.3kStAR plasmid using PCR amplification approach. Each 5′ primer (listed in Table 1) contained the XhoI site and therefore the deletion PCR products amplified by 5′ primer and GL primer 3 (counterclockwise primer on pGL3-basic) contained XhoI site at 5′ end and HindII site at StAR gene. The mutagenesis was performed by two rounds of PCR. In brief, the primers (mF and mR) used to introduce mutation were designed to be complementary and contain mismatch bases located in the middle of each primer (refer to Table 1). In the first round PCR, 20 ng of 327 StAR/luc plasmids were amplified in two separate amplification reactions with mF/GLprimer2 (primer on pGL3-basic) and −327StAR/mR primers to generate overlapping DNA fragments. Twenty cycles of amplification reaction was performed (30 seconds denaturation at 95°C, 30 seconds annealing at 50°C, and 30 seconds elongation at 72°C) followed by elongation at 72°C for 10 minutes. The two amplified fragments were separated by 2% agarose gel and purified by gel extraction. In the second round PCR, 20 ng of each amplified fragments were mixed and amplified with −327StAR/GLprimer2 as first round PCR. The PCR products containing mutation bases were purified, digested, and inserted between HindIII/XhoI sites of pGL3-basic as described above.
Table 1.
List of Primers and Their Sequences
| Name | Sequence |
|---|---|
| RVprimer3 | 5′-CTAGCAAAATAGGCTGTCCC-3′ |
| GLprimer2 | 5′-CTTTATGTTTTTGGCGTCTTCCA-3′ |
| StAR_-868 | 5′-CCGCTCGAGAGGGTCCTCACCCAGA AGAA-3′ |
| StAR_-327 | 5′-CCGCTCGAGGAGCCTCAATTCCAGA TCAGC-3′ |
| StAR_-109 | 5′-CCGCTCGAGGCTCTATCCTTGACCC CTTCC-3′ |
| StAR-92 | 5′-CCGCTCGAGTTCCTTTGCACAGTGA GTGATG-3′ |
| StAR-53 | 5′-CCGCTCGAGTGATGATGCACAGCCT TCAG-3′ |
| StAR_-37 | 5′-CCGCTCGAGTCAGCGGGGGACATTT AAG-3′ |
| mSP1_F | 5′-CTCCCaaGCCCCAAG-3′ |
| mSP1_R | 5′-CTTGGGGCttGGGAG-3′ |
| mC/EBP_F | 5′-AGTGTGAGtttATCGCTCTATC-3′ |
| mC/EBP_R | 5′-GATAGAGCGATaaaCTCACACT-3′ |
| mSF-1_m_F | 5′-GCTCTATCCcTGACCCCTTC-3′ |
| mSF-1_m_R | 5′-GAAGGGGTCAgGGATAGAGC-3′ |
| mCRE1_F | 5′-CTCTATCCTTccgCCCTTCCTTT-3′ |
| mCRE1_R | 5′-AAAGGAAGGGcggAAGGATAGAG-3′ |
| mCRE1-2_F | 5′-CTCTATCCTaagCCCCTTCCTTT-3′ |
| mCRE1-2_R | 5′-AAAGGAAGGGGcttAGGATAGAG-3′ |
| mGATA-4_2F | 5′-TGGCGTTTTTAcCTCCTGATGA-3′ |
| mGATA-4_2R | 5′-TCATCAGGAGgTAAAAACGCCA-3′ |
Underline: XhoI restriction site; bold lowercase: mutated bases.
Transient Transfection and Luciferase Assays
Endometriotic stromal cells were plated on 24-well plates for the luciferase/renilla assays. Plasmids were transfected using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA). Transfections were followed by rinsing and incubation in serum-free Dulbecco’s modified Eagle’s medium/F12 medium for 12 hours. After the medium was changed, cells were then treated with PGE2 (1 μmol/L) for another 12 hours. After treatment, the medium was removed and wells were rinsed with phosphate-buffered saline (PBS) to remove detached cells and residual growth medium. Then 200 μl of 1× passive lysis buffer, provided by a dual-luciferase reporter assay system (Promega Corp.), was added per well. The plate was shaken at 180 rpm for 15 minutes at room temperature. Then cells were detached from the well by pipetting and freeze/thaw cycle to accomplish complete lysis of cells. After centrifugation at 15,000 × g, the cleared cell lysate was transfer to another 1.5-ml microcetrifuge tube. Luciferase assays were performed using a dual luciferase reporter assay system according to the manufacturer’s instructions (Promega). Briefly, 100 μl of luciferase substrate was added to 20 μl of lysate, and luciferase activity was measured using a 20/20 luminometer (Turner Designs, Sunnyvale, CA). Luciferase data were expressed as the means ± SEM. Each luciferase assay experiment was performed in duplicate and repeated for the number of times indicated in the figure legends using different batches of cells.
Preparation of Nuclear Extract
After treatment, cells were washed by cold 1× PBS, and then the cells were scraped with 400 μl of buffer A (10 mmol/L HEPES-KOH, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μg/ml pepstatin A, 0.5 mmol/L dithiothreitol, 50 mmol/L NaF, and 1 mmol/L Na3VO4). After incubation on ice for 10 minutes, the cells were vortexed for 20 seconds at high speed, and the nuclei were pelleted by centrifugation for 15 seconds at 12,000 × g and resuspended in buffer C (20 mmol/L HEPES-NaOH, 25% glycerol, 1.5 mmol/L MgCl2, 0.2 mmol/L ethylenediamintetraacetic acid, 420 mmol/L NaCl, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 2 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μg/ml pepstatin A, 0.5 mmol/L dithiothreitol, 50 mmol/L NaF, and 1 mmol/L Na3VO4). After incubation on ice for 20 minutes, the nuclear extract was centrifuged for 2 minutes at 12,000 × g and the supernatant was transferred to a new microcetrifuge tube and stored at −80°C.
Electrophoretic Mobility Shift Assay
Double-stranded oligonucleotides corresponding to C/EBP binding sites (−140 to −118, 5′-6-FAM-CAGCAGTGTGAGAGGCAATCGCTCT-3′) in the human StAR promoter were synthesized by Qiagen (Valencia, CA) and annealed in 10 mmol/L Tris-HCl (pH 7.5), 1 mmol/L ethylenediaminetetraacetic acid, 25 mmol/L NaCl, 10 mmol/L MgCl2, and 1 mmol/L dithiothreitol. The oligonucleotide probes were labeled using FAM fluorescence dye. Unlabeled oligonucleotides (50-fold excess) were used as competitors in some experiments, and antibodies against C/EBPα, C/EBPβ, and CREB were used for supershift analysis. A total of 10 μg of nuclear extracts were incubated in the presence or absence of competitor or antibodies for 20 minutes at 10°C in binding buffer. The FAM-labeled probe was added, and the incubation was continued for an additional 20 minutes. The DNA/protein complexes were resolved on a 5% nondenaturing acrylamide gel in TGE buffer (0.25 mol/L Tris, 1.9 mol/L glycine, and 10 mmol/L ethylenediaminetetraacetic acid final), which was subsequently dried and exposed to fluorescent Phosphor Imager FLA3000 (Fuji, Tokyo, Japan).
Chromatin Immunoprecipitation-PCR (ChIP) Assay
The protocol used was as described before.26,27 In brief, proteins and DNA in primary culture endometriotic stromal cells (1 × 106 cells) were cross-linked by incubation for 10 minutes at 37°C with a final concentration of 1% formaldehyde. Cells were washed twice with ice-cold PBS containing protease inhibitors, scraped into a conical tube, centrifuged, resuspended in 400 μl of lysis buffer [1% sodium dodecyl sulfate (SDS), 10 mmol/L ethylenediaminetetraacetic acid, 50 mmol/L Tris-HCl, pH 8.1], and placed on ice for 10 minutes. Genomic DNA was sheared to lengths of 0.5 to 1 kb by sonication. Anti-CREB, C/EBPα, or C/EBPβ, antibodies, or mouse IgG (for negative control) was added to the supernatant fraction and the mixture was incubated overnight at 4°C with rotation, then incubated with 60 μl of salmon sperm DNA/Protein A agarose slurry for another 1 hour at 4°C with rotation. The pellet was then washed sequentially (5 minutes per wash) on a rotating platform with washing buffers and 1× TE buffer. After the final wash, the pellet was eluted by resuspension in freshly made elution buffer (1% SDS and 50 mmol/L NaHCO3) followed by centrifugation. The DNA was released from protein-DNA complex and 2 μl of ChIP DNA was drawn to perform the first round of PCR with StARP first primer pair (5′-GTTTCTGAGCCTCAATTCCAG-3′ and 5′-CGACTTCCGACACGTAGTAG-3′). After a 20-cycle amplification (30 seconds denaturation at 95°C, 30 seconds annealing at 57°C, and 30 seconds elongation at 72°C), the PCR product was diluted and 2 μl of diluted PCR product was subjected to nested PCR amplification with StARP second primer paired (5′-TGGCCCTGTCCTCCCTAC-3′ and 5′-TGAAGGCTGTGCATCATCATC-3′). After PCR amplification (30 seconds denaturation at 95°C, 30 seconds annealing at 58°C, and 30 seconds elongation at 72°C followed by elongation at 72°C for 10 minutes), the PCR product was resolved in agarose gel for evaluation.
Western Blot
The detailed procedure of Western blotting was described previously.8,28 In brief, endometriotic stromal cells were stimulated with PGE2 (1 μmol/L) for various times and equal amounts of proteins were resolved on SDS-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and incubated with specific antibodies. After washing and incubating with horseradish peroxidase-conjugated second antibodies, signals were detected by a enhanced chemiluminescence system. The blots were then stripped and reprobed with different antibodies.
Statistical Analysis
All data were expressed as means ± SEM. The data were analyzed by one-way analysis of variance in commercial statistical software (GraphPad Prism 4.02; GraphPad Software, San Diego, CA). Tukey’s procedure was used to test differences between groups found significant using the F-test. Significant differences were accepted when two-tailed analysis yielded P < 0.05.
Results
PGE2-Induced StAR Promoter Activation
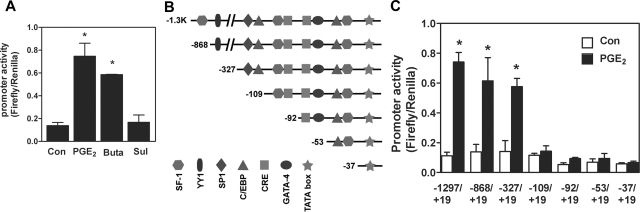
To determine the cis-element responsible for PGE2-induced, PKA-dependent StAR promoter activity, a 1.3-kb human StAR gene promoter was cloned and serially deleted constructs were created. PGE2 and its EP2 receptor agonist, butaprost, induced full activity of StAR promoter whereas the EP3 receptor agonist, sulprostone, failed to exert any observable promoter activity (Figure 1A). Bioinformatic annotation indicated that there are two SF-1 (−118/−109 and −56/−49), two C/EBP (−132/−124 and −64/−55), two CRE half sites (−112/−109 and −92/−89), one GATA (−77/−72), and one SP-1 (−173/−164) sites within the −327/+19 region of human StAR promoter. In addition, the hairpin structure, which serves as a binding sited for DAX-1 (−61/−27), is also reserved within this −327 StAR promoter (Figure 1B). Deletion of the −1297 to −327 of StAR promoter had no substantial effect on basal and PGE2-induced promoter activity (Figure 1C). Deletion of −327 to −109 significantly reduced PGE2-induced StAR promoter activity whereas deletion to the −92 bp completely abolished basal and PGE2-induced StAR promoter activity (Figure 1C).
Figure 1.
Transcriptional activation of StAR promoter activity by PGE2. A: Endometriotic stromal cells were transfected with 1.3 kb of human StAR reporter construct and treated with vehicle (Con), PGE2, EP2 receptor agonist (butaprost, Buta), or EP3 receptor agonist (sulprosterone, Sul) for 12 hours. Data show means of five independent experiments performed in duplicates using different batches of cells. *Significant difference compared to control group at P < 0.05. B: Schematic drawing of human StAR gene promoter shows several important transcription factor binding sites. C: Endometriotic stromal cells were transfected with serial-deleted StAR reporter constructs, treated with vehicle (Con) or PGE2 for another 12 hours, and harvested for luciferase activity assay. Data show means of four independent experiments performed in duplicates using different batches of cells. *Significant difference compared to individual control group at P < 0.05.
DAX-1 Is Associated with Basal but Not PGE2-Induced StAR Expression
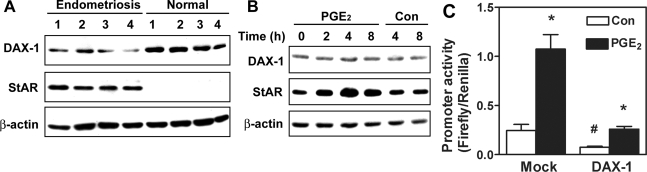
We next sought to determine which cis-element within this region is important for PGE2-induced StAR gene activity. It has been reported that DAX-1 is an important repressor in regulating aromatase expression in endometriotic stromal cells.29 To characterize whether DAX-1 is involved in PGE2-induced StAR expression in human endometriotic stromal cell, we first determined the expression of DAX-1 in nuclear extract of normal endometrium and ectopic endometriotic implant. Expression of DAX-1 in normal uterine endometrium was greater than that in ectopic endometriotic tissues, which was inversely correlated with the expression of StAR protein (Figure 2A). Treatment of endometriotic stromal cells with PGE2, however, failed to affect the nuclear concentration of DAX-1, suggesting that DAX-1 may not be involved in PGE2-induced StAR expression (Figure 2B). Transient transfection of human DAX-1 in endometriotic stromal cell decreased basal StAR promoter activity but had no substantial effect on PGE2-induced StAR promoter as evidenced by the similar induction folds (Figure 2C). Taken together, these data strongly suggest that PGE2-induced StAR expression in endometriotic stromal cells is DAX-1-independent.
Figure 2.
DAX-1 controls basal but not PGE2-induced StAR gene expression. A: Expression of DAX-1 and StAR protein in endometrial tissues collected from four normal and endometriosis patients, respectively. Equal amounts of total cell lysates from homogenized tissues were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies against DAX-1 (top), StAR (middle), and β-actin (bottom), respectively. B: Representative Western blot shows the expression of DAX-1 and StAR protein at different time points after PGE2 treatment. Endometriotic stromal cells were treated with or without 1 μmol/L PGE2 for 2, 4, or 8 hours. Equal amounts of total cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies against DAX-1 (top), StAR (middle), and β-actin (bottom), respectively. This experiment was repeated for five times using different batches of cells and the pattern was the same. C: Endometriotic stromal cells were transiently co-transfected with empty vector (mock) or plasmids containing human DAX-1 cDNA (DAX-1) and −327StAR reporter plasmids. Data show means and standard errors of four experiments using different batches of cells. *Difference between PGE2-treated and control groups. #Difference between mock- and DAX-1-transfected groups at P < 0.05.
CCAAT Enhancer Binding Site Is Critical for PGE2-Induced StAR Expression
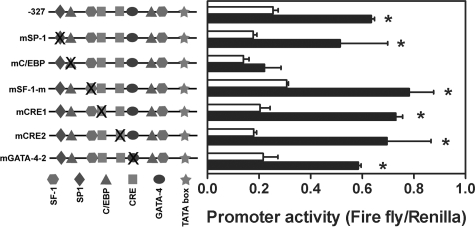
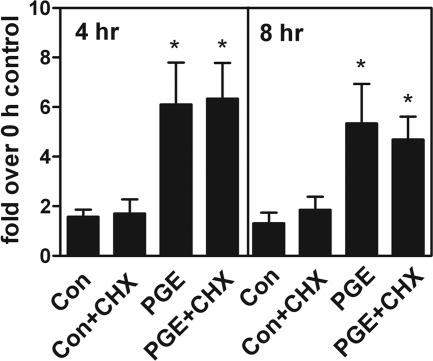
Because DAX-1 is not involved in PGE2-induced StAR promoter activity, we then focused on other transcription activators. Site-mutated constructs of several cis-elements were transiently transfected into primary cultured endometriotic stromal cells and the promoter activity induced by PGE2 treatment was determined. Mutation of two SF-1 sites, two CRE half sites, or the GATA-4 site had no substantial effect on PGE2-induced StAR promoter activity (Figure 3). In contrast, mutation of C/EBP site (−132/−124) completely abolished PGE2-induced StAR promoter activity (Figure 3). These results indicated that the CCAAT site of StAR promoter is critical for transactivation of StAR gene activity by PGE2. Because CREB is indispensable for PGE2-induced StAR expression and only C/EBP mutated element affects StAR promoter activity, it is possible that PGE2-induced StAR expression is mediated via CREB-dependent up-regulation of C/EBP protein. To test this hypothesis, cycloheximide (1 μmol/L) was added to block de novo protein biosynthesis and the effect of PGE2 on StAR mRNA expression was examined. Real-time RT-PCR results demonstrated that PGE2 significantly induced StAR mRNA expression whereas pretreatment with cycloheximide did not inhibit this effect (Figure 4).
Figure 3.
CCAAT enhancer binding site is critical for PGE2-induced StAR promoter activity. Cells were transfected with reporter constructs harboring human StAR promoter (−327 StAR-luc) with site-directed mutations at specific transcription factor binding sites as indicated. Data show means and standard errors of three independent experiments using different batches of cells. *Significant difference between control and PGE2-treated groups at P < 0.05.
Figure 4.
PGE2-induced StAR mRNA expression does not involve de novo synthesis of new protein. Endometriotic stromal cells were pretreated without or with cycloheximide (CHX) for 1 hour followed by treatment with vehicle (con) or PGE2 for 4 and 8 hours. Total RNA was isolated and subjected to real-time RT-PCR quantification. Data show means and standard errors of four independent experiments using different batches of cells. *Significant difference from control group at P < 0.05.
Binding of CREB to the C/EBP Site of StAR Promoter
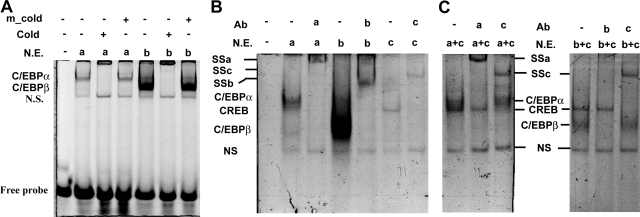
To determine which transcription factors bind to the putative C/EBP element identified within the human StAR promoter, electrophoretic mobility shift assay were performed using oligonucleotides corresponding to the middle C/EBP element (−140/−118) with nuclear extracts. Specific bands corresponding to C/EBPα and C/EBPβ were identified and could be competed away by adding excess unlabeled consensus C/EBP probe but not mutated probe (Figure 5A). Addition of specific antibodies against C/EBPα or/and C/EBPβ further confirmed that both C/EBPα and C/EBPβ bound to this element (Figure 5B). The anti-C/EBPα antibody failed to affect the binding of C/EBPβ and vice versa and demonstrated that these two factors bind to the CCAAT site independently (data not shown). Next, we sought to determine whether CREB can bind to this C/EBP cis-element on human StAR promoter. Concordant with the functional role of CREB in PGE2-induced StAR promoter activity, the results demonstrated that CREB also bound to the C/EBP probe, which lacks the consensus CRE half site (Figure 5B). This binding was supershifted by incubation with specific CREB antibody (Figure 5B) indicating the binding is specific. We next tested whether the binding of CREB to the C/EBP site is attributable to formation of heterodimeric CREB-C/EBP complex instead of direct binding by CREB protein. Incubation of double-stranded oligonucleotide corresponding to CCAAT site with nuclear extract obtained from cells that overexpressed C/EBPα or C/EBPβ, and CREB clearly showed two distinct bands, which can be supershifted by CREB antibody and C/EBPα or C/EBPβ antibody, respectively, without affecting each other’s binding (Figure 5C). Co-immunoprecipitation experiments using anti-CREB or anti-C/EBPβ antibody revealed that CREB and C/EBP were not physically associated with each other (data not shown). Taking together, these data show that CREB can bind to CCAAT site without the involvement of C/EBP.
Figure 5.
In vitro binding of C/EBPα, C/EBPβ, and CREB to the CCAAT site of StAR promoter. A: Nuclear extracts (N.E.) collected from cells overexpressing C/EBPα (a) or C/EBPβ (b) were incubated with FAM-labeled oligonucleotides corresponding to −140/−118 of human StAR promoter in the presence or absence of excess unlabeled probes (cold) or C/EBP site-mutated unlabeled probe (m_cold). Specific protein/DNA complexes were indicated. NS, nonspecific binding. B: Binding of C/EBPα, C/EBPβ, and CREB to the CCAAT site of human StAR promoter. SSa, supershifted protein/DNA complex using antibody against C/EBPα; SSb, supershifted protein/DNA complex using antibody against C/EBPβ; SSc, supershifted protein/DNA complex using antibody against CREB. a: C/EBPα; b: C/EBPβ; c: CREB. C: Nuclear extracts collected from cells overexpressing C/EBPα and CREB (a + c) or C/EBPβ and CREB (b + c) were incubated with FAM-labeled oligonucleotide corresponding to −140/−118 of human StAR promoter in the presence or absence of antibodies against C/EBPα (a), C/EBPβ (b), or CREB (c).
Transactivation of StAR Promoter by CREB and C/EBP
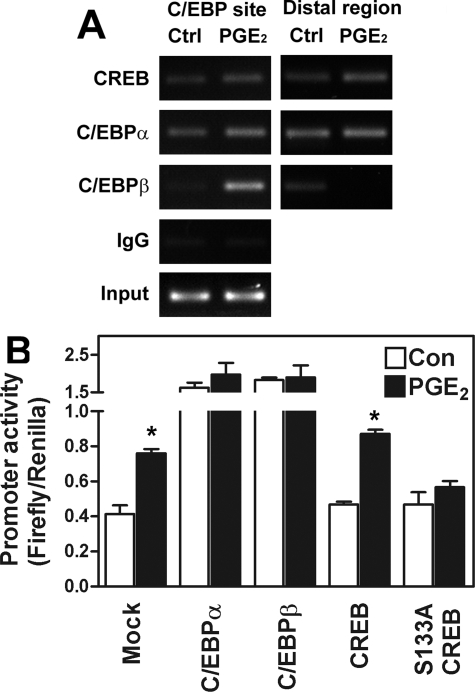
Because both CREB and C/EBP bound to the CCAAT site independently, we sought to determine the mechanism of PGE2-induced StAR promoter transactivation. Consistent with the notion that ectopic endometriotic stroma aberrantly expresses StAR protein, a basal binding of CREB, C/EBPα, and C/EBPβ to the StAR promoter was observed by ChIP assay (Figure 6A). The binding of CREB to the CCAAT site of StAR promoter was not changed after PGE2 treatment and is consistent with the notion that it was regulated at the phosphorylation status. In contrast, the binding of C/EBPβ to this region was markedly increased after the addition of PGE2 (Figure 6A). Binding of C/EBPα was not as evident as that of C/EBPβ, which is consistent with the result of electrophoretic mobility shift assay.
Figure 6.
Binding of C/EBPβ to StAR promoter is enhanced by PGE2 treatment and is sufficient to induce StAR promoter activity. A: Representative picture of ChIP assay demonstrates that in vivo binding of C/EBPβ to StAR promoter (C/EBP site) is increased after PGE2 treatment whereas binding of CREB and C/EBPα was not changed. A primer set that amplifying a distal region was also used to serve as internal control. The experiment was done without overexpression of any plasmids. This experiment was repeated three more times using different batches of cells and the pattern was the same. B: Forced expression of C/EBPα and C/EBPβ but not CREB is sufficient to up-regulate StAR promoter activity. Stromal cells were transiently transfected with −327 StAR-luc reporter plasmid and C/EBPα, C/EBPβ, CREB, or serine 133-mutated CREB (S133A CREB) and then treated with or without PGE2 for 16 hours. Data show means and standard errors of two independent experiments performed in duplicate using different batches of cells.
Finally, cells were transiently transfected with plasmids containing C/EBPα, C/EBPβ, CREB, or S133A-mutated CREB and the promoter activity of human StAR gene was determined. Transfection of CREB did not result in activation of human StAR promoter unless PGE2 was added (Figure 6B). In contrast, forced expression of C/EBPα and C/EBPβ alone was sufficient to induce StAR promoter activity (Figure 6B). That transfection with plasmids containing S133A-mutated CREB abolished PGE2-induced StAR promoter activity suggests again that the presence of functionally competent CREB is a requisite for transactivation of StAR gene activity by PGE2.
Discussion
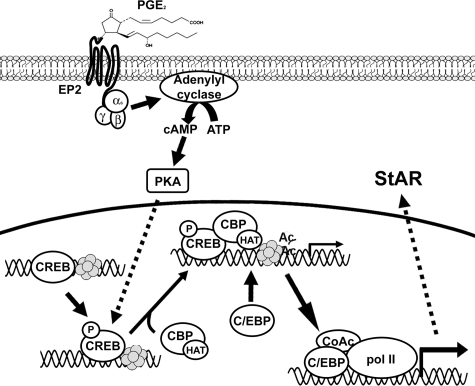
Aberrant expression of StAR and aromatase induced by PGE2 plays an important role in maintaining continuous support of estrogen in ectopic endometriotic tissue.30 PGE2-induced StAR expression is mediated by EP2 receptor-coupled cAMP/PKA-dependent pathway.8 It has been shown that CREB plays an indispensable role in PKA-mediated StAR gene transcription although no consensus CRE has ever been identified in human StAR promoter. In this study, we provide compelling evidence to demonstrate that PGE2-induced StAR promoter involves binding of CREB and C/EBPβ to the CCAAT site. CREB constitutively binds to the CCAAT/enhancer binding site, which might serve as an anchor position for formation of transcriptional initiation complex. On activation, phosphorylated CREB recruits CBP leading to histone acetylation and chromatin remodeling,8 which enhances the binding of its bona fide transcription factor, C/EBP, especially C/EBPβ to this region. The binding of C/EBPβ recruits basic transcriptional machinery to form an initiation complex, which further enhances the transcription of StAR gene (Figure 7). Our results provide a likely explanation of how the CRE-lacking human StAR gene responds to PGE2-mediated promoter activation in ectopic endometriotic stromal cells and may be applicable to other cell types.
Figure 7.
A proposed model illustrates that PGE2 induces StAR gene expression in endometriotic stromal cells (see text for details). Only one histone octomer is shown here for representation. Ac, acetylated histone; CoAc, transcriptional co-activators; Pol II, RNA polymerase II; HAT, histone acetyl transferase activity.
Many transcription factors including SF-1, GATA factors, SP1, C/EBP, and members of the CREB family have been implicated to play important roles in up-regulation of StAR gene expression. In human endometrium, the amounts of SF-1 transcripts were rare whereas SF-1 transcripts were detected in all endometriotic tissues.31 Overexpression of SF-1 in eutopic endometrial and ectopic endometriotic cells increases baseline and cAMP-induced activities of aromatase promoter II construct.31 This indicates that expression of SF-1 may be associated with aberrant expression of aromatase in endometriotic lesion. In this study, our results indicate that a similar effect might not apply to the up-regulation of StAR gene expression in ectopic endometriotic stromal cells because mutation of the SF-1 binding site does not affect StAR promoter activity. Recently, it has been shown that GATA-4 binds to the StAR promoter and contributes to ∼20% of the cAMP-dependent induction of StAR gene expression in mouse MA-10 cell.32 In the current study, that mutation of GATA-4 binding site in human StAR promoter does not alter the reporter gene expression indicates that GATA-4 is not involved in PGE2-induecd StAR expression in ectopic endometriotic stromal cells. Mutation of the SP-1 site in human StAR promoter also shows no effect on PGE2-induced StAR promoter activity and thus eliminates the role of SP-1 in the aberrant expression of StAR in endometriotic stromal cells.
The orphan nuclear receptor DAX-1 has been identified to block steroid synthesis by impairing the expression of StAR gene in adrenal gland, testis, and ovary. The DNA binding site for DAX-1 is a hairpin structure with minimal stem length of 10 nucleotides,16 which is distinct from all other nuclear receptors. It has been reported that binding of DAX-1 resulted in decreasing basal and cAMP-induced StAR promoter activity in Y-1 adrenocortical cells or R2C rat tumor cells.16,33 Our results support part of the previous findings while contradicting with the rest by showing DAX-1 only involves inhibition of basal but not PGE2-induced StAR expression. In the current study, we conclude that DAX-1 inhibits basal StAR expression by direct and indirect evidence. That levels of DAX-1 inversely correlate with StAR expression in normal and endometriotic tissues represents the indirect evidence whereas force expression of DAX-1 in endometriotic stromal cells reduces basal StAR expression provides the direct one. However, PGE2 does not affect DAX-1 expression in endometriotic stromal cells and force expression of DAX-1 also fails to block PGE2-induced StAR expression provide other lines of evidence to demonstrate that DAX-1 is not responsible for up-regulation of StAR by PGE2. The discrepancy between our current finding and previous report is not clear but could be attributable to cell type-specific and ligand-specific effects.
A plethora of evidence has demonstrated that up-regulation of StAR is a cAMP-PKA-dependent event. The intriguing part is that no consensus CRE has been identified in the StAR gene promoter. In mouse StAR promoter, a CRE half-site was identified and proven functional in cAMP-induced StAR gene expression.12 The CRE half-site is not functional in human StAR promoter whereas the C/EBP site provides a different story in regulating StAR gene expression induced by PGE2. In agreement with report by Christenson and colleagues,11 we found this C/EBP element could be bound by C/EBPα and C/EBPβ. In addition, our results further demonstrate that CREB also binds to this response element. We have previously shown that phosphorylation of CREB induced by PGE2, which causes CBP recruitment and histone H3 acetylation, is associated with PGE2-induced StAR expression.8 In this study, we further demonstrated that blockage of CREB phosphorylation by overexpression of S133A-mutated CREB abolishes PGE2-induced StAR expression. By identifying that CREB actually binds to the C/EBP site in human StAR promoter, we provide a physical link between PKA-induced CREB phosphorylation and its functional role in StAR promoter activity.
The molecular mechanism of how CREB regulates PGE2-induced, PKA-mediated StAR promoter activity remains unclear. Our current and previous8 results suggest that CREB might play as an initiator to unfold the DNA-histone binding, which then provides space for C/EBP binding to the StAR promoter. CREB constitutively binds to the StAR promoter as has been proposed before.34 On phosphorylation by PKA, phosphorylated CREB recruits CBP, which then causes histone acetylation. Acetylation of histone by CBP may result in its dissociation from DNA, which provides space for C/EBP binding. Although both C/EBPα and C/EBPβ can bind to this region and forced expression of both proteins is sufficient to induce StAR promoter activity, the increased binding capacity of C/EBPβ after PGE2 treatment suggests it might play a more prominent role than C/EBPα. The identification of CREB binding to the C/EBP site of StAR promoter provides a likely explanation why the conventional CRE-lacking StAR promoter would respond to PKA action. Further investigation is warranted to elucidate whether other stimuli (for example, luteinizing hormone and adrenocorticotropic hormone) that activate StAR gene promoter via PKA-dependent signaling pathway is mediated via a similar molecular paradigm.
Taking together our previous and current findings, we have demonstrated, for the first time, that PGE2-induced up-regulation of StAR gene activity involves phosphorylation of CREB, recruitment of CBP, acetylation of histone, and binding of C/EBPβ to the StAR promoter. Considering the overexpression of COX-2 gene leading to elevation of PGE2 in peritoneal fluid of women with endometriosis21,22 and the critical role of estrogen in the development of endometriosis, our model provides a molecular framework that helps explain the etiology of this disease. These findings also provide novel information for future therapeutic consideration that aims at disruption of steroidogenic capacity of PGE2.
Footnotes
Address reprint requests to Shaw-Jenq Tsai, Ph.D., Department of Physiology, National Cheng Kung University Medical College, Tainan 701, Taiwan, Republic of China. E-mail: seantsai@mail.ncku.edu.tw.
Supported by the National Science Council of Taiwan (grant NSC 95-2320-B-006-047) and the National Research Program of Genomic Medicine (grant NSC94-3112-B-006-010).
References
- Wu MH, Shoji Y, Chuang PC, Tsai SJ. Endometriosis: disease pathophysiology and the role of prostaglandins. Expert Rev Mol Med. 2007;9:1–20. doi: 10.1017/S146239940700021X. [DOI] [PubMed] [Google Scholar]
- Waller KG, Lindsay P, Curtis P, Shaw RW. The prevalence of endometriosis in women with infertile partners. Eur J Obstet Gynecol Reprod Biol. 1993;48:135–139. doi: 10.1016/0028-2243(93)90254-a. [DOI] [PubMed] [Google Scholar]
- Wu MH, Shoji Y, Wu MC, Chuang PC, Lin CC, Huang MF, Tsai SJ. Suppression of matrix metalloproteinase-9 by prostaglandin E(2) in peritoneal macrophage is associated with severity of endometriosis. Am J Pathol. 2005;167:1061–1069. doi: 10.1016/S0002-9440(10)61195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble LS, Simpson ER, Johns A, Bulun SE. Aromatase expression in endometriosis. J Clin Endocrinol Metab. 1996;81:174–179. doi: 10.1210/jcem.81.1.8550748. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Noble LS, Takayama K, Michael MD, Agarwal V, Fisher C, Zhao Y, Hinshelwood MM, Ito Y, Simpson ER. Endocrine disorders associated with inappropriately high aromatase expression. J Steroid Biochem Mol Biol. 1997;61:133–139. [PubMed] [Google Scholar]
- Noble LS, Takayama K, Zeitoun KM, Putman JM, Johns DA, Hinshelwood MM, Agarwal VR, Zhao Y, Carr BR, Bulun SE. Prostaglandin E2 stimulates aromatase expression in endometriosis-derived stromal cells. J Clin Endocrinol Metab. 1997;82:600–606. doi: 10.1210/jcem.82.2.3783. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Wu MH, Lin CC, Sun HS, Chen SM. Regulation of steroidogenic acute regulatory protein expression and progesterone production in endometriotic stromal cells. J Clin Endocrinol Metab. 2001;86:5765–5773. doi: 10.1210/jcem.86.12.8082. [DOI] [PubMed] [Google Scholar]
- Sun HS, Hsiao KY, Hsu CC, Wu MH, Tsai SJ. Transactivation of steroidogenic acute regulatory protein in human endometriotic stromal cells is mediated by the prostaglandin EP2 receptor. Endocrinology. 2003;144:3934–3942. doi: 10.1210/en.2003-0289. [DOI] [PubMed] [Google Scholar]
- Manna PR, Wang XJ, Stocco DM. Involvement of multiple transcription factors in the regulation of steroidogenic acute regulatory protein gene expression. Steroids. 2003;68:1125–1134. doi: 10.1016/j.steroids.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Reinhart AJ, Williams SC, Stocco DM. Transcriptional regulation of the StAR gene. Mol Cell Endocrinol. 1999;151:161–169. doi: 10.1016/s0303-7207(98)00257-3. [DOI] [PubMed] [Google Scholar]
- Christenson LK, Johnson PF, McAllister JM, Strauss JF., III CCAAT/enhancer-binding proteins regulate expression of the human steroidogenic acute regulatory protein (StAR) gene. J Biol Chem. 1999;274:26591–26598. doi: 10.1074/jbc.274.37.26591. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, Zeleznik AJ, Stocco DM. Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol Endocrinol. 2002;16:184–199. doi: 10.1210/mend.16.1.0759. [DOI] [PubMed] [Google Scholar]
- Silverman E, Eimerl S, Orly J. CCAAT enhancer-binding protein beta and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J Biol Chem. 1999;274:17987–17996. doi: 10.1074/jbc.274.25.17987. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Saito M, Fujimoto S. Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression. Endocrinology. 2000;141:2895–2903. doi: 10.1210/endo.141.8.7602. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Viger RS. Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol Reprod. 2001;64:1191–1199. doi: 10.1095/biolreprod64.4.1191. [DOI] [PubMed] [Google Scholar]
- Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature. 1997;390:311–315. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- Nackley AC, Shea-Eaton W, Lopez D, McLean MP. Repression of the steroidogenic acute regulatory gene by the multifunctional transcription factor yin yang 1. Endocrinology. 2002;143:1085–1096. doi: 10.1210/endo.143.3.8668. [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Jeffs B, Ito M, Achermann JC, Yu RN, Hales DB, Jameson JL. Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. Proc Natl Acad Sci USA. 2001;98:7988–7993. doi: 10.1073/pnas.141543298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Hamel F, Viger RS. Protein kinase A-dependent cooperation between GATA and CCAAT/enhancer-binding protein transcription factors regulates steroidogenic acute regulatory protein promoter activity. Endocrinology. 2002;143:3935–3945. doi: 10.1210/en.2002-220413. [DOI] [PubMed] [Google Scholar]
- Wu MH, Sun HS, Lin CC, Hsiao KY, Chuang PC, Pan HA, Tsai SJ. Distinct mechanisms regulate cyclooxygenase-1 and -2 in peritoneal macrophages of women with and without endometriosis. Mol Hum Reprod. 2002;8:1103–1110. doi: 10.1093/molehr/8.12.1103. [DOI] [PubMed] [Google Scholar]
- Wu MH, Wang CA, Lin CC, Chen LC, Chang WC, Tsai SJ. Distinct regulation of cyclooxygenase-2 by interleukin-1beta in normal and endometriotic stromal cells. J Clin Endocrinol Metab. 2005;90:286–295. doi: 10.1210/jc.2004-1612. [DOI] [PubMed] [Google Scholar]
- Wu MH, Chuang PC, Chen SM, Lin CC, Tsai SJ. Increased leptin expression in endometriosis cells is associated with endometrial stromal cell proliferation and leptin gene-upregulation. Mol Hum Reprod. 2002;8:456–464. doi: 10.1093/molehr/8.5.456. [DOI] [PubMed] [Google Scholar]
- Wing L-YC, Chuang P-C, Wu M-H, Chen H-M, Tsai S-J. Expression and mitogenic effect of fibroblast growth factor-9 in human endometriotic implant is regulated by aberrant production of estrogen. J Clin Endocrinol Metab. 2003;88:5547–5554. doi: 10.1210/jc.2003-030597. [DOI] [PubMed] [Google Scholar]
- Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–821. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- Chen KF, Lai YY, Sun HS, Tsai SJ. Transcriptional repression of human cad gene by hypoxia inducible factor-1alpha. Nucleic Acids Res. 2005;33:5190–5198. doi: 10.1093/nar/gki839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Chen KF, Lin SC, Lgu CW, Tsai SJ. Aberrant expression of leptin in human endometriotic stromal cells is induced by elevated levels of hypoxia inducible factor-1alpha. Am J Pathol. 2007;170:590–598. doi: 10.2353/ajpath.2007.060477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SJ, Wu MH, Chuang PC, Chen HM. Distinct regulation of gene expression by prostaglandin F2α (PGF2α) is associated with PGF2α resistance or susceptibility in human granulosa-luteal cells. Mol Hum Reprod. 2001;7:415–423. doi: 10.1093/molehr/7.5.415. [DOI] [PubMed] [Google Scholar]
- Gurates B, Sebastian S, Yang S, Zhou J, Tamura M, Fang Z, Suzuki T, Sasano H, Bulun SE. WT1 and DAX-1 inhibit aromatase P450 expression in human endometrial and endometriotic stromal cells. J Clin Endocrinol Metab. 2002;87:4369–4377. doi: 10.1210/jc.2002-020522. [DOI] [PubMed] [Google Scholar]
- Attar E, Bulun SE. Aromatase and other steroidogenic genes in endometriosis: translational aspects. Hum Reprod Update. 2006;12:49–56. doi: 10.1093/humupd/dmi034. [DOI] [PubMed] [Google Scholar]
- Zeitoun K, Takayama K, Michael MD, Bulun SE. Stimulation of aromatase P450 promoter (II) activity in endometriosis and its inhibition in endometrium are regulated by competitive binding of steroidogenic factor-1 and chicken ovalbumin upstream promoter transcription factor to the same cis-acting element. Mol Endocrinol. 1999;13:239–253. doi: 10.1210/mend.13.2.0229. [DOI] [PubMed] [Google Scholar]
- Wooton-Kee CR, Clark BJ. Steroidogenic factor-1 influences protein-deoxyribonucleic acid interactions within the cyclic adenosine 3,5-monophosphate-responsive regions of the murine steroidogenic acute regulatory protein gene. Endocrinology. 2000;141:1345–1355. doi: 10.1210/endo.141.4.7412. [DOI] [PubMed] [Google Scholar]
- Jo Y, Stocco DM. Regulation of steroidogenesis and steroidogenic acute regulatory protein in R2C cells by DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene-1). Endocrinology. 2004;145:5629–5637. doi: 10.1210/en.2004-0941. [DOI] [PubMed] [Google Scholar]
- Hiroi H, Christenson LK, Chang L, Sammel MD, Berger SL, Strauss JF., III Temporal and spatial changes in transcription factor binding and histone modifications at the steroidogenic acute regulatory protein (StAR) locus associated with StAR transcription. Mol Endocrinol. 2004;18:791–806. doi: 10.1210/me.2003-0305. [DOI] [PubMed] [Google Scholar]