Abstract
The morphological patterns of glioma cell invasion are known as the secondary structures of Scherer. In this report, we propose a biologically based mechanism for the nonrandom formation of Scherer’s secondary structures based on the differential expression of stromal cell-derived factor (SDF)-1α and CXCR4 at the invading edge of glioblastomas. The chemokine SDF-1α was highly expressed in neurons, blood vessels, subpial regions, and white matter tracts that form the basis of Scherer’s secondary structures. In contrast, the SDF-1α receptor, CXCR4, was highly expressed in invading glioma cells organized around neurons and blood vessels, in subpial regions, and along white matter tracts. Neuronal and endothelial cells exposed to vascular endothelial growth factor up-regulated the expression of SDF-1α. CXCR4-positive tumor cells migrated toward a SDF-1α gradient in vitro, whereas inhibition of CXCR4 expression decreased their migration. Similarly, inhibition of CXCR4 decreased levels of SDF-1α-induced phosphorylation of FAK, AKT, and ERK1/2, suggesting CXCR4 involvement in glioma invasion signaling. These studies offer one plausible molecular basis and explanation of the formation of Scherer’s structures in glioma patients.
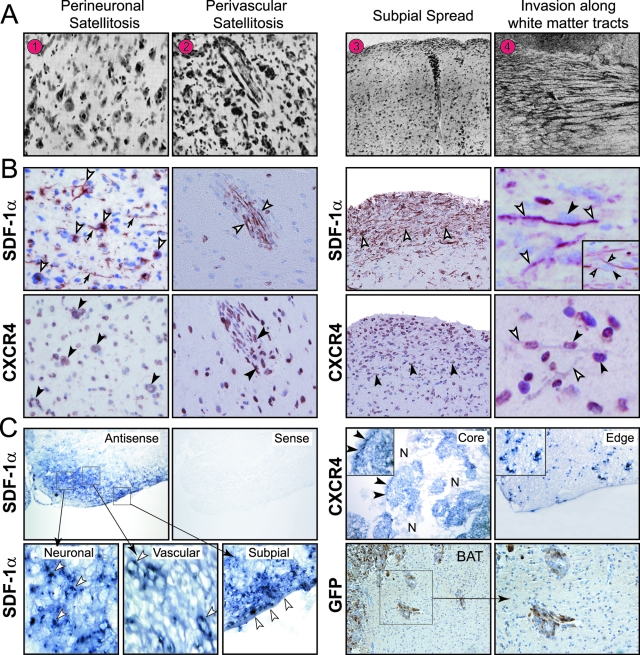
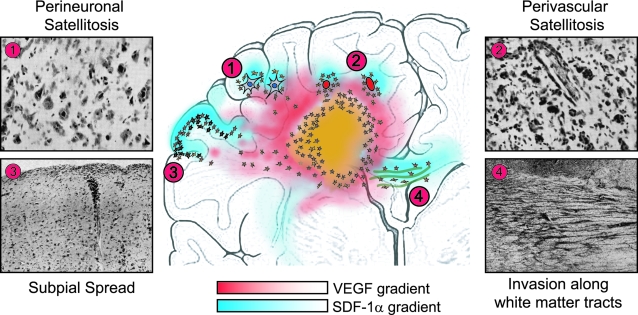
In 1938, Hans Joachim Scherer1 described the diffuse invasion of glioblastoma multiforme (GBMs) from the main tumor mass into the adjacent brain parenchyma. The morphological patterns of glioma cell invasion have since been referred to as the “secondary structures of Scherer.” Although he named eight distinct types of secondary structures, this report will focus on only four of them as shown in his original manuscript (Figure 1A). These secondary structures, as described in his words,1 include: i) “perineural and neurophagic growth: collections of glioma cells about all or a great many nerve cells”; ii) “perivascular growth that is the arrangement of the glioma cells about many of the pre-existing vessels and with a tendency of concentrically arranged layers of cells”; iii) “subpial surface growth that consists in a more or less thick layer of gliomatous cells in the molecular layer of the cortex”; and iv) “secondary structure related to the migration of glioma cells along the white matter, intrafascicular spread.” Today, these secondary structures of Scherer are referred to as i) perineuronal satellitosis, ii) perivascular satellitosis, iii) subpial spread, and iv) invasion along the white matter tracts.2
Figure 1.
SDF-1α and CXCR4 expression in secondary structures. A: Secondary structures of Scherer. As described in Dr. Scherer’s own words1: 1) “Precocious perineural structures forming capsules about nerve cells. Case 6/36 Nissl.” 2) “Precocious perivascular and perineural structures in the putamen, imitating perfectly the normal architecture of the corpus striatum. Case 204/35 Nissl.” 3) “Combination of precocious superficial, perivascular, and perineural structures. Case 27.37. Van Gieson. The perineural growth is seen only in the deep layers of the cortex.” 4) “Typical perifascicular structures, cut transversely. Case 75/35. Nissl.” (Reproduced from Scherer HJ: Structural development in gliomas. Am J Cancer 1938, 34:333–351, with permission from Columbia University).1 B: SDF-1α and CXCR4 expression in the secondary structures of Scherer in human GBMs. Left: SDF-1α and CXCR4 expression in perineuronal and perivascular satellitosis. SDF-1α staining in neurons and blood vessels (white arrowheads) and neuronal processes (arrows). CXCR4-positive tumor cells clustered around neurons and blood vessels (black arrowheads). Isolated CXCR4-immunoreactive invading glioma cells are also seen. Right: SDF-1α and CXCR4 expression in subpial spread and in white matter tracts. Intense SDF-1α staining in subpial region and in myelinated tracts (white arrowheads). Strong CXCR4 staining of collections of tumor cells in subpial regions and along white matter tracts (black arrowheads). Invading glioma cells (black arrowheads) are contiguous to SDF-1α-expressing white matter tracts (white arrowheads). Inset: A tumor cell (black arrowhead) is flanked by two separate positive SDF-1α white matter tracts (white arrowheads). Immunohistochemistry was done on serial sections. C: In situ hybridization for SDF-1α and CXCR4 in the experimental GL261 murine glioma. Left: SDF-1α mRNA signal was detected in neurons, blood vessels, and subpial region at the invading tumor edge (arrowheads). No mRNA signal was detected with the sense probe. Top right: Strong CXCR4 mRNA signal was detected in pseudopalisading cells (black arrowheads), invading glioma cells, in tumor core and tumor edge, respectively. N, necrosis. Inset: CXCR4 mRNA signal detected in invading cells. Bottom right: Perivascular GFP-positive GL261 glioma cells seen at the invading edge of the tumor into the brain adjacent to tumor (BAT). Original magnifications: ×50 (C, top and bottom right); ×100 (B, right; C, bottom); ×200 (B, left; C, bottom and inset); ×400 (B, right).
In his original article, Scherer defined the secondary structures based on his extensive morphological analysis of 100 GBMs and proposed the possibility of a “selective migration of tumor cells toward pre-existing structures,” eg, neurons or blood vessels. In this report, now 7 decades after his morphological-based hypothesis, we propose a biological-based mechanism that may help explain the formation of the secondary structures. We analyzed a series of 31 GBMs for patterns of glioma cell invasion related to the expression of the chemokine stromal cell-derived factor-1 (SDF-1)α and its receptor CXCR4. We found that SDF-1α was highly expressed in neuronal cells, blood vessels, subpial regions and white matter tracts that form the basis of Scherer’s secondary structures. In contrast, CXCR4, the receptor for SDF-1α, was highly expressed in invading glioma cells organized around neurons (perineuronal satellitosis), blood vessels (perivascular satellitosis), in the subpial region (subpial spread), and along white matter tracts (invasion along the white matter tracts).
Based on our in vivo results, we hypothesized that the secondary structures of Scherer may arise, in part, based on a SDF-1α gradient originating from structures expressing SDF-1α and the subsequent migration of CXCR4-positive glioma cells toward pre-existing structures. To help explain the basis for the formation of the secondary structures of Scherer, we evaluated several potential biological-based mechanisms using in vivo and in vitro assays. In vivo studies included in situ hybridization for SDF-1α and CXCR4 mRNA using the experimental GL261 murine glioma. In vitro assays evaluated expression of SDF-1α in neuronal and endothelial cell cultures exposed to the angiogenic factor vascular endothelial growth factor (VEGF). Human glioma cell lines expressing different levels of CXCR4 were used to determine the effect of a SDF-1α gradient on their migration potential in vitro. We then tested the effect of two inhibitory approaches for CXCR4: AMD3100, a specific CXCR4 inhibitor, and knockdown of CXCR4 expression using shRNA expression vectors directed against CXCR4. Finally, we analyzed the effect of SDF-1α on the level of expression of phosphorylated FAK, AKT, and ERK1/2 in glioma cells, all critical signaling molecules in glioma invasion. Based on our in vivo and in vitro observations, we propose that the differential expression of SDF-1α and CXCR4 at the invading edge of GBMs may represent one plausible explanation for the formation of the well-described secondary structures of Scherer.
Materials and Methods
Tissue Samples
This study was conducted under a protocol approved by the Institutional Review Board of New York University School of Medicine. Thirty-one GBMs classified according to World Health Organization criteria were studied.3 The tumors were from 18 female and 13 male patients. All tumors were supratentorial. Their location was frontal (13 cases), temporal (14 cases), parietal (2 cases), and occipital (2 cases) lobes. The age range was from 34 to 80 years. Three samples of normal brain removed in the course of surgical exposure were used as controls. When present, normal tissue adjacent to the tumor was used as an internal control.
Immunohistochemistry in Human GBMs
Immunohistochemistry for SDF-1α and CXCR4 was performed as previously described.4,5,6 Serial sections from 31 GBMs and 3 samples of normal brain were immunostained for SDF-1α or CXCR4. SDF-1α was detected with a mouse monoclonal antibody (clone 7801B; R&D Systems, Minneapolis, MN) diluted 1:50 and CXCR4 was detected with mouse monoclonal antibody (clone 44716, R&D Systems) diluted 1:150. Heat-induced epitope retrieval was done by boiling the deparaffinized tissue sections in 10 mmol/L citrate buffer (pH 6.0) for 20 minutes for SDF-1α and CXCR4 in a 1200 W microwave oven at 90% output. The sections were allowed to cool to room temperature for 30 minutes and then incubated with primary antibodies at room temperature overnight on a NexES (Ventana Medical Systems, Tucson, AZ) automated immunostainer. SDF-1α and CXCR4 were detected with a prediluted biotinylated goat anti-mouse secondary antibody (Ventana Medical Systems, Tucson, AZ). Flanking sections were stained for α-smooth muscle actin (SMA) with a mouse monoclonal antibody directed against the amino terminal decapeptide of SMA (Biogenex Laboratories, San Ramon, CA), at 1:100 dilution.7,8 Neu-N immunostaining9 (Chemicon, Temecula, CA) was used to identify neurons at 1:2000 dilution. In each case, the secondary antibody was applied for 30 minutes followed by horseradish peroxidase-conjugated streptavidin with 3,3′-diaminobenzidine as the chromogen.
For double-label immunohistochemistry we first used anti-CXCR4 antibody incubated overnight at room temperature and detection with a biotinylated goat anti-mouse antibody followed by application of streptavidin-horseradish-peroxidase conjugate, visualized with 3,3′-diaminobenzidine (brown). Second, we used anti-SDF-1α antibody incubated overnight at room temperature and detected with a biotinylated goat anti-mouse antibody, followed by application of alkaline phosphatase-streptavidin conjugate, visualized with Fast Red (red). Nuclei were lightly counterstained with hematoxylin, and slides dehydrated and mounted with permanent medium. Control procedures included isotype-matched murine monoclonal antibody for SDF-1α and CXCR4.
Histological Assessment
The GBMs used in this study were selected because each specimen included brain parenchyma adjacent to the tumor. This allowed analysis of tumor cells at the edge of the tumor invading into the brain tissue. We identified from the files of the Division of Neuropathology at New York University, cases in which at least one of the secondary structures was recognized. Two pathologists (D.Z. and H.Y.) independently evaluated the immunostaining results. When an evaluation differed, the final decision was made by consensus. The immunohistochemical analysis of SDF-1α and CXCR4 was scored as follows: 0, no staining; +, staining of less than 1% of structures; ++, staining of 1 to 10% of structures; +++, staining of 10 to 50% of structures; ++++, staining of greater than 50% of structures.4 In each GBM specimen, immunostaining for SDF-1α in neurons, blood vessels, subpial regions, and white matter tracts and immunostaining for CXCR4 in invading glioma cells was assessed separately.
Intracranial Tumor Model
This study was conducted under a protocol approved by the Institutional Animal Care and Use Committee of New York University School of Medicine. The animal model used green fluorescence protein (GFP)-tagged GL261 glioma cells implanted in the brains of syngeneic C57BL/6 mice as previously described.10 This experimental system allows us to detect GL261-invading glioma cells using immunohistochemistry for GFP as described.10
In Situ Hybridization in the Murine GL261 Glioma
Frozen tissue sections of tumor-containing cryoprotected mouse brains from mice 28 days after implantation and two normal brains from uninjected mice were used. The RNA probes for SDF-1α and CXCR4 were kindly provided by Dr. Y. Shen in Dr. D. Littman’s laboratory at New York University. To obtain an antisense SDF-1α probe, a plasmid carrying 880 bp of the SDF-1α cDNA was linearized with XhoI and transcribed with SP6 RNA polymerase. To obtain a sense SDF-1α probe, the plasmid was linearized with HindIII and transcribed with T7 RNA polymerase. To obtain an antisense CXCR4 probe, a plasmid carrying 800 bp of the CXCR4 cDNA was linearized with SalI and transcribed with T3 RNA polymerase. To obtain a sense CXCR4 probe, the plasmid was linearized with XbaI and transcribed with T7 RNA polymerase. In situ hybridization was performed on 20-μm-thick coronal cryostat frozen sections, using digoxigenin-labeled sense and anti-sense RNA probes against SDF-1α and CXCR4 as previously described.11 Briefly, slides were fixed in 4% paraformaldehyde (PFA) for 10 minutes at room temperature, rinsed three times in phosphate-buffered saline (PBS), treated with proteinase K for 5 minutes, refixed with 4% PFA for 10 minutes at room temperature, and washed three times for 5 minutes each in PBS. After acetylation, the sections were incubated with hybridization buffer containing sense or antisense RNA probes at 55°C for 18 hours in a moist chamber. After hybridization, slides were washed under high stringency conditions and treated with ribonuclease A to remove unhybridized probes, followed by incubation with the anti-digoxigenin antibody and visualization with BM-Purple AP substrate, according to the manufacturer’s instructions (Roche Biosciences, Palo Alto, CA). Serial brain sections were also evaluated using immunohistochemistry for GFP, performed as previously described.10 Three independent experiments were performed.
Cells and Reagents
The human glioma cell lines, LN229 and LN308 (kindly provided by Dr. F. Furnari from the Ludwig Institute for Cancer Research, University of California at San Diego, San Diego, CA), the U87MG cell line (originally obtained from the American Type Culture Collection, Manassas, VA) and the human embryonic kidney 293T cells, used for lentivirus production studies (kindly provided by Dr. M. Pagano, New York University) were cultured in 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin, and 2 mmol/L glutamine (Life Technologies, Inc., Grand Island, NY). SDF-1α and VEGF were purchased from R&D Systems and a stock solution (100 μg/ml and 10 μg/ml, respectively) was prepared in 0.1% bovine serum albumin (BSA) in PBS and kept at −20°C until used. AMD3100, a specific CXCR4 inhibitor,12 (kindly provided by Dr. J.B. Rubin, Washington University, St. Louis, MO), was prepared in PBS (5 mg/ml) and kept at 4°C until used. For hypoxic exposure, cells were placed in a sealed modular incubator chamber (Billups-Rothenberg Inc., Del Mar, CA) flushed with 1% O2, 5% CO2, and 94% N2 and incubated at 37°C for the appropriate time interval. Human cortical neurons (HCN-1A), herein after referred to as HCNs, were purchased from the American Type Culture Collection and cultured in DMEM supplemented with 10% FBS. Human brain microvascular endothelial cells (HBMECs) were purchased from Cell Systems (Kirkland, WA) and cultured in CS-C Complete medium (Cell Systems).
Immunofluorescence Microscopy of Glioma Cells
LN229, LN308, and U87MG glioma cells (3 × 104) were seeded onto poly-d-lysine-coated glass coverslips and incubated overnight. Cells were grown under normoxia or hypoxia for 12 hours and processed for immunofluorescence. For SDF-1α staining, cells were incubated with a rabbit polyclonal anti-human SDF-1α antibody (2.5 μg/ml) (Peprotech, Inc., Rocky Hill, NJ) for 1 hour at 4°C, followed by a secondary anti-rabbit fluorescein isothiocyanate-conjugated antibody (Jackson ImmunoResearch, West Grove, PA) for 30 minutes at 4°C, washed, and fixed with 1% PFA. For CXCR4 staining, cells were first fixed in 0.25% PFA in PBS for 30 minutes and permeabilized for 20 minutes in 0.01% saponin/PBS (Sigma-Aldrich, Saint Louis, MO) at room temperature. Nonspecific binding was blocked by incubation in blocking buffer containing 2% BSA for 30 minutes at room temperature. Cells were incubated overnight at 4°C with primary anti-CXCR4 rabbit polyclonal antibody (IMG-537; Imgenex, San Diego, CA) diluted 1:500. Cells were washed in blocking buffer three times for 5 minutes each before incubation with secondary goat anti-rabbit fluorescein isothiocyanate-conjugated antibody (Jackson ImmunoResearch) diluted 1:500 for 2 hours at room temperature in the dark. Cells were counterstained with propidium iodide (PI, 20 μg/ml) for 20 minutes at room temperature, washed in PBS, mounted onto glass slides, and examined using a Nikon (Melville, NY) fluorescence microscope. Images were adjusted using Adobe Photoshop 9.0 software (Adobe, San Jose, CA). Four independent experiments were performed. Control cells were incubated with irrelevant polyclonal antibody or after omission of the primary antibodies.
Immunofluorescence Studies in Neuronal and Endothelial Cells
Cells (2 × 106) were seeded onto poly-d-lysine-coated coverslips (HCNs) or attachment factor-coated coverslips (HBMECs) and grown in DMEM supplemented with 10% FBS or CS-C complete medium, respectively. After 48 hours, the medium was changed to serum-free medium, containing 50 mmol/L HEPES. After 4 hours of serum starvation, cells were exposed to serum-containing media with or without 50 ng/ml of VEGF for 8 hours in normoxic conditions and stained for SDF-1α as described above for glioma cells. The percentage of SDF-1α-positive cells was counted in five random fields at ×200 magnification. Three independent experiments were performed and pooled for statistical analysis.
Flow Cytometry
HCNs and HBMECs were collected from the culture dishes and incubated with a rabbit polyclonal anti-human SDF-1α antibody (Peprotech, Inc.) diluted 1:500 for 1 hour at 4°C. After washing, cells were incubated for 30 minutes at 4°C with a secondary anti-rabbit fluorescein isothiocyanate-conjugated antibody (Jackson ImmunoResearch), diluted 1:1000. Cells were examined using a LSRII flow cytometer (Becton Dickinson Biosciences, San Jose, CA) and data were analyzed by FlowJo software (Tree Star Inc., Ashland, OR). Three independent experiments were performed. Results were expressed as a ratio of the mean fluorescence intensity of cells stained with the SDF-1α antibody after VEGF exposure versus cells exposed to control media. Representative flow histograms are presented.
Western Blot Analysis
LN229, LN308, and U87MG glioma cells (2 × 106) were seeded in 10-cm dishes in complete growth medium. After 48 hours, the medium was changed to DMEM containing 1% FBS and 50 mmol/L HEPES (pH 7.4). Cells were exposed to normoxic or hypoxic conditions for 24 hours. Cells were lysed in RIPA buffer (150 mmol/L NaCl, 1% Nonidet P-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate, 10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L ethylenediaminetetraacetic acid, pH 8.0) supplemented with protease inhibitors.5 Protein quantitation and electrophoresis was performed as previously described.5 Western blot analysis was performed with the following antibodies: rabbit anti-CXCR4 polyclonal antibody (IMG-537, Imgenex), rabbit anti-total FAK polyclonal antibody (Abcam, Cambridge, MA), rabbit anti-p-FAK polyclonal antibody (Abcam), rabbit anti-total AKT polyclonal antibody (Cell Signaling Technology, Inc., Danvers, MA), rabbit anti-p-AKT polyclonal antibody (Cell Signaling Technology, Inc.), rabbit anti-total ERK1/2 polyclonal antibody (Cell Signaling Technology, Inc.), and mouse anti-pERK1/2 monoclonal antibody (sc-7383; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Anti-FAK, anti-AKT, and anti-ERK1/2 antibodies were all used at 1:1000 dilutions whereas the anti-CXCR4 was used at 1:500 dilution. We also used a mouse anti-actin monoclonal antibody at 1:20,000 dilution (mAb 1501, clone C4; Chemicon International, Inc., Temecula, CA). Sheep anti-mouse and donkey anti-rabbit IgG (Amersham Life Pharmacia Biotech, Piscataway, NJ) horseradish peroxidase-conjugated secondary antibodies were used at 1:2500 dilution. Immunodetection was performed with the Supersignal West Pico chemiluminescent substrate enhanced chemiluminescence detection system (Pierce Biotechnology Inc., Rockford, IL) followed by visualization and densitometry using NIH Image software (National Institutes of Health, Bethesda, MD). Two independent experiments were performed.
Migration Assay
BD Biocoat chambers (BD Biosciences Discovery Labware, Bedford, MA) with 8-μm pore size polystyrene filter inserts were used according to the manufacturer’s instructions and as described.5 Briefly, cells (5 × 104) in 400 μl of DMEM with 10% FBS were seeded onto the upper compartment of each chamber and placed into wells containing 600 μl of complete medium in the lower chamber in the absence or presence of SDF-1α (0 ng/ml or 100 ng/ml). Cells were allowed to adhere for 1.5 hours, and then the medium in the upper chamber was replaced with complete medium with or without varying concentrations of SDF-1α (0, 1, 10, 100 ng/ml), in the absence or presence of 100 nmol/L of AMD3100. The migration chambers were incubated for 24 hours in normoxic or hypoxic conditions at 37°C. After incubation, the inserts were fixed and stained and the number of migrating cells was counted as described.5 Two independent experiments, done in triplicate, were performed.
Lentivirus Production and Infection of Glioma Cells
Five different shRNA sequences directed against CXCR4 were purchased from Open Biosystems (Huntsville, AL). Recombinant lentiviruses were produced by co-transfecting human embryonic kidney 293T cells with the lentivirus expression vector (pLKO.1 puro), either empty or expressing a CXCR4 shRNA sequence and packaging plasmids (Δ8.9 and vsv-g) using Fugene 6 (Roche Diagnostics, Indianapolis, IN) as a transfection reagent.13 Infectious lentiviruses were collected at 24, 48, and 72 hours after transfection and the pooled supernatants centrifuged to remove cell debris and filtered through a 0.45-μm filtration unit. Cells were infected and stable transfectants were selected in puromycin for 7 days. Three of the five sequences down-regulated CXCR4 expression in glioma cells based on Western blot analysis and were used for further investigations.
Statistical Analysis
The potential association between detection of SDF-1α and CXCR4 expression by immunohistochemistry and the presence of a given secondary structure of Scherer was statistically analyzed using two different methods. The Fisher exact test with Bonferroni correction with a stringent statistical significance level of 0.0125 was used because of multiple testing. A logistic regression model evaluated the probability of observing a given secondary structure in glioma specimens based on the detection of SDF-1α and/or CXCR4 immunoreactivity. Using this model, the reference group was defined as cases in which both SDF-1α and CXCR4 were detected. We first analyzed cases in which SDF-1α or CXCR4 was present and asked whether it was predictive of detecting a given secondary structure. We then studied cases in which neither SDF-1α nor CXCR4 were present and asked whether it was predictive of detecting a given secondary structure. A P value and an odds ratio were calculated. The log odds ratio (LOR) was defined as the natural log of the ratio of the odds of a specific secondary structure occurring in one group compared to the odds of it occurring in the reference group. For in vitro studies, experiments were performed two or three times. Results are expressed as mean ± SD. All analyses for the conditions being compared were performed using a two-sided Student’s t-test for significance (P < 0.05). All statistical analysis was performed using statistical software R 2.5.1.
Results
SDF-1α and CXCR4 Patterns of Expression Associated with the Secondary Structures of Scherer in GBMs
As a first step to investigate the role of SDF-1α and CXCR4 expression in the formation of the secondary structures of Scherer, we performed immunohistochemical analysis in 31 GBM cases. In each GBM specimen we analyzed the invading edge of the tumors. The results of the analyses are summarized in Table 1. Patterns of expression for SDF-1α and CXCR4 are illustrated in Figure 1B and Figure 2A.
Table 1.
Immunoreactivity for SDF-1α and CXCR4 in the Secondary Structures of Schere
| Case number | PNS
|
PVS
|
SPS
|
IAWMT
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neurons SDF-1α | Glioma cells CXCR4 | Blood vessels SDF-1α | Glioma cells CXCR4 | Subpial region SDF-1α | Glioma cells CXCR4 | White matter tracts SDF-1α | Glioma cells CXCR4 | ||||||
| 1 | + | ++ | ++ | + | ++ | ++++ | ++++ | +++ | |||||
| 2 | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | 0 | ++ | |||||
| 3 | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | |||||
| 4 | 0 | ++ | +++ | ++ | ++ | ++ | ++ | ++++ | |||||
| 5 | +++ | ++++ | ++++ | ++++ | ++++ | +++ | 0 | 0 | |||||
| 6 | 0 | ++ | +++ | ++++ | +++ | ++ | + | + | |||||
| 7 | + | + | + | 0 | ++++ | +++ | +++ | ++ | |||||
| 8 | + | 0 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |||||
| 9 | 0 | + | ++++ | ++ | +++ | 0 | ++++ | ++++ | |||||
| 10 | ++++ | ++++ | ++ | 0 | ++ | 0 | ++++ | +++ | |||||
| 11 | 0 | + | 0 | + | ++++ | ++++ | ++++ | ++ | |||||
| 12 | ++ | ++ | +++ | +++ | 0 | ++ | ++ | ++ | |||||
| 13 | ++++ | ++++ | ++++ | ++++ | 0 | ++ | ++++ | ++++ | |||||
| 14 | 0 | ++ | ++++ | +++ | ++++ | ++++ | + | 0 | |||||
| 15 | ++++ | ++ | +++ | ++++ | 0 | + | +++ | ++++ | |||||
| 16 | ++++ | ++++ | 0 | + | 0 | 0 | ++++ | ++ | |||||
| 17 | 0 | ++ | 0 | + | ++++ | ++++ | ++++ | ++++ | |||||
| 18 | +++ | +++ | 0 | 0 | ++++ | ++++ | +++ | ++++ | |||||
| 19 | ++ | ++++ | 0 | ++ | ++++ | ++ | 0 | 0 | |||||
| 20 | +++ | +++ | 0 | +++ | ++++ | ++ | ++++ | ++ | |||||
| 21 | 0 | 0 | 0 | ++ | 0 | 0 | 0 | + | |||||
| 22 | ++++ | ++++ | ++++ | +++ | 0 | 0 | ++ | +++ | |||||
| 23 | ++++ | +++ | ++++ | ++++ | 0 | 0 | 0 | 0 | |||||
| 24 | 0 | 0 | ++ | ++++ | 0 | 0 | 0 | 0 | |||||
| 25 | ++++ | ++++ | 0 | +++ | +++ | +++ | 0 | ++ | |||||
| 26 | ++++ | ++ | 0 | 0 | 0 | 0 | ++++ | ++++ | |||||
| 27 | 0 | 0 | ++ | 0 | +++ | ++++ | 0 | 0 | |||||
| 28 | +++ | +++ | ++++ | ++ | 0 | 0 | ++++ | ++++ | |||||
| 29 | ++++ | ++ | ++++ | ++++ | ++++ | +++ | ++++ | +++ | |||||
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | ++ | 0 | |||||
| 31
|
++
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|||||
|
|
PNS
|
|
PVS
|
|
SPS
|
|
IAWMT
|
||||||
|
|
D
|
ND
|
D
|
ND
|
D
|
ND
|
D
|
ND
|
|||||
| SDF-1α | CXCR4 | 21 | 10 | 19 | 12 | 17 | 14 | 23 | 8 | ||||
| + | + | 17 | 2 | 16 | 1 | 16 | 1 | 18 | 2 | ||||
| + | − | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | ||||
| − | + | 2 | 4 | 1 | 6 | 0 | 3 | 2 | 1 | ||||
| − | − | 1 | 3 | 1 | 3 | 0 | 9 | 2 | 4 | ||||
Serial sections from 31 GBMs and 3 samples of normal brain were immunostained for SDF-1α or CXCR4. The patterns of SDF-1α and CXCR4 staining relating to each of the secondary structures of Scherer included 21 (68%) cases with perineuronal satellitosis (PNS), 19 (61%) with perivascular satellitosis (PVS), 17 (55%) cases with subpial spread (SPS), and 23 (74%) cases with invasion along white matter tracts (IAWMT). Within each column for the secondary structure, SDF-1α results refer to staining in neurons, blood vessels, subpial regions, and white matter tracts. CXCR4 results refer to staining in invading glioma cells. The immunohistochemical analysis of SDF-1α and CXCR4 was scored as follows: 0, no staining; +, staining of less than 1% of structures; ++, staining of 1 to 10% of structures; +++, staining of 10 to 50% of structures; ++++, staining of greater than 50% of structures. The bold portions of the table correspond to the cases where a given secondary structure was not detected. For the bottom portion of the table: D, number of cases where a secondary structure was detected; ND, number of cases where a secondary structure was not detected. Further, for this section, SDF-1α + indicates the number of cases where SDF-1α was detected and SDF-1α − indicates the number of cases where SDF-1α was not detected. CXCR4 + indicates the number of cases where CXCR4 was detected and CXCR4 − indicates the number of cases where CXCR4 was not detected.
Figure 2.
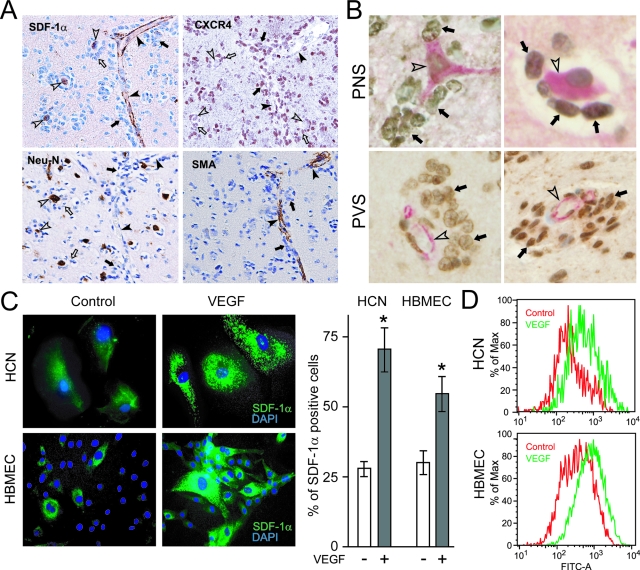
VEGF up-regulates SDF-1α in neurons and vascular cells. A: Perineuronal and perivascular satellitosis at the edge of GBMs. Top left: Immunoreactivity for SDF-1α is seen in neurons (open arrowheads) and in blood vessels (black arrowheads). Both perineuronal (open arrows) and perivascular (black arrows) satellitosis are evident as aggregates of tumor cells are seen around neurons and blood vessels, respectively. Top right: Perineuronal (open arrows) and perivascular (black arrows) CXCR4-immunopositive glioma cells are seen. Bottom left: Neu-N immunoreactivity decorates neuronal cells. Right bottom panel: Blood vessels at the edge of GBMs are highlighted by SMA immunoreactivity (black arrowheads). B: Double-label immunohistochemistry for SDF-1α and CXCR4 show SDF-1α-immunoreactive neurons and vascular channels (red, open arrowheads) and perineuronal and perivascular CXCR4-immunoreactive tumor cells (brown, black arrows). C: Immunofluorescence for SDF-1α in HCNs and HBMECs. In the absence of VEGF exposure, HCNs and HBMECs demonstrated weak and scant SDF-1α immunostaining. VEGF-treated cells showed strong immunoreactivity of SDF-1α (green fluorescence). DAPI: nuclear blue fluorescence. Bar graphs represent data pooled from three independent experiments for statistical analysis comparing the number of cells positive for SDF-1α expression in different groups. (*P < 0.05). D: Flow cytometry for SDF-1α in HCNs and HBMECs. After VEGF exposure, increasing levels of SDF-1α expression were found on HCNs and HBMECs. The relative mean fluorescence intensity increased 50% for HCNs and 100% in HBMECs. The results are representative of three independent experiments (*P < 0.05). Original magnifications: ×200 (A); ×400 (B); ×40 (C).
The patterns of SDF-1α and CXCR4 staining relating to each of the secondary structures of Scherer included 21 (68%) cases with perineuronal satellitosis, 19 (61%) cases with perivascular satellitosis, 17 (55%) cases with subpial spread, and 23 (74%) cases with invasion along white matter tracts. In some cases mixed features were seen as described by Scherer in his original report.1 In this study the most common combination we observed was the presence of perineuronal and perivascular satellitosis and subpial spread in seven cases, a combination previously described by Scherer1 (Figure 1A).
SDF-1α and CXCR4 immunoreactivity was present in each of the 31 GBMs, but to a variable degree. For SDF-1α, 22 cases showed nuclear, cytoplasmic, and membranous staining; 6 cases showed nuclear and cytoplasmic staining without any membranous staining; and 3 cases showed only nuclear or cytoplasmic staining but not both. Overall staining for SDF-1α in tumor cells was scant, consistent with previous reports.14,15 For CXCR4, 20 cases showed nuclear, cytoplasmic, and membranous staining; 7 cases showed nuclear and cytoplasmic staining without any membranous staining; and 4 cases showed only nuclear or cytoplasmic staining, but not both. Nuclear localization of CXCR4, reported in prostate16 and breast cancer17 has been correlated with invasive and metastatic spread.18 Immunohistochemistry for Neu-N and SMA was performed to identify neurons and blood vessels, respectively. Immunoreactivity for SMA was variably expressed in blood vessels, whereas Neu-N immunoreactivity was seen in neurons (Figure 2A).
Perineuronal Satellitosis (PNS)
The first of the secondary structures was seen in 21 of 31 cases (Table 1). In these positive cases, the number of neurons surrounded by tumor cells varied from only a few neurons to almost all of the neurons present in the specimen. In addition, glioma cells forming the perineuronal clusters also varied in number (Figure 1B). In 17 of the 21 cases, SDF-1α immunoreactivity was present in neurons, whereas CXCR4 immunoreactivity was observed in the glioma cells forming clusters around the neurons. The perinuclear cytoplasm of cortical neurons was highlighted by SDF-1α immunoreactivity, whereas neuronal cell processes were stained to a lesser degree (Figure 1B). The percentage of neurons expressing SDF-1α was scored as 4+ in 10 cases, 3+ in 5 cases, and 2+ in 2 cases. Regarding the percentage of CXCR4 expression in glioma cells, eight cases were scored as 4+, five as 3+, and four as 2+. Of the 21 cases with PNS there were 4 exceptions: 1 case showed SDF-1α-positive neurons (2+) surrounded by CXCR4-negative glioma cells; 2 cases showed CXCR4-positive glioma cells (2+, 2+) surrounding SDF-1α-negative neurons; and 1 case lacked both SDF-1α and CXCR4 immunoreactivity in neurons and perineuronal glioma cells, respectively. In addition, there were 10 cases without PNS. Three cases lacked neurons and tumor cells positive for SDF-1α and CXCR4, respectively. The remaining seven cases, in general, contained less than 10% of SDF-1α immunopositive neurons or CXCR4-positive glioma cells (Table 1).
Perivascular Satellitosis (PVS)
The second of the secondary structures was seen in 19 of 31 cases (Table 1). In these positive cases, the number of blood vessels surrounded by cuffs of glioma cells varied from field to field. Similarly, the number of glioma cells present in the thin or thick perivascular aggregates was variable (Figure 1B). In 16 of the 19 cases, SDF-1α immunoreactivity was present in blood vessels whereas CXCR4 immunoreactivity was observed in glioma cells surrounding the blood vessels (Figure 1B). The percentage of blood vessels expressing SDF-1α was scored as 4+ in 11 cases, 3+ in 4 cases, and 2+ in 1 case. Regarding the percentage of CXCR4 expression in glioma cells, 10 cases were scored as 4+, 3 as 3+, and 3 as 2+. Of the 19 cases with PVS there were 3 exceptions: 1 case showed SDF-1α-positive blood vessels (2+) surrounded by CXCR4-negative glioma cells; 1 case showed CXCR4-positive glioma cells (3+) surrounding SDF-1α-negative blood vessels; and 1 lacked both SDF-1α and CXCR4 immunoreactivity in blood vessels and perivascular glioma cells, respectively. In addition, there were 12 cases without PVS. Three cases lacked blood vessels and tumor cells positive for SDF-1α and CXCR4, respectively. The remaining nine cases, in general, contained less than 10% of SDF-1α immunopositive blood vessels or CXCR4-positive glioma cells (Table 1).
Subpial Spread (SPS)
The third of the secondary structures was seen in 17 of 31 cases (Table 1). In these positive cases, the zone just below the pial margin was the site of strong immunoreactivity for SDF-1α (Figure 1B) and included the glia limitans in all 17 cases. Glioma cells were observed as closely packed collections of CXCR4 immunopositive tumor cells located beneath the pia in the subpial regions in 16 of the 17 cases (Figure 1B). The expression of SDF-1α in subpial regions was scored as 4+ in 12 cases, 3+ in 3 cases, and 2+ in 1 case. Regarding the percentage of CXCR4 expression in glioma cells, nine cases were scored as 4+, four as 3+, and three as 2+. Of the 17 cases with SPS there was 1 exception where SDF-1α-positive subpial regions (2+) with CXCR4-negative aggregates of glioma cells were seen. There was no case in which CXCR4 was detected in the absence of SDF-1α expression or in which both SDF-1α and CXCR4 were absent. In addition, there were 14 cases without SPS. Nine cases lacked subpial regions and tumor cells positive for SDF-1α and CXCR4, respectively. The remaining five cases, in general, contained less than 10% of SDF-1α immunopositive subpial regions or CXCR4-positive glioma cells (Table 1).
Invasion Along White Matter Tracts (IAWMT)
Finally, the fourth of the secondary structures was seen in 23 of 31 cases (Table 1). In these positive cases the number of tracts with contiguous tumor cells varied with the proximity to the tumor edge, ie, these tracts were more numerous closer to the tumor edge. Here white matter tracts were highlighted by the immunoreactivity for SDF-1α (Figure 1B), whereas CXCR4 immunoreactivity was observed in invading glioma cells seen just contiguous to white matter tracts in 18 of the 23 cases (Figure 1B). The percentage of white matter tracts expressing SDF-1α was scored as 4+ in 13 cases, 3+ in 3 cases, and 2+ in 2 cases. Regarding the percentage of CXCR4 expression in glioma cells, nine cases were scored as 4+, five as 3+, and four as 2+. Of the 23 cases with IAWMT there were 5 exceptions: 1 case showed SDF-1α-positive white matter tracts (2+) with adjacent CXCR4-negative glioma cells; 2 cases showed CXCR4-positive glioma cells (2+, 2+) adjacent to SDF-1α-negative white matter tracts; and 2 cases lacked both SDF-1α and CXCR4 immunoreactivity in white matter tracts and invading glioma cells, respectively. In addition, there were eight cases without IAWMT. Four cases lacked white matter tracts and glioma cells positive for SDF-1α and CXCR4, respectively. The remaining four cases, in general, contained less than 10% SDF-1α immunopositive white matter tracts or CXCR4-positive tumor cells (Table 1).
In the three samples of normal brain, neurons showed weak immunoreactivity for SDF-1α and blood vessels showed weak immunoreactivity for SDF-1α and CXCR4.14,19 In all cases, no SDF-1α and CXCR4 staining was observed when isotype-matched murine monoclonal antibody was used. To show the relation between SDF-1α and CXCR4 expression, we performed double-label immunohistochemistry. Perineuronal and perivascular CXCR4 immunoreactive tumor cells (brown) were aggregated around SDF-1α immunoreactive neurons (red) and vascular channels (red) (Figure 2B). In addition, CXCR4-immunoreactive tumor cells were seen contiguous to SDF-1α-immunoreactive white matter tracts and subpial regions (data not shown).
Association Studies of the Occurrence of a Given Secondary Structure of Scherer with the Expression of SDF-1α and CXCR4
Using the Fisher exact test with Bonferroni correction, the detection of SDF-1α and/or CXCR4 immunoreactivity in a given secondary structure of Scherer, except for IAWMT, which had borderline significance, all of Scherer’s structures (PNS, PVS, and SPS) showed a statistically significant association. The P values were: PNS, P = 0.003; PVS, P < 0.001; SPS, P < 0.001; and IAWMT, P < 0.019. Using a logistic regression model, the reference group was defined as cases in which both SDF-1α and CXCR4 immunoreactivity were present. We first analyzed cases in which SDF-1α or CXCR4 was present and asked whether it was predictive of detecting a given secondary structure. In the first analysis, the detection of either SDF-1α or CXCR4 immunoreactivity had a significant log odds ratio (LOR) for the presence of the secondary structures PNS, PVS, and SPS, as compared to the reference group. In contrast, for the presence of IAWMT, the detection of either SDF-1α or CXCR4 immunoreactivity had no significant LOR. The P values and LORs were: PNS, P = 0.011, LOR = −2.65; PVS, P = 0.001, LOR = −4.16; SPS, P = 0.006, LOR = −2.74; IAWMT, P = 0.128, LOR = −1.79. Negative LORs indicate that the likelihood of the presence of immunoreactivity for SDF-1α or CXCR4 detected in a given secondary structure is low compared to the reference group.
We then studied cases in which neither SDF-1α nor CXCR4 were present and asked whether it was predictive of detecting a given secondary structure. In the second analysis of cases in which no SDF-1α or CXCR4 immunoreactivity was detected, there was a marked decrease in the probability of the presence of any of the secondary structures compared to the reference group. For SPS, there was no case in which both SDF-1α and CXCR4 were absent precluding the analysis by the logistic regression model. For the other secondary structures, the P values and LORs were: PNS, P = 0.019, LOR = −3.24; PVS, P = 0.012, LOR = −3.87; SPS, not done, LOR = −4.97; IAWMT, P = 0.011, LOR = −2.89. Thus, the absence of both SDF-1α and CXCR4 immunoreactivity predicted the absence of a secondary structure.
SDF-1α and CXCR4 Are Expressed at the Invading Edge of the Murine GL261 Glioma
Because the classic secondary structures of Scherer occur in the murine GL261 glioma model,10 we performed in situ hybridization to detect SDF-1α and CXCR4 mRNA expression patterns in the brains of mice with a GL261 glioma. SDF-1α mRNA was expressed in neurons, blood vessels, and in subpial regions at the tumor edge (Figure 1C), but not in white matter tracts. Strong CXCR4 mRNA signal was detected in invading GL261 glioma cells and in blood vessels at the tumor edge (Figure 1C). CXCR4 mRNA was also up-regulated in pseudopalisading cells within the tumor core (Figure 1C), consistent with our previous report in human GBMs.5 In normal mouse brain, weak SDF-1α and CXCR4 mRNA signals were detected in cortical neurons and in dentate gyrus, respectively.20,21 No signal was detected with the sense probe (Figure 1C). Using immunohistochemistry for GFP-tagged GL261 glioma cells, we detected secondary structures. For example, GFP-tagged tumor cells were seen clustered around blood vessels, ie, perivascular satellitosis, at the advancing edge of the tumor (Figure 1C).
VEGF Up-Regulates SDF-1α Expression in Neuronal and Vascular Cells
Based on previous reports showing that VEGF up-regulates SDF-1α in many cell types,22 we tested the hypothesis that VEGF might also be responsible for the up-regulation of SDF-1α observed in neurons and blood vessels at the edge of GBMs that form the basis of Scherer’s secondary structures. Neuronal (HCNs) and endothelial (HBMECs) cells were exposed to VEGF for 8 hours and SDF-1α expression was analyzed by immunostaining and flow cytometry (Figure 2, C and D). As visualized by immunostaining, stimulation with VEGF resulted in a significant increase in the percentage of SDF-1α-positive HCNs and HBMECs (Figure 2C). The percentage of SDF-1α-positive HCNs was 71.4 ± 16.2 after VEGF exposure compared to 25.9 ± 4.1 in untreated cells (P = 0.02). The percentage of SDF-1α-positive HBMECs was 54.9 ± 13.4 after VEGF exposure compared to 26.5 ± 5.5 in untreated cells (P = 0.007). In addition, we noted that cells exposed to VEGF showed increased staining intensity of SDF-1α (Figure 2C), which was confirmed by flow cytometry (Figure 2D).
CXCR4 Expression in Glioma Cells and Migration toward a SDF-1α Gradient
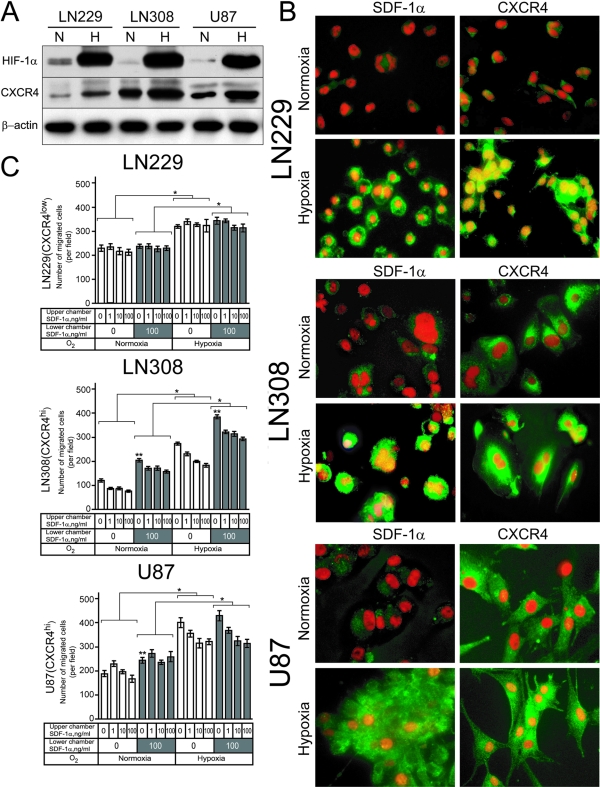
Based on our in vivo observations in human GBMs, we hypothesized that the secondary structures of Scherer might be associated with the migration of CXCR4-expressing glioma tumor cells toward brain structures expressing SDF-1α, ie, the source of a SDF-1α gradient. To test this hypothesis, we screened several glioma cell lines for their basal level of CXCR4 expression. We selected three cell lines that differed in CXCR4 expression by Western blot analysis: low-CXCR4-expressing LN229 (CXCR4lo), high-CXCR4-expressing LN308 (CXCR4hi), and U87MG (CXCR4hi) (Figure 3A). These Western blot results were consistent with the reported mRNA levels in these cell lines.23 Previously, we demonstrated that hypoxia and HIF-1 up-regulated CXCR4 expression in glioma cells.5 Therefore, we first tested the effect of hypoxia on the expression of CXCR4 protein in LN229, LN308, and U87MG glioma cells. Hypoxia increased the protein levels of HIF-1α and CXCR4 in all cell lines by approximately threefold (Figure 3A). However, the level of hypoxia-induced CXCR4 expression in LN229 (CXCR4lo) glioma cells remained low compared with the endogenous level of CXCR4 in LN308 (CXCR4hi) and U87MG (CXCR4hi) glioma cells in normoxic conditions.
Figure 3.
SDF-1α and CXCR4 in LN229, LN308, and U87MG glioma cells. A: CXCR4 expression in LN229, LN308, and U87MG glioma cell lines under normoxic and hypoxic conditions. The results are representative of two independent experiments. β-Actin was used as loading control. B: Detection of SDF-1α and CXCR4 by immunofluorescence in LN229, LN308, and U87MG glioma cells grown under normoxic and hypoxic conditions. The yellow color results from the overlay of the counterstaining with propidium iodide (red fluorescence), performed to visualize the nuclei, and the green fluorescence from the SDF-1α or CXCR4 immunoreactivity indicating nuclear translocation under hypoxic conditions. The results are representative of three independent experiments. C: CXCR4 expression in glioma cells and migration toward a SDF-1α gradient. Hypoxia exposure increased the migration of LN229 and LN308 and U87MG glioma cells compared to normoxic cultures (*P < 0.001). Under both normoxic and hypoxic conditions, the SDF-1α gradient affected the migration of LN308 and U87MG glioma cells (**P < 0.001). Bar graphs represent data from two independent experiments pooled for statistical analysis. Original magnifications, ×40 (B).
Using immunofluorescence, we investigated the levels of SDF-1α and CXCR4 expression in LN229, LN308, and U87MG cells grown in normoxic and hypoxic conditions. Under normoxic conditions, weak staining for SDF-1α and CXCR4 was observed in LN229 (CXCR4lo) cells whereas membranous or diffuse cytoplasmic pattern of expression was seen in LN308 (CXCR4hi) and U87MG (CXCR4hi) glioma cells. However, after 12 hours of exposure to hypoxia, LN229, LN308, and U87MG glioma cells up-regulated the expression of SDF-1α and CXCR4 in the nucleus (Figure 3B). This localization of SDF-1α, a secreted cytokine, and CXCR4, a cell membrane receptor to the nucleus, was consistent with our in vivo observations of GBM tumor cells where both SDF-1α and CXCR4 were observed in the nucleus (Figure 1B).
Next we evaluated the effects of a SDF-1α gradient on the migration potential of LN229 (CXCR4lo), LN308 (CXCR4hi), and U87MG (CXCR4hi) glioma cells. Cells were cultured for 24 hours under normoxic or hypoxic conditions and evaluated for their capacity to migrate in the Boyden chamber assay in the absence or presence of a SDF-1α gradient. Results from two independent experiments are shown in Figure 3C. The migration of LN229 (CXCR4lo), LN308 (CXCR4hi), and U87MG (CXCR4hi) glioma cells increased under hypoxic compared to normoxic conditions (P < 0.001). However, under both normoxic and hypoxic conditions, the migration of LN229 (CXCR4lo) glioma cells was not affected by the presence of SDF-1α in the lower chamber or as the concentration of SDF-1α increased in the upper chamber. In sharp contrast, under both normoxic and hypoxic culture conditions, the migration of LN308 (CXCR4hi) and U87MG (CXCR4hi) cells increased in the presence of SDF-1α in the lower chamber (P < 0.001) and decreased as the concentration of SDF-1α increased in the upper chamber (P < 0.01).
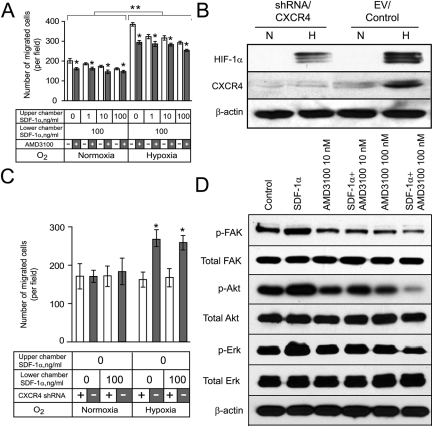
Blocking CXCR4 Expression Inhibits Migration of LN308 (CXCR4hi) Glioma Cells toward a SDF-1α Gradient
To further support the concept that CXCR4 expression by glioma cells potentiates their migration toward SDF-1α, we tested two separate approaches to block CXCR4 expression in LN308 (CXCR4hi) cells. First we used a specific CXCR4 inhibitor, AMD3100 (Figure 4A) and second we used knockdown of CXCR4 expression using shRNA expression vectors directed against CXCR4 (Figure 4B). AMD3100 inhibited the hypoxia- and SDF-1α-induced migration of LN308 cells compared with control cultures (P < 0.001). When repeated in the presence or absence of 10 mmol/L hydroxyurea, which blocks proliferation,24 the effect of AMD3100 on the migration of LN308 was unchanged (data not shown). When AMD3100-treated cells were analyzed for annexin V staining, a marker of early apoptosis,25 no increase in the number of annexin V-positive cells was observed compared with staurosporine, used as a positive control (data not shown). Thus, the inhibition of migration by AMD3100 was not because of inhibition of proliferation or induction of apoptosis. Hypoxic LN308 (CXCR4hi) glioma cells infected with an shRNA vector against CXCR4 specifically decreased expression of CXCR4 but not of HIF-1α (Figure 4B). After knockdown of CXCR4 expression, these cells failed to show increased migration toward SDF-1α under hypoxic conditions compared with cells infected with an empty vector (P < 0.001). These experiments were repeated using two other shRNA sequences producing the same results. A representative experiment obtained with the sequence 5′-CCGGGAGAAGCATGACGGACAAGTACTCGAGTACTTGTCCGTCATGCTTCTCTTTTT-3′ is shown in Figure 4C.
Figure 4.
Blocking CXCR4 expression inhibits migration of LN308 (CXCR4hi) glioma cells toward a SDF-1α gradient. A: Effect of a SDF-1α gradient in the presence or absence of AMD3100. Hypoxia exposure significantly increased the migration of LN308 glioma cells compared to control normoxic cultures (**P < 0.001). Addition of AMD3100 significantly inhibited the migration of LN308 glioma cells (*P < 0.001). Bar graphs represent data pooled from three independent experiments for statistical analysis. B: CXCR4 expression in LN308 glioma cells after knockdown of CXCR4 under normoxic (N) or hypoxic (H) conditions. The results are representative of two independent experiments. β-Actin expression was used as a loading control. C: Effects of knockdown of CXCR4 expression on the migration of LN308 glioma cells under normoxic or hypoxic conditions. Knockdown of CXCR4 expression diminished the hypoxia-induced increased migration of LN308 glioma cells (*P < 0.01). Bar graphs represent data pooled from two independent experiments for statistical analysis. D: AMD3100 inhibits the SDF-1α-induced phosphorylation of FAK, AKT, and ERK1/2 in LN308 glioma cells. Data represent one of two independent experiments. β-Actin expression was used as a loading control.
SDF-1α Induces Phosphorylation of FAK, AKT, and ERK1/2 in LN308 Glioma Cells
We and others26,27 have shown that FAK overexpression occurs in invading glioma cells at the edge of GBMs. Thus, we tested whether exposure of glioma cells to SDF-1α would induce phosphorylation of FAK and the signaling molecules AKT and ERK1/2 in LN308 glioma cells that demonstrated CXCR4-dependent migration toward SDF-1α (Figure 3C). LN308 glioma cells were exposed to SDF-1α for 15 minutes and analyzed for total and phosphorylated FAK, AKT, and ERK1/2 by Western blot analysis. In LN308 glioma cells SDF-1α increased the levels of expression of phosphorylated FAK, AKT, and ERK1/2, threefold, twofold, and twofold, respectively, (Figure 4D). Exposure to SDF-1α in the presence of AMD3100 decreased levels of expression of phosphorylated FAK, AKT, and ERK1/2, 10-, 3-, and 6-fold, respectively.
Discussion
The primary mechanism of spread for GBMs is different from that of other cancers. Unlike the hematogenous or lymphatic spread of carcinoma cells from their primary location to metastatic sites, GBMs invade locally throughout the brain tissue adjacent to the tumor. This highly invasive nature of GBMs renders most therapies, including radiotherapy and local drug delivery ineffective, resulting in the failure of conventional therapy to control glioma growth.28 The various patterns of local spread of GBMs were described by Scherer1 and are known as the secondary structures of Scherer. These include perineuronal and perivascular satellitosis, subpial spread, and invasion along white matter tracts. This study was performed because we hypothesized that invasion of glioma cells at the edge of human GBMs was not random. We postulated that the SDF-1α/CXCR4 axis was important in the directional invasion of glioma cells into the neighboring parenchyma and could represent a molecular basis for the formation of the secondary structures of Scherer. To test this hypothesis, we investigated the relative patterns of expression of the SDF-1α ligand and its receptor CXCR4 at the invading edge of human GBMs using immunohistochemistry and in murine gliomas using in situ hybridization. We then performed in vitro experiments to validate our in vivo findings.
Our results support the hypothesis that migration of glioma cells at the edge of GBMs is not random (Figure 5). Hypoxia induces local migration by up-regulating CXCR4 in glioma cells,5 and by inducing VEGF secretion, which in turn leads to up-regulation of SDF-1α by neurons and blood vessels. Therefore, turning on SDF-1α expression and its secretion from these pre-existing structures would form a SDF-1α gradient that would attract CXCR4-positive glioma cells to migrate toward SDF-1α-positive pre-existing structures, eg, neurons and blood vessels. Scherer1 called these structures “secondary” because their formation depended on underlying normal brain structures, as opposed to “primary” structures, such as pseudopalisading cells, necrosis, and microvascular proliferation. Our report gives new meaning to the designation of these structures as secondary because they may result, in part, from the differential expression of SDF-1α and CXCR4 in the structures and tumor cells, respectively, at the edge of GBMs inducing local invasion.
Figure 5.
Molecular basis for the formation of the secondary structures of Scherer: 1) perineuronal satellitosis, 2) perivascular satellitosis, 3) subpial spread, and 4) invasion along the white matter tracts. Our immunohistochemical data in clinical specimens show that CXCR4 is expressed in invading glioma cells whereas SDF-1α is expressed in neurons, blood vessels, subpial regions, and in white matter tracts. Our results support the hypothesis that migration of glioma cells at the edge of GBMs is not random. Our data in vivo and our findings in vitro suggest that the differential expression of SDF-1α and CXCR4 at the advancing edge of GBMs may lead to the formation of the secondary structures of Scherer. Hypoxia up-regulates CXCR4 in glioma cells, and induces VEGF release by the tumor (red gradient), which in turn could be responsible, at least in part, for the overexpression of SDF-1α in pre-existing structures, eg, neuronal cells and angiogenic blood vessels. Therefore, turning on SDF-1α expression and its secretion from these pre-existing structures, eg, neurons and blood vessels would form a SDF-1α gradient (blue gradient) that would attract CXCR4-positive glioma cells from the tumor core to migrate into the brain adjacent to tumor.
The subcellular localization of CXCR4 in tumor specimens is variable and has been reported as nuclear, cytoplasmic, or membranous in a wide variety of cancers.4,16,17,18,29,30,31,32,33,34 Staining for CXCR4 in hepatocellular,29 human colorectal carcinomas,30 and metastatic melanomas31 was primarily cytoplasmic whereas a few cases showed weak membranous staining. Pancreatic32 and cervical carcinomas33,34 were found to express variable membranous staining. In this study we found 20 of 31 GBMs displayed CXCR4 membrane staining. Previously, we have described differential CXCR4 staining in renal cell carcinomas and hemangioblastomas in which 8 of 15 renal cell carcinomas and 2 of 22 cases of hemangioblastomas showed membranous staining.4
Strong SDF-1α immunoreactivity was observed in neuronal cells and in blood vessels at the edge of GBMs compared with weak staining of neurons and blood vessels of normal human brain. Similarly, in the mouse GL261 glioma model, a strong signal for SDF-1α mRNA was present in neuronal cells and in blood vessels adjacent to the invading tumor edge compared with weak mRNA signal in normal mouse brain. VEGF, a critical angiogenic factor in gliomas,6 has been reported to up-regulate SDF-1α in many cell types,22 for example, in perivascular fibroblasts35 and in U251MG and U87MG glioma cells.36 Thus, we hypothesized that the increased expression of SDF-1α at the invasive edge of glial tumors was potentially linked to VEGF. After exposure of neuronal and endothelial cells to VEGF in vitro, we observed increased expression of SDF-1α under normoxic conditions by immunofluorescence and flow cytometry. Taken together, these results implicate the chemokine VEGF in modulating SDF-1α expression in neurons and blood vessels that form the basis of two of the secondary structures of Scherer.
Our hypothesis that the chemoattractant SDF-1α mobilizes the migration of CXCR4-expressing tumor cells toward the pre-existing structures requires that SDF-1α be present in neurons and blood vessels before the tumor cells arrive. In support of this argument, we observed several GBM cases in which SDF-1α-immunopositive neurons and blood vessels were present without CXCR4-immunopositive tumor cells surrounding them. This observation may suggest that some neurons and blood vessels express SDF-1α before colonization by CXCR4-positive tumor cells to form the secondary structures known as perineuronal or perivascular satellitosis.
Previous studies by our group and others have shown that hypoxia stimulates glioma cell migration in vitro and invasion in orthotopic glioma models and human gliomas in vivo.5,37,38,39 However, the mechanisms of the hypoxia-mediated invasion in gliomas have not been fully elucidated. We and others have shown that SDF-1α and CXCR4 are up-regulated in GBMs in vivo.14,15,36,40,41 Our results, as well as the findings of several others, suggest that the hypoxia-induced up-regulation of CXCR4 in glioma cells and their responsiveness to a SDF-1α gradient can potentiate glioma cell invasion.5,36,41 For example, CXCR4-expressing human and rodent glioma cells demonstrate enhanced invasive capacity as compared to low-CXCR4-expressing tumor cells in vitro and in vivo.41
Although CXCR4 plays a major role in cell migration, the effects of hypoxia on migration that we observed in glioma cells suggested that CXCR4 expression was only partially responsible for increased chemotaxis. Hypoxia increased migration of both the low-CXCR4-expressing (LN229) and high-CXCR4-expressing (LN308 and U87MG) glioma cells. However, only the LN308 and U87MG (CXCR4hi) glioma cells migrated in response to a SDF-1α gradient. The role of CXCR4 in migration was further supported by blocking CXCR4 expression. Under our conditions, migration of glioma cells was only partially inhibited indicating the possibility of a CXCR4-independent component of hypoxia-induced migration. For example, hypoxia up-regulates the receptor c-met in glioma cells and the hepatocyte growth factor/c-met pathway is well-known to play an essential role in glioma cell invasion.42 Other factors have been reported at the edge of GBMs. For example, stem cell factor, a hypoxia-induced ligand43 is overexpressed by neurons in regions of the brain infiltrated by glioma cells.44 Stem cell factor induces release of SDF-1α45 and chemotaxis of cells bearing its cognate receptor, c-kit.46 Although our results suggest an important role for the SDF-1α/CXCR4 axis and VEGF in the formation of the secondary structures of Scherer, they do not exclude other pathways, potentially hypoxia-induced, that may also play a role in the formation of these structures.
There is abundant experimental evidence that SDF-1α stimulates the invasion of CXCR4-positive malignant cells including ovarian,47 prostate,48 and small cell lung cancer cells.49 SDF-1α/CXCR4 interaction has also been shown to play a role in the growth pattern of optic pathway glioma.18 CXCR4 is expressed in several embryonic cell types that all follow a SDF-1α gradient, including pluripotent stem cells and neural stem cells.50,51 In addition, CXCR4-expressing neural stem cells, cancer stem cells, pluripotent stem cells, and glioma cells sense and migrate toward a SDF-1α gradient.35,50,51,52 Migration of CXCR4-expressing cells can be blocked by an anti-CXCR4 monoclonal neutralizing antibody41,53 or by the CXCR4 inhibitor, AMD3100.12 For example, the ability of neural stem cells to target established tumors in the brain can be inhibited by neutralization of CXCR4.52 The effect of a SDF-1α gradient on glioma cells documented in this study suggests that the invasive behavior of the glioma cells may correspond to the reacquisition of primitive migratory behavior occurring during central nervous system embryogenesis.
Treatment of LN308 glioma cells with SDF-1α enhanced FAK, AKT, and ERK1/2 phosphorylation. Although similar results have been reported in a variety of other cancers,23,50,54 to our knowledge, SDF-1α-induced phosphorylation of FAK in glioma cells has not been previously reported. Phosphorylated FAK,27 AKT,55 and ERK1/256 play a critical role in glioma cell migration. Thus, SDF-1α secretion from pre-existing structures would induce phosphorylation of these signaling molecules in CXCR4-expressing glioma cells promoting invasion. Other molecular events have been linked to the effects of SDF-1α on cellular migration, such as increased actin polymerization required for the formation of stress fiber and pseudopodia that are implicated in cell migration and invasion.57 Taken together, these findings suggest that SDF-1α/CXCR4 ligand receptor system activates signaling pathways and cellular events known to be essential for invasion of glioma cells.
This study demonstrates that i) in the human brain, CXCR4-immunoreactive invading glioma cells were closely organized around and along SDF-1α-positive structures; ii) in the mouse brain invading GL261 murine glioma cells expressing CXCR4 mRNA were closely associated with neurons, blood vessels, and subpial regions that expressed SDF-1α mRNA in the brain adjacent to the tumor; iii) VEGF up-regulated SDF-1α expression in human neuronal and endothelial cells; iv) CXCR4-high-expressing glioma cells, but not CXCR4-low-expressing glioma cells, responded to a SDF-1α gradient, a response enhanced by hypoxia; v) AMD3100, a selective CXCR4 inhibitor, or the use of a knockdown approach directed against CXCR4 decreased the migration of CXCR4-high-expressing glioma cells toward a SDF-1α gradient; and vi) SDF-1α treatment of glioma cells increased phosphorylation of FAK, AKT, and ERK1/2, which was decreased by AMD3100. Taken together, our studies suggest a biological-based mechanism that may help explain the formation of the secondary structures of Scherer. Based on our results, we propose that the hypoxia-induced expression of functional CXCR4 by glioma cells together with the VEGF-induced SDF-1α up-regulation in pre-existing structures at the edge of GBMs may represent one molecular basis for the well-described secondary structures of Scherer. Moreover, our results suggest that inhibiting the SDF-1α/CXCR4 interactions by small molecule antagonists, such as AMD3100, could potentially control the invasive behavior of glioma cells, decrease tumor spread into vital regions of the brain, and improve the prognosis of patients with malignant gliomas.
Footnotes
Address reprint requests to David Zagzag, M.D., Ph.D., Department of Pathology, MSB-130, New York University School of Medicine, 550 First Ave., New York, NY 10016. E-mail: dz4@nyu.edu.
Supported by grants to D.Z. from the National Institutes of Health (grant R01 CA100426) and the Goldhirsh Foundation.
References
- Scherer HJ. Structural development in gliomas. Am J Cancer. 1938;34:333–351. [Google Scholar]
- Burger PC, Scheithauer BW, Vogel FS. Burger PC, Scheithauer BW, Vogel FS, editors. New York: Churchill Livingstone,; Surgical Pathology of the Nervous System and Its Coverings, (ed 4) 2002:p 225. [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. Lyon: International Agency for Research on Cancer,; WHO Classification of Tumours of the Central Nervous System, (ed 4) 2007 [Google Scholar]
- Zagzag D, Krishnamachary B, Yee H, Okuyama H, Chiriboga L, Ali MA, Melamed J, Semenza GL. Stromal cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Res. 2005;65:6178–6188. doi: 10.1158/0008-5472.CAN-04-4406. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Lukyanov Y, Lan L, Ali MA, Esencay M, Mendez O, Yee H, Voura EB, Newcomb EW. Hypoxia-inducible factor-1 and VEGF upregulate CXCR4 in glioblastoma: implications for angiogenesis and glioma cell invasion. Lab Invest. 2006;86:1221–1232. doi: 10.1038/labinvest.3700482. [DOI] [PubMed] [Google Scholar]
- Plate KH. Mechanisms of angiogenesis in the brain. J Neuropathol Exp Neurol. 1999;58:313–320. doi: 10.1097/00005072-199904000-00001. [DOI] [PubMed] [Google Scholar]
- Skalli O, Ropraz P, Tzociak A, Benzonano G, Gillessen D, Gabbiani G. A mAb against alpha-smooth muscle actin. A new probe for smooth muscle differentiation. J Cell Biol. 1986;103:22787–22796. doi: 10.1083/jcb.103.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagzag D, Hooper A, Friedlander DR, Chan W, Holash J, Wiegand SJ, Yancopoulos GD, Grumet M. In situ expression of angiopoietins in astrocytomas identifies angiopoietin-2 as an early marker of tumor angiogenesis. Exp Neurol. 1999;159:391–400. doi: 10.1006/exnr.1999.7162. [DOI] [PubMed] [Google Scholar]
- Wolf HK, Buslei R, Schmidt-Kastner R, Schmidt-Kastner PK, Pietsch T, Wiestler OD, Blumcke I. NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 1996;44:1167–1171. doi: 10.1177/44.10.8813082. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Miller DC, Chiriboga L, Yee H, Newcomb EW. Green fluorescent protein immunohistochemistry as a novel experimental tool for the detection of glioma cell invasion in vivo. Brain Pathol. 2003;13:34–37. doi: 10.1111/j.1750-3639.2003.tb00004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- Liao W, Nguyen MT, Imamura T, Singer O, Verma IM, Olefsky JM. Lentiviral short hairpin ribonucleic acid-mediated knockdown of GLUT4 in 3T3–L1 adipocytes. Endocrinology. 2006;147:2245–2252. doi: 10.1210/en.2005-1638. [DOI] [PubMed] [Google Scholar]
- Rempel SA, Dudas S, Ge S, Gutiérrez JA. Identification and localization of the cytokine SDF1 and its receptor, CXC chemokine receptor 4, to regions of necrosis and angiogenesis in human glioblastoma. Clin Cancer Res. 2000;6:102–111. [PubMed] [Google Scholar]
- Woerner BM, Warrington NM, Kung AL, Perry A, Rubin JB. Widespread CXCR4 activation in astrocytomas revealed by phospho-CXCR4-specific antibodies. Cancer Res. 2005;65:11392–11399. doi: 10.1158/0008-5472.CAN-05-0847. [DOI] [PubMed] [Google Scholar]
- Sun YX, Wang J, Shelburne CE, Lopatin DE, Chinnaiyan AM, Rubin MA, Pienta KJ, Taichman RS. Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem. 2003;89:462–473. doi: 10.1002/jcb.10522. [DOI] [PubMed] [Google Scholar]
- Kato M, Kitayama J, Kazama S, Nagawa H. Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res. 2003;5:144–150. doi: 10.1186/bcr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SU, Bae JW, Kim CH, Lee JB, Koo BW. A significant correlation between nuclear CXCR4 expression and axillary lymph node metastasis in hormonal receptor negative breast cancer. Ann Surg Oncol. 2008;15:281–285. doi: 10.1245/s10434-007-9595-1. [DOI] [PubMed] [Google Scholar]
- Warrington NM, Woerner BM, Daginakatte GC, Dasgupta B, Perry A, Gutmann DH, Rubin JB. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res. 2007;67:8588–8595. doi: 10.1158/0008-5472.CAN-06-2220. [DOI] [PubMed] [Google Scholar]
- Stumm RK, Rummel J, Junker V, Culmsee C, Pfeiffer M, Krieglstein J, Höllt V, Schulz S. A dual role for the SDF-1/CXCR4 chemokine receptor system in adult brain: isoform-selective regulation of SDF-1 expression modulates CXCR4-dependent neuronal plasticity and cerebral leukocyte recruitment after focal ischemia. J Neurosci. 2002;22:5865–5878. doi: 10.1523/JNEUROSCI.22-14-05865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Wang CE, Goffinet AM. Expression of the chemokine receptor CXCR4 mRNA during mouse brain development. Brain Res Dev Brain Res. 2004;149:63–71. doi: 10.1016/j.devbrainres.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol. 2007;28:299–307. doi: 10.1016/j.it.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Larsen PH, Hao C, Yong VW. CXCR4 is a major chemokine receptor on glioma cells and mediates their survival. J Biol Chem. 2002;277:49481–49487. doi: 10.1074/jbc.M206222200. [DOI] [PubMed] [Google Scholar]
- Newcomb EW, Lukyanov Y, Schnee T, Esencay M, Fischer I, Hong D, Shao Y, Zagzag D. The geldanamycin analogue 17-allylamino-17-demethoxygeldanamycin inhibits the growth of GL261 glioma cells in vitro and in vivo. Anticancer Drugs. 2007;18:875–882. doi: 10.1097/CAD.0b013e3281430df8. [DOI] [PubMed] [Google Scholar]
- Newcomb EW, Tamasdan C, Entzminger Y, Alonso J, Friedlander D, Crisan D, Miller DC, Zagzag D. Flavopiridol induces mitochondrial-mediated apoptosis in murine glioma GL261 cells via release of cytochrome c and apoptosis inducing factor. Cell Cycle. 2003;2:243–250. [PubMed] [Google Scholar]
- Zagzag D, Friedlander DR, Margolis B, Grumet M, Semenza GL, Zhong H, Simons JW, Holash J, Wiegand SJ, Yancopoulos GD. Molecular events implicated in brain tumor angiogenesis and invasion. Pediatr Neurosurg. 2000;33:49–55. doi: 10.1159/000028975. [DOI] [PubMed] [Google Scholar]
- Cox BD, Natarajan M, Stettner MR, Gladson CL. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J Cell Biochem. 2006;99:35–52. doi: 10.1002/jcb.20956. [DOI] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Schimanski CC, Bahre R, Gockel I, Müller A, Frerichs K, Hörner V, Teufel A, Simiantonaki N, Biesterfeld S, Wehler T, Schuler M, Achenbach T, Junginger T, Galle PR, Moehler M. Dissemination of hepatocellular carcinoma is mediated via chemokine receptor CXCR4. Br J Cancer. 2006;95:210–217. doi: 10.1038/sj.bjc.6603251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski CC, Schwald S, Simiantonaki N, Jayasinghe C, Gönner U, Wilsberg V, Junginger T, Berger MR, Galle PR, Moehler M. Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res. 2005;11:1743–1750. doi: 10.1158/1078-0432.CCR-04-1195. [DOI] [PubMed] [Google Scholar]
- Scala S, Giuliano P, Ascierto PA, Ieranò C, Franco R, Napolitano M, Ottaiano A, Lombardi ML, Luongo M, Simeone E, Castiglia D, Mauro F, De Michele I, Calemma R, Botti G, Caracò C, Nicoletti G, Satriano RA, Castello G. Human melanoma metastases express functional CXCR4. Clin Cancer Res. 2006;12:2427–2433. doi: 10.1158/1078-0432.CCR-05-1940. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Hosotani R, Miyamoto Y, Ida J, Tsuji S, Nakajima S, Kawaguchi M, Kobayashi H, Doi R, Hori T, Fujii N, Imamura M. Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: a possible role for tumor progression. Clin Cancer Res. 2000;9:3530–3535. [PubMed] [Google Scholar]
- Yang YC, Lee ZY, Wu CC, Chen TC, Chang CL, Chen CP. CXCR4 expression is associated with pelvic lymph node metastasis in cervical adenocarcinoma. Int J Gynecol Cancer. 2007;17:676–686. doi: 10.1111/j.1525-1438.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Lu WG, Ye F, Chen HZ, Zhou CY, Xie X. Study on CXCR4/SDF-1alpha axis in lymph node metastasis of cervical squamous cell carcinoma. Int J Gynecol Cancer. 2007;17:478–483. doi: 10.1111/j.1525-1438.2007.00786.x. [DOI] [PubMed] [Google Scholar]
- Grunewald M, Avraham I, Dor Y, Bachar-Lustig E, Itin A, Jung S, Chimenti S, Landsman L, Abramovitch R, Keshet E. VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell. 2006;124:175–189. doi: 10.1016/j.cell.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Hong X, Jiang F, Kalkanis SN, Zhang ZG, Zhang XP, DeCarvalho AC, Katakowski M, Bobbitt K, Mikkelsen T, Chopp M. SDF-1 and CXCR4 are up-regulated by VEGF and contribute to glioma cell invasion. Cancer Lett. 2006;236:39–45. doi: 10.1016/j.canlet.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1α in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–2618. [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Brat DJ, Castellano-Sanchez AA, Hunter SB, Pecot M, Cohen C, Hammond EH, Devi SN, Kaur B, Van Meir EG. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64:920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- Barbero S, Bonavia R, Bajetto A, Porcile C, Pirani P, Ravetti JL, Zona GL, Spaziante R, Florio T, Schettini G. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003;63:1969–1974. [PubMed] [Google Scholar]
- Ehtesham M, Winston JA, Kabos P, Thompson RC. CXCR4 expression mediates glioma cell invasiveness. Oncogene. 2006;25:2801–2806. doi: 10.1038/sj.onc.1209302. [DOI] [PubMed] [Google Scholar]
- Eckerich C, Zapf S, Fillbrandt R, Loges S, Westphal M, Lamszus K. Hypoxia can induce c-Met expression in glioma cells and enhance SF/HGF-induced cell migration. Int J Cancer. 2007;121:276–283. doi: 10.1002/ijc.22679. [DOI] [PubMed] [Google Scholar]
- Jin DK, Shido K, Kopp HG, Petit I, Shmelkov SV, Young LM, Hooper AT, Amano H, Avecilla ST, Heissig B, Hattori K, Zhang F, Hicklin DJ, Wu Y, Zhu Z, Dunn A, Salari H, Werb Z, Hackett NR, Crystal RG, Lyden D, Rafii S. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat Med. 2006;12:557–567. doi: 10.1038/nm1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, Crystal RG, Besmer P, Lyden D, Moore MA, Werb Z, Rafii S. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;10:625–637. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lee J, Fine HA. Neuronally expressed stem cell factor induces neural stem cell migration to areas of brain injury. J Clin Invest. 2004;113:1364–1374. doi: 10.1172/JCI20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotton CJ, Wilson JL, Scott K, Stamp G, Wilbanks GD, Fricker S, Bridger G, Balkwill FR. Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Cancer Res. 2002;2:5930–5938. [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;2:1832–1837. [PubMed] [Google Scholar]
- Kijima T, Maulik G, Ma PC, Tibaldi EV, Turner RE, Rollins B, Sattler M, Johnson BE, Salgia R. Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c-Kit in small cell lung cancer cells. Cancer Res. 2002;2:6304–6311. [PubMed] [Google Scholar]
- Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- Ehtesham M, Yuan X, Kabos P, Chung NH, Liu G, Akasaki Y, Black KL, Yu JS. Glioma tropic neural stem cells consist of astrocytic precursors and their migratory capacity is mediated by CXCR4. Neoplasia. 2004;6:287–293. doi: 10.1593/neo.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissinotto E, Cavalloni G, Leone F, Fonsato V, Mitola S, Grignani G, Surrenti N, Sangiolo D, Bussolino F, Piacibello W, Aglietta M. Involvement of chemokine receptor 4/stromal cell-derived factor 1 system during osteosarcoma tumor progression. Clin Cancer Res. 2005;11:490–497. [PubMed] [Google Scholar]
- Sutton A, Friand V, Brulé-Donneger S, Chaigneau T, Ziol M, Sainte-Catherine O, Poiré A, Saffar L, Kraemer M, Vassy J, Nahon P, Salzmann JL, Gattegno L, Charnaux N. Stromal cell-derived factor-1/chemokine (C-X-C motif) ligand 12 stimulates human hepatoma cell growth, migration, and invasion. Mol Cancer Res. 2007;5:21–33. doi: 10.1158/1541-7786.MCR-06-0103. [DOI] [PubMed] [Google Scholar]
- Chandrasekar N, Mohanam S, Gujrati M, Olivero WC, Dinh DH, Rao JS. Downregulation of uPA inhibits migration and PI3k/Akt signaling in glioblastoma cells. Oncogene. 2003;22:392–400. doi: 10.1038/sj.onc.1206164. [DOI] [PubMed] [Google Scholar]
- Lakka SS, Jasti SL, Gondi C, Boyd D, Chandrasekar N, Dinh DH, Olivero WC, Gujrati M, Rao JS. Downregulation of MMP-9 in ERK-mutated stable transfectants inhibits glioma invasion in vitro. Oncogene. 2002;21:5601–5608. doi: 10.1038/sj.onc.1205646. [DOI] [PubMed] [Google Scholar]
- Mori T, Doi R, Koizumi M, Toyoda E, Ito D, Kami K, Masui T, Fujimoto K, Tamamura H, Hiramatsu K, Fujii N, Imamura M. CXCR4 antagonist inhibits stromal cell-derived factor 1-induced migration and invasion of human pancreatic cancer. Mol Cancer Ther. 2004;3:29–37. [PubMed] [Google Scholar]