Abstract
Xath3 encodes a Xenopus neuronal-specific basic helix–loop–helix transcription factor related to the Drosophila proneural factor atonal. We show here that Xath3 acts downstream of X-ngnr-1 during neuronal differentiation in the neural plate and retina and that its expression and activity are modulated by Notch signaling. X-ngnr-1 activates Xath3 and NeuroD by different mechanisms, and the latter two genes crossactivate each other. In the ectoderm, X-ngnr-1 and Xath3 have similar activities, inducing ectopic sensory neurons. Among the sensory-specific markers tested, only those that label cranial neurons were found to be ectopically activated. By contrast, in the retina, X-ngnr-1 and Xath3 overexpression promote the development of overlapping but distinct subtypes of retinal neurons. Together, these data suggest that X-ngnr-1 and Xath3 regulate successive stages of early neuronal differentiation and that, in addition to their general proneural properties, they may contribute, in a context-dependent manner, to some aspect of neuronal identity.
Neurogenesis in vertebrates is primarily controlled by the actions of basic helix–loop–helix (bHLH) proneural factors, homologues of the Drosophila atonal and achaete-scute complex genes. Many of the vertebrate bHLH proneural genes are expressed in specific subsets of neurons. Mash1 and ngn1/2 are complementarily expressed in the developing central nervous system and in distinct sublineages of the peripheral nervous system (1), ngn1 and ngn2 are transiently expressed in different subsets of cranial sensory ganglia (2), and Xath5 is expressed only in retinal precursors and olfactory placodes (3). Moreover, when expressed within the same subset of neurons, these bHLH genes are often activated at distinct stages during neurogenesis. In Xenopus, X-ngnr-1 expression precedes that of NeuroD in primary neurons (4). Similarly, Mash1, Math4C/neurogenin1, and NeuroD genes mark distinct stages of development of mouse olfactory neuron precursors (5). Overexpression experiments in Xenopus and loss-of-function analysis in mouse have confirmed these bHLH gene expression cascades. Overexpression of X-ngnr-1 promotes ectopic NeuroD expression, but not reciprocally (4). In Mash1 mutant embryos, Math4C/neurogenin1 and NeuroD fail to be expressed in some developing olfactory neurons (5).
A role for bHLH proteins as determinant of neuronal identity has been described in Drosophila, where the genes achaete and scute confer the competence to form external sense organs, while the gene atonal provides the competence to form chordotonal organs and photoreceptors (6). Whether in vertebrates the different proneural genes are also involved in specifying different neural subtypes has only recently received attention. Evidence has been obtained in the mouse by the analysis of specific bHLH knockout mutants. Mice lacking Mash1 product have a loss of olfactory and autonomic neurons as well as a delay in differentiation of retinal neurons (7). Knockouts of ngn1, ngn2, and Math1 show that these genes are essential for the development of different subsets of neurons (8–10). In chick, misexpression of ngns biases premigratory crest cells to the dorsal root sensory ganglia (11). ngn1 in zebrafish has also been suggested to induce ectopic neurons of dorsal character, based on a single lateral marker (12). In Xenopus, many of the bHLH genes have been characterized as capable of promoting ectopic neuronal differentiation in the ectoderm (3, 4, 13–16). However, little is known about the identity of these ectopic neurons and the extent to which these factors specify neuronal subtypes. Ectopic neurons induced by X-ngnr-1 do not respond to glutamate and do not form functional synapses with myocytes, suggesting that those cells are either Rohon–Beard-like sensory neurons or are not fully differentiated (17). Misexpression of Xath5 in the retina has shown that it can promote retinal ganglion cell fate (3), but it is not known whether other bHLH factors would have a similar effect.
In an attempt to define the role of the recently identified bHLH factor Xath3 (16) during neurogenesis, we have first positioned it in the proneural genetic cascade. Our results indicate that Xath3 acts downstream of X-ngnr-1 and that its expression and function are sensitive to Notch inhibition. To investigate potential differences between Xath3 and the other Xenopus bHLH proneural factors, we compared its inducing properties in relation to X-ngnr-1. When ectopically expressed in the nonneural ectoderm of neurula embryos, Xath3 and X-ngnr-1 are able to induce similarly sensory neurons. In contrast, in the retina, X-ngnr-1 and Xath3 overexpression promotes the development of distinct subtypes of retinal neurons. Together, these data suggest that these proneural genes may possess intrinsic functional differences, allowing them to play a role, in a context-dependent manner, in the differentiation of distinct neuron subtypes.
Materials and Methods
Microinjection of mRNA and Animal Caps.
Capped X-ngnr-1 (4), Xath3 (16), X-Delta-1Stu (18), X-NotchICD (19), X-MyT1 (20), and NLS LacZ (18) RNAs were prepared with a Message Machine kit (Ambion, Austin, TX). RNAs (5 nl) were injected on one side at the two-cell stage at 50–100 pg/nl. Caps were dissected at blastula stage. To block protein synthesis, explants were cultivated in the presence of cycloheximide (CHX) at 10 μg/ml. Dexamethasone (dex) was added at 10 μM.
In Vivo Lipofection.
The coding region of Xath3 cDNA was subcloned into EcoRI of CS2 vector (14). CS2 Xath3 or CS2 X-ngnr-1 (4) DNA was transfected into the presumptive retina as described (3). CS2 green fluorescent protein (GFP) DNA was cotransfected to mark transfected cells. Embryos were fixed at stage 40 and cryostat-sectioned (10 μm). GFP-positive cells were counted, and cell types were identified based on their laminar position and morphology (3).
In Situ Hybridization.
Whole-mount in situ hybridizations were performed as described (20) with digoxigenin-labeled antisense RNA probes generated from the X-ngnr-1 (4), Xash3 (14), NeuroD (15), Xath3 (16), N-tubulin (18), X-Delta-1 (18), X-MyT1 (20), Xiro3 (21), Xcoe2 (22), Xlim3 (23), VegT (24), XHox11L2 (25), and Pax2 (26) clones. The Xaml (27) and HB9 (28) probes were generated from a cloned PCR fragment (primer sequences are available on request). Double in situ hybridizations were performed as described (3).
Immunohistochemistry.
Immunohistochemistry was performed as described (3). To stain ganglion, rod, and cone cells, we used an anti-islet, an anti-calbindin (Sigma), and an anti-rhodopsin (R2–12N) Ab, respectively; as a neurofilament marker, we used the Ab RMO 270.7.
RNase Protection Assay.
The coding region of X-ngnr-1 was amplified by PCR. Products were digested with BamHI and NotI and inserted in frame to the ligand-binding domain of the human glucocorticoid receptor (GR) into the BamHI and NotI sites of the p3′HGR vector (constructed by K. Ryan, Wellcome/Cancer Research Campaign, Cambridge, U.K.). RNAs encoding X-ngnr-1-GR were injected at the two-cell stage in the animal region of each blastomere. Eight caps were pooled for each assay. RNA was prepared with the phenol/NETS method. RNase protection templates for NeuroD and Xath3 were generated by PstI and NdeI digestion, respectively, and the antisense probes were synthesized with T7 polymerase. The protected NeuroD and Xath3 fragments are 339 and 147 bases long, respectively.
Results
Xath3 Acts Downstream of X-ngnr-1 During Both Primary and Retinal Neurogenesis.
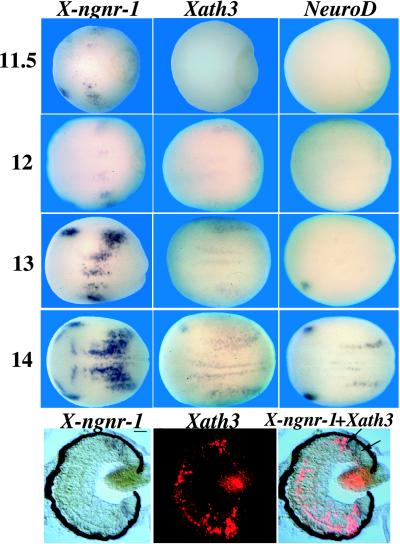
The bHLH factors Xath3, X-ngnr-1, and NeuroD are expressed during primary neurogenesis (4, 15, 16). To compare their timing of expression, we performed in situ hybridizations at different stages (Fig. 1 Upper). At stage 11.5, patches of X-ngnr-1 expression are observed corresponding to the prospective primary neurons and the trigeminal placodes, while no NeuroD and Xath3 expression is yet detected. Xath3 expression can be detected weakly within the domains of X-ngnr-1 expression, starting from stage 12. NeuroD expression can only be observed starting from stage 13, the strongest expression being detected in the trigeminal placodes. At stage 14, X-ngnr-1 expression can also be detected in a group of cells associated with the olfactory placodes and in transverse stripes of cells in anterior parts of the neural plate. No Xath3 and NeuroD expression is detected in these territories at that stage. In the neural tube of stage-20 embryos, Xath3, like NeuroD, is expressed in the lateral part of the spinal cord, while X-ngnr-1 expression is detected in the ventricular layer (data not shown). In the ciliary marginal zone (CMZ) of the retina, X-ngnr-1 starts to be expressed closer to the periphery than Xath3, which appears at the same level as NeuroD (Fig. 1 Lower, and ref. 29). Thus, expression of Xath3 follows X-ngnr-1 expression, but slightly precedes NeuroD during primary neurogenesis. In the neural tube and in the CMZ of the retina, Xath3 also follows X-ngnr-1 expression, but Xath3 and NeuroD have similar expression characteristics, suggesting that they function at closely related stages of neuronal differentiation.
Figure 1.
Sequential expression of X-ngnr-1, Xath3, and NeuroD during primary and retinal neurogenesis. (Upper) Whole-mount in situ hybridizations on stage-11.5, -12, -13, and -14 embryos with probes as indicated on the top of each column. (Lower) Double in situ hybridizations on sections of stage-40 Xenopus retina with X-ngnr-1 (blue) and Xath3 (red) probes. The three panels show the same section. Arrows indicate the positions where X-ngnr-1 and Xath3 expressions appear in the CMZ. (Scale bar = 20 μm.)
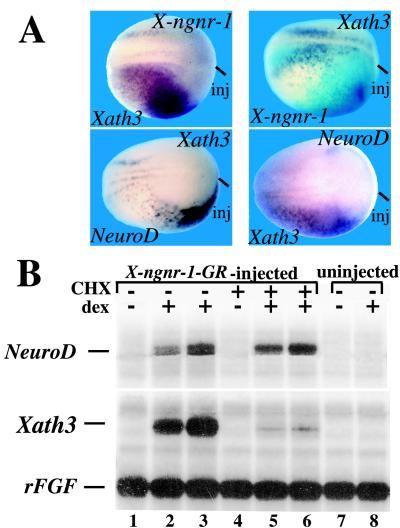
To determine whether Xath3 expression could be regulated by X-ngnr-1, we unilaterally injected X-ngnr-1 mRNA into two-cell stage embryos and analyzed its effects on Xath3 expression (Fig. 2A). Overexpression of X-ngnr-1 mRNA activates ectopic expression of Xath3 (44/50 embryos). By contrast, injection of Xath3 mRNA did not induce the expression of X-ngnr-1 (59/59 embryos; Fig. 2A). Together, these results suggest that Xath3 is a downstream target of X-ngnr-1.
Figure 2.
Xath3 participates in a network of bHLH genes. (A) Embryos were injected with the indicated RNA (top of each panel). Dorsal views of stage-14 embryos probed by in situ hybridization as indicated (bottom of each panel) are shown. The injected side is indicated (inj) or visualized by light blue staining for LacZ expression. (B) Embryos were injected with RNA encoding a X-ngnr-1-GR fusion protein at the two-cell stage. Blastula animal caps were isolated from injected embryos and were left untreated (lanes 1–3), or were incubated in the presence of CHX (lanes 4–6). After 1 hr of culture, dex was added for 2 (lanes 2 and 5) or 3 (lanes 3 and 6) hr. RNA was prepared and analyzed by RNase protection. RNAs isolated from caps derived from uninjected embryos plus or minus dex are shown in lanes 7 and 8. The addition of CHX did not block the activation of NeuroD expression, but, in contrast, abolished Xath3 expression induced by X-ngnr-1-GR (compare lanes 2 and 3 with 5 and 6). rFGF, fibroblast growth factor receptor.
To further analyze the relationship between X-ngnr-1 and the two downstream genes, Xath3 and NeuroD, we constructed a glucocorticoid-inducible X-ngnr-1 fusion protein by inserting the ligand-binding domain of the GR into the X-ngnr-1-coding region. We first asked whether the fusion protein elicited hormone-dependent ectopic neurogenesis. Embryos were injected with X-ngnr-1-GR mRNA. Blastula animal caps from X-ngnr-1-GR-injected embryos were incubated with or without dex at stage 11 for 2 hr and tested for Xath3 and NeuroD expressions by an RNase protection assay (Fig. 2B). Without dex, animal caps did not show any activation of Xath3 and NeuroD expressions. In contrast, when the caps were treated with dex, we observed a strong activation of the Xath3 and NeuroD genes. Thus, the X-ngnr-1-GR fusion protein could efficiently activate neurogenesis in animal caps, and the activity of the protein was regulated by dex. We next asked whether Xath3 and NeuroD could be activated by X-ngnr-1-GR in the absence of protein synthesis. X-ngnr-1-GR-injected animal caps were preincubated in the presence of CHX, which inhibits protein synthesis, for 1 hr and then treated with CHX plus or minus dex for 2–3 hr. Control caps showed that this preincubation with CHX before induction with dex reduced protein synthesis approximately 3.5-fold, as measured by incorporation of [35S]methionine into acid-insoluble material. We found that incubation in CHX plus dex inhibits Xath3 mRNA accumulation. In contrast, NeuroD was still strongly activated (Fig. 2B). These results suggest that NeuroD and Xath3 are differentially regulated by X-ngnr-1.
Because both NeuroD and Xath3 appear to act downstream of X-ngnr-1, we tested whether they regulate each other's expression in embryos injected unilaterally at the two-cell stage (Fig. 2A). Overexpression of NeuroD mRNA caused the induction of Xath3 expression (21/27 embryos). Similarly, injection of Xath3 caused expansion of NeuroD expression (55/60 embryos). Thus, NeuroD and Xath3 crossactivate each other's expression.
Xath3 Induces X-Delta-1 and Is Regulated by Lateral Inhibition.
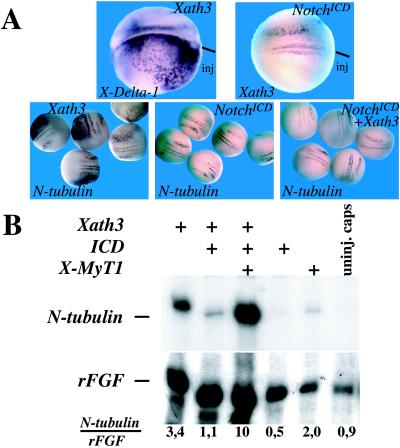
X-ngnr-1 promotes lateral inhibition by inducing X-Delta-1 expression (4). To determine whether Xath3 plays a role in the induction or maintenance of X-Delta-1 expression, which starts from stage 12 onwards, we overexpressed Xath3 in embryos, as described above, and analyzed them for X-Delta-1 expression by in situ hybridization. In 38/41 cases, we saw an induction of X-Delta-1 expression on the injected side, indicating that Xath3 is indeed an activator of X-Delta-1 (Fig. 3A). We then asked whether Notch signaling has an inhibitory effect on Xath3 (Fig. 3A). We found that overexpression of mRNAs encoding a dominant-active form of X-Notch-1 (X-NotchICD; ref. 19), which blocks X-ngnr-1 expression, also represses Xath3 expression (25/30 embryos). Coinjection of X-NotchICD mRNA together with Xath3 mRNAs prevented Xath3 from inducing ectopic expression of the neuronal marker N-tubulin: none of 32 Xath3/X-NotchICD-injected embryos showed ectopic tubulin expression, whereas 66/70 embryos injected with Xath3 alone showed ectopic N-tubulin staining (Fig. 3A). Finally, we wanted to know whether the zinc finger transcription factor X-MyT1, which starts to be expressed at about the same time as Xath3 and has been shown to synergize with the earlier bHLH factors X-ngnr-1 and Xash3 to induce neuronal differentiation and to allow cells to escape lateral inhibition (20), could also cooperate with Xath3 in the presence of X-NotchICD. As observed in embryos, overexpression of Xath3 alone induces N-tubulin expression in animal caps, although at much lower levels compared with NeuroD or X-ngnr-1, and this induction is inhibited by coinjection of X-Notch ICD (Fig. 3B, and data not shown). Addition of X-MyT1, which by itself has only a weak inductive effect on N-tubulin expression, strongly increases the ability of Xath3 to induce N-tubulin expression (Fig. 3B). Thus, Xath3 can cooperate with X-MyT1 to induce neuronal differentiation, and its expression and function, like that of X-ngnr-1, are inhibited by the Notch pathway.
Figure 3.
Relationships between Xath3 and lateral inhibition. (A) Embryos were injected with the indicated RNA (top of each panel). Dorsal views of stage-14 embryos probed by in situ hybridization as indicated (bottom of each panel) are shown. (B) X-MyT1 can function with Xath3 to stimulate N-tubulin expression in the presence of Notch signaling. Animal caps were derived from embryos injected with RNA as indicated and analyzed for N-tubulin expression by RNase protection. rFGF, fibroblast growth factor receptor. Numbers below lanes indicate rFGF-normalized N-tubulin level with maximum level detected set to 10.
Xath3- and X-ngnr-1-Misexpressing Cells Have Characteristics of Sensory Neurons.
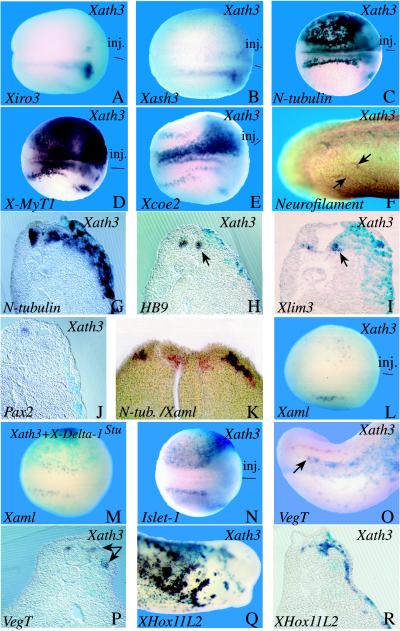
To further analyze the action of Xath3 on neural development, we tested in Xath3-injected embryos the expression of several neural/neuronal markers (Fig. 4). Overexpression of Xath3 inhibits the expression of Xiro3 (14/36 embryos), a homeobox gene that is expressed early in a subset of the posterior neural plate and that defines an early neural precursor state (21) as well as that of Xash3 (14/27 embryos), a bHLH factor that is similarly expressed early in a restricted area of the posterior neural plate and in retinal progenitor cells (14). In contrast, overexpression of Xath3 leads to ectopic activation of the N-tubulin pan-neuronal gene (18) and of the earlier general neuronal markers X-MyT1 (31/31 embryos) and Xcoe2 (28/29 embryos), which are activated at about the same time as Xath3 in prospective primary neurons and cranial ganglia, data not shown, and refs. 20 and 22). Generic markers of late neurons, such as neurofilament (6/9 embryos), were also activated in response to Xath3 overexpression. X-ngnr-1 misexpression also induces ectopic expression of these general neuronal markers but to higher levels than Xath3 misexpression (neurofilament: 7/7 embryos; data not shown, and refs. 4, 21, and 22).
Figure 4.
Misexpression of Xath3 causes ectopic development of distinct types of neurons. Embryos were injected with the indicated RNA (top of each panel). Embryos were analyzed by in situ hybridization with several different markers, as indicated (bottom of each panel), or by immunostaining with an anti-neurofilament marker (F). The injected side is indicated (inj) or visualized by light blue staining for LacZ expression. (A–E) and (L–N) Dorsal views of neurula-stage embryos. (F, O, and Q) Lateral views (F and O, posterior region; Q, anterior region) of tailbud embryos. (G–J, P, and R) Transverse sections of tailbud embryos (stage 30). (K) Double-labeling Xaml (black) and N-tubulin (red) on a section in the posterior part of a stage-15 embryo. Arrows indicate stained ectopic fibers in F, the endogenous signal on the injected side in H, J, and O, and the expanded signal in P.
To address the question of the nature of the ectopically induced neurons, we analyzed in Xath3- or X-ngnr-1-injected embryos the expression of markers for specific subtypes of neurons (Fig. 4). In contrast to the strong ectopic expression of N-tubulin or the generic neuronal markers described above, overexpression of Xath3 did not induce ectopic activation of HB9 (9/9 embryos) and Xlim3 (13/13 embryos), which are expressed specifically in primary motor neurons starting at late neurula/early tailbud stages (28, 23). Similarly, no ectopic expression of HB9 (3/3 embryos) and Xlim3 (14/14 embryos) was detected in X-ngnr-1-injected embryos (data not shown).
To look for induction of interneurons, we used the Pax2 gene, which has been shown to be expressed in interneurons in chick (30). A very similar expression pattern was observed with Xenopus Pax2 (26). Overexpression of neither Xath3 nor X-ngnr-1 was able to induce ectopic Pax2 expression (Xath3, 16 embryos; X-ngnr-1, 10 embryos; Fig. 4, and data not shown). In most embryos, Pax2 expression was even reduced on the injected side, suggesting that Xath3 overexpression may alter the fate of interneuron precursors.
To determine whether Xath3 is able to induce dorsal neural tube-type neurons, we first used the RUNT domain gene Xaml, which is expressed at neurula stage in a lateral stripe of cells in the neural plate (27). To confirm that these Xaml-positive cells are indeed differentiating Rohon–Beard neurons, we first performed double-labeling in situ hybridization. Scattered cells that express Xaml also express N-tubulin and are located only within the lateral N-tubulin-positive stripe (Fig. 4). In addition, as expected for a bona fide neuronal marker, increasing Notch signaling by expressing X-NotchICD (19) inhibited the expression of Xaml (18/20 embryos), whereas injecting embryos with an antimorphic form of X-Delta-1 (X-Delta-1stu), which blocks lateral inhibition (18), increased the density of Xaml-expressing cells (22/38 embryos) (data not shown). As these results indicate that Xaml is indeed an early dorsal-specific neuronal marker, we analyzed its expression in Xath3 and X-ngnr-1 injected embryos. Overexpression of neither of these two factors was able to induce ectopic Xaml expression (Xath3, 44 embryos; X-ngnr-1, 32 embryos). When the injected area corresponded to the lateral neural plate, the expression of Xaml was often more diffuse and partially or totally inhibited (Fig. 4, and data not shown). As overexpression of bHLH proneural genes induces ectopic X-Delta-1 expression, we hypothesized that the inhibition of Xaml expression might be the result of X-Delta-1 activation of lateral inhibition. However, coinjection of X-Delta-1stu with Xath3 (8/8 embryos) or X-ngnr-1 (21/21 embryos) did not rescue the inhibition, showing that this is not the case (Fig. 4, and data not shown). We were also unable to detect in Xath3 or X-ngnr-1 injected embryos any ectopic activation of the VegT gene (24), which starts to be expressed in tailbud embryos in a subset of posterior Rohon–Beard neurons, although some increase of expression was observed in the expanded neural tube (Xath3, 32 embryos; X-ngnr-1, 34 embryos) (Fig. 4, and data not shown). In contrast, overexpression of Xath3 and X-ngnr-1 induces a strong ectopic expression of XHox11L2 (Xath3: 30/32 embryos; X-ngnr-1: 29/33 embryos), which is expressed in the cranial sensory ganglia and the Rohon–Beard neurons (25). Ectopic activation of Islet-1, which is expressed in sensory cranial ganglia and Rohon–Beard cells, but also in motorneurons (ref. 31, and C. Kintner, personal communication), was also ectopically activated in neurula ectoderm (Xath3: 24/24 embryos; X-ngnr-1: 7/8 embryos; Fig. 4, and data not shown). Together, these results suggest that Xath3 and X-ngnr-1 expressing cells have characteristics of sensory neurons.
Xath3 and X-ngnr-1 Promote the Development of Distinct Subtypes of Retinal Neurons.
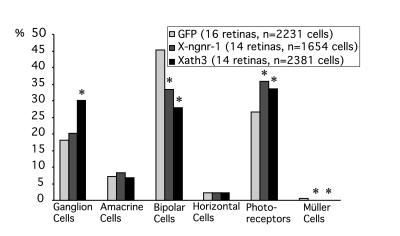
The different cell types of the retina are produced from proliferative precursor neuroepithelial cells at different overlapping periods during retinal neurogenesis. Ganglion, cone photoreceptor, and horizontal cells are born first, followed by amacrine and rod photoreceptor cells, while bipolar and Müller cells are born last (32). To know whether X-ngnr-1 and Xath3 play a specific role in regulating the differentiation of these cells, we targeted their expression to the developing retina by in vivo lipofection at stage 18 (Fig. 5). We found that transfection of Xath3 produced about twice the number of ganglion cells (P < 0.0001) and more photoreceptor cells (P = 0.0022) than control transfection with GFP alone, at the expense of bipolar (P < 0.0001) and Müller glia cells (P = 0.0001). Transfection of X-ngnr-1 also produced more photoreceptor cells (P = 0.001), at the expense of bipolar (P < 0.001) and Müller cells (P = 0.0050), but no change was observed in the number of ganglion cells. We confirmed that the Xath3 and X-ngnr-1 transfected cells observed in the ganglion and photoreceptor cell layers are indeed differentiated ganglion and photoreceptor cells by staining with anti-islet-1 (anti-ganglion cell), anti-calbindin (anti-cone), and anti-rhodopsin (anti-rod) Abs (data not shown).
Figure 5.
Xath3 and X-ngnr-1 lipofections lead to different effects on retinal cell fate decisions. Percentage of retinal cell types generated by GFP alone, or GFP plus Xath3, or GFP plus X-ngnr-1 lipofection. The percentage of each cell type was calculated as a weighted average. The statistical analysis was performed using Student's t tests, except for the Müller cell, for which we used a χ2 test because of the very low number of Müller cells in each sample. *, P < 0.005.
Discussion
We showed here that Xath3 lies downstream of X-ngnr-1 and that Xath3 and NeuroD are differently regulated by X-ngnr-1. Xath3 and X-ngnr-1 induce in ectodermal cells the ectopic expression of markers that label sensory neurons. In the retina, targeted expression of Xath3 and X-ngnr-1 in progenitor cells promotes the development of distinct neuronal subtypes. Together, our data suggest that Xath3 and X-ngnr-1, in addition to their generic neuronal-inducing properties, may contribute, together with additional factors, to the specification of neuronal identity.
Xath3 Is a Target of X-ngnr-1.
During primary neurogenesis Xath3 expression is activated after X-ngnr-1 and before NeuroD. Consistent with our data, NeuroM, a putative homologue of Xath3, is expressed before NeuroD in the chicken developing nervous system (33). However, in the neural tube of tailbud-stage embryos and in the CMZ of the retina, we found no offset of expression between NeuroD and Xath3. We do not know yet whether this reflects a problem in the sensitivity of the technique or whether it reflects a real difference between primary and secondary neurogenesis. Also, Xath3 transcription is activated by X-ngnr-1 but not vice versa. This indicates that Xath3 is one target of X-ngnr-1. Consistent with this, ngn1 and ngn2 are essential for the expression of Math3 in mouse cranial sensory neurons (8, 9). In Xenopus, X-ngnr-1 also activates NeuroD expression in a unidirectional cascade (4). So far, we do not know whether the interactions between X-ngnr-1 and these two targets are direct or not. We showed here that X-ngnr-1 is sufficient to induce NeuroD expression in the absence of protein synthesis, whereas additional factors might be required to induce Xath3 expression. The activation of Xath3 by X-ngnr-1 might therefore be indirect. However, we cannot exclude that Xath3 is a direct target of X-ngnr-1 and that different levels of X-ngnr-1 or X-ngnr-1 plus some cofactors are required to induce it. We also found that Xath3 and NeuroD can activate each other's expression. Whether these bHLH genes crossactivate themselves directly or not has to be further analyzed.
Xath3 Interacts with the Lateral Inhibition Pathway.
X-ngnr-1 activates the expression of X-Delta-1, and X-ngnr-1 expression and function are blocked in the presence of activated X-Notch-1 (4). Our results indicate that Xath3 overexpression also induces X-Delta-1, and that both Xath3 expression and function are inhibited by the X-Notch-1 pathway. We cannot rule out that the NotchICD inhibition of Xath3 expression is indirect because of its inhibition of X-ngnr-1 expression. As Xath3 expression in neurula embryos does not appear before that of X-Delta-1, Xath3 induction of X-Delta-1 expression may correspond to a positive-feedback mechanism. Interestingly, NeuroD function has been reported to be less sensitive to Notch inhibition than Xash3, an earlier expressed bHLH gene (19), suggesting that downstream bHLH factors like NeuroD might be less sensitive to lateral inhibition than are more “upstream” genes. However, in animal cap, by coinjection of increasing amount of NotchICD and testing for N-tubulin induction by an RNase protection assay, we were unable to detect any clear difference in sensitivity between X-ngnr-1, NeuroD, and Xath3 bHLH factors (data not shown). We also found that the zinc-finger transcription factor X-MyT1, which is thought to play a role in the resistance to Notch inhibition (20), can cooperate with all three bHLH factors (this study, and data not shown). How X-MyT1 synergizes with these factors to promote neuronal differentiation remains to be determined.
Xath3 and X-ngnr-1 Overexpression in the Ectoderm Induces the Expression of Pan-Neuronal and Sensory-Specific Genes.
In agreement with previous reports, we found that overexpression of Xath3 and X-ngnr-1 leads to ectopic expression of general neuronal markers (4, 16). As two of the general markers tested, X-MyT1 and Xcoe2, have timing of expression similar to Xath3, their ectopic expression may reflect a possible role for Xath3 in the maintenance of their expression rather than in their activation. X-ngnr-1-misexpressing ectopic neurons, although showing typical action potentials and classic neuronal morphology, are insensitive to glutamate and do not cause miniature endplate currents in contacted myocytes, suggesting that these cells are either incompletely differentiated or lateral/dorsal neurons (17). Our observations that both Xath3 and X-ngnr-1 induce some dorsal but not ventral neuronal markers confirm these previous findings and are also in accordance with results obtained in other species. In zebrafish, ngn-1 misexpression does not induce motorneuron markers, but promotes restricted activation of the Zash1 marker that presumably labels some interneurons (12). Targeted inactivation of ngn-1 and ngn-2 in mouse has indicated that they are required for the generation of different subsets of cranial sensory neurons (8, 9). Ectopic expression of ngns in chicken bias migrating neural crest cells to a sensory neuronal fate (11). Interestingly, Rohon–Beard-specific markers, such as Xaml and VegT, were found not to be ectopically induced, while, in contrast, early (Islet-1) and late (XHox11L2) markers that label both cranial and Rohon–Beard sensory neurons were found to be ectopically expressed. One interpretation of these results may be that Xath3- and X-ngnr-1-misexpressing ectodermal cells follow a pathway more characteristic of cranial sensory neurons. However, as Xaml expression is only transient in Rohon–Beard sensory neurons, one cannot exclude the possibility that Xath3 and X-ngnr-1 push cells through a stage of differentiation where it is down-regulated. Also, as VegT expression is restricted to only a subset of posterior Rohon–Beard sensory neurons, it is also possible that Xath3 and X-ngnr-1 are involved in the differentiation of other classes of Rohon–Beard sensory neurons. In mouse, ngn1 and ngn2 are required for the determination of distinct types of sensory neurons in dorsal root ganglia (34).
Overexpression of ngn in chicken is sufficient to induce sensory markers in mesodermal derivatives (11). No ectopic expression of XHox11L2 and of the pan-neuronal markers was observed in the mesoderm of the injected Xenopus embryos. Whether this lack of effect of Xath3 and X-ngnr-1 in mesodermal tissue reflects a difference in responsiveness of the mesoderm between the two organisms or a difference in the inducing properties of the misexpressed genes remains to be tested.
If Xath3 and X-ngnr-1 can contribute to the specification of a sensory subtype, such contribution must, however, be context-dependent because both Xath3 and X-ngnr-1 are also expressed in ventral regions of the early neural tube. One possibility is that X-ngnr-1 in the neural plate cooperates with other regionally localized factors to give neurons their subtype identity. Sonic hedgehog signaling has been shown to modulate ngn1 activity in zebrafish and to allow development of ectopic motorneurons (12). The homeodomain transcription factor MNR2, which is induced by Sonic Hedgehog, is one such factor that specifies multiple aspects of motorneuron identity (35). As bone morphogenetic proteins (BMPs) have been implicated in the induction of dorsal neurons in the neural tube (36), it is tempting to speculate that X-ngnr-1 activity may also be modulated by BMP signals to give neurons of dorsal character. Among the potential candidates for factors that cooperate with bHLH proteins to give neurons a dorsal phenotype is the XGli3 gene, which is expressed in the lateral neural plate. We have tested whether XGli3 can cooperate with X-ngnr-1 to induce Xaml expression, but were unable to detect any ectopic activation of Xaml expression in XGli3/X-ngnr-1-coinjected embryos (data not shown). Another good candidate is the Pax-3 gene, whose expression is restricted to the posterior dorsal neural tube. Interestingly, overexpression of Pax-3 in ascidians induces ectopic expression of the dorsal neural marker tyrosinase in neurulae (37).
X-ngnr-1 and Xath3 Have Distinct Effects on Retinal Neuron Differentiation.
In lipofection experiments of retinal precursor cells, Xath3 and X-ngnr-1 overexpression affects normal cell-type distribution, producing more early-born cells at the expense of late-born cells. Interestingly, we observed that Xath3 promotes both ganglion and photoreceptor cells, while X-ngnr-1 promotes only photoreceptor cells. A bias toward ganglion cells but without increase in photoreceptors has been previously observed with Xath5 retinal transfections (3). These results suggest that Xath3, Xath5, and X-ngnr-1 may have intrinsic differences and play a role in the specification of neuronal identity. Alternatively, the ability of these bHLH factors to promote the differentiation of early-born cells could be because of their general neuronal differentiation properties, which cause premature differentiation of multipotent cells at a time when signals for early-born neuron types are present, thereby reducing the number of multipotent progenitors available for production of later-generated cell types. In this case, the apparent difference in their ability to promote different retinal cell types could be explained by a difference in sensitivity to inhibitors of neuronal differentiation. According to this hypothesis, Xath5, which promotes only the earliest cell type, ganglion cells (3), would be the less sensitive and X-ngnr-1 the most sensitive to inhibitors of neuronal differentiation. However, in our animal cap assay, we have not been able to detect such a difference in sensitivity between X-ngnr-1 and Xath3 with respect to Notch inhibition. In the mouse retina, Mash1 and NeuroD are also important for proper ratio of retinal cell types (38, 39).
The fact that we obtain distinct effects upon overexpression in the nonneural ectoderm and in retinal progenitor cells (for example, ectopic expression of Islet-1 by X-ngnr-1 when overexpressed in early embryos, but not when overexpressed in the retina) most probably reflects the dependence of the activity of the bHLH factors on the context in which they are active. Positional information has been shown, for example, to be important in the case of scute, which promotes the formation of different types of external sensory organs in different places, because of the local expression of genes such as poxn or the BarH genes (40). It will be important to discover what determines the functional differences observed between these bHLH factors, i.e., site-recognition specificity or cofactor binding, by performing domain-swapping experiments and to identify the factors that cooperate with them to give neurons their specific properties.
Acknowledgments
We thank Dr. K. Takebayashi, I. David, L. Dubois, Q. Ma, J. P. Gergen, M. L. King, P. Krieg, and A. W. Brandli for providing us with the Xath3, Xlim3, Xcoe2, X-ngnr-1, Xaml, VegT, XHox11L2, and Pax2 clones, respectively. We thank S. Thor, N. Colley, and V. Lee for providing us with the anti-islet, anti-rhodopsin, and anti-neurofilament Abs, respectively. We thank C. Kintner for allowing us to use the Islet-1 clone before publication. We thank Bernard Avalosse for excellent technical assistance, Vincent Taelman for help with vibratome sectioning, and Monica Vetter for critical reading of the manuscript. Part of this work was carried out when E.J.B. was working in the laboratory of J. B. Gurdon. This work was supported by grants from the Fund for Scientific Medical Research and the International Brachet Shiftung (to E.J.B.) and by a grant from the European Commission and the Wellcome Trust (to W.A.H.). M.P. is a European Marie-Curie Fellow.
Abbreviations
- bHLH
basic helix–loop–helix
- CMZ
ciliary marginal zone
- GR
glucocorticoid receptor
- dex
dexamethasone
- GFP
green fluorescent protein
- CHX
cycloheximide
References
- 1.Ma Q, Sommer L, Cserjesi P, Anderson D J. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer L, Ma Q, Anderson D J. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 3.Kanekar S, Perron M, Dorsky R, Harris W A, Jan L Y, Jan Y N, Vetter M L. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- 4.Ma Q, Kintner C, Anderson D J. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 5.Cau E, Gradwohl G, Fode C, Guillemot F. Development (Cambridge, UK) 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 6.Jarman A P, Ahmed I. Mech Dev. 1998;76:117–125. doi: 10.1016/s0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 7.Guillemot F. Biol Cell. 1995;84:3–6. doi: 10.1016/0248-4900(96)81312-8. [DOI] [PubMed] [Google Scholar]
- 8.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa J L, Anderson D J. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 9.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, Zoghbi H Y. Nature (London) 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 11.Perez S E, Rebelo S, Anderson D J. Development (Cambridge, UK) 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- 12.Blader P, Fisher N, Gradwohl G, Guillemot F, Sträle U. Development (Cambridge, UK) 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- 13.Ferreiro B, Kintner C, Zimmerman K, Anderson D, Harris W A. Development (Cambridge, UK) 1994;120:3649–3655. doi: 10.1242/dev.120.12.3649. [DOI] [PubMed] [Google Scholar]
- 14.Turner D L, Weintraub H. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 15.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 16.Takebayashi K, Takahashi S, Yokota C, Tsuda H, Nakanishi S, Asashima M, Kageyama R. EMBO J. 1997;16:384–395. doi: 10.1093/emboj/16.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson E C, Schinder A F, Dantzker J L, Marcus E A, Spitzer N C, Harris W A. Mol Cell Neurosci. 1998;12:281–299. doi: 10.1006/mcne.1998.0712. [DOI] [PubMed] [Google Scholar]
- 18.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Nature (London) 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 19.Chitnis A, Kintner C. Development (Cambridge, UK) 1996;122:2295–2301. doi: 10.1242/dev.122.7.2295. [DOI] [PubMed] [Google Scholar]
- 20.Bellefroid E J, Bourguignon C, Hollemann T, Ma Q, Anderson D J, Kintner C, Pieler T. Cell. 1996;87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- 21.Bellefroid E J, Kobbe A, Gruss P, Pieler T, Gurdon J B, Papalopulu N. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubois L, Bally-Cuif L, Crozatier M, Moreau J, Paquereau L, Vincent A. Curr Biol. 1998;8:199–209. doi: 10.1016/s0960-9822(98)70084-3. [DOI] [PubMed] [Google Scholar]
- 23.Taira M, Hayes W P, Otani H, Dawid I B. Dev Biol. 1993;159:245–256. doi: 10.1006/dbio.1993.1237. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, King M L. Development (Cambridge, UK) 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- 25.Patterson K D, Krieg P A. Dev Dyn. 1999;214:34–43. doi: 10.1002/(SICI)1097-0177(199901)214:1<34::AID-DVDY4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 26.Heller N, Brandli A W. Mech Dev. 1997;69:83–104. doi: 10.1016/s0925-4773(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 27.Tracey W D, Jr, Pepling M E, Horb M E, Thomsen G H, Gergen J P. Development (Cambridge, UK) 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- 28.Saha M S, Miles R R, Grainger R M. Dev Biol. 1997;187:209–223. doi: 10.1006/dbio.1997.8625. [DOI] [PubMed] [Google Scholar]
- 29.Perron M, Kanekar S, Vetter M L, Harris W A. Dev Biol. 1998;199:185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- 30.Mikkola I, Fjose A, Kuwada J Y, Wilson S, Guddal P H, Krauss S J. J Neurobiol. 1992;23:933–946. doi: 10.1002/neu.480230802. [DOI] [PubMed] [Google Scholar]
- 31.Thor S, Ericson J, Brännström T, Edlund T. Neuron. 1991;7:881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- 32.Reh T A. In: Determinants of Neuronal Identity. Shanland M, Macagno E R, editors. New York: Academic; 1992. pp. 433–467. [Google Scholar]
- 33.Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter J M. Development (Cambridge, UK) 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Fode C, Guillemot F, Anderson D J. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanabe Y, William C, Jessell T M. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 36.Liem K F, Tremml G, Roelink H, Jessell T M. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 37.Wada H, Holland P H, Sato S, Yamamoto H, Satoh N. Dev Biol. 1997;187:240–252. doi: 10.1006/dbio.1997.8626. [DOI] [PubMed] [Google Scholar]
- 38.Tomita K, Nakanishi S, Guillemot F, Kageyama R. Genes Cells. 1996;1:765–774. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 39.Morrow E M, Furukawa T, Lee J E, Cepko C L. Development (Cambridge, UK) 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 40.Ghysen A, Dambly-Chaudière C. BioEssays. 1993;15:293–298. doi: 10.1002/bies.950150502. [DOI] [PubMed] [Google Scholar]