Abstract
Members of the transforming growth factor-β family play critical roles in body patterning, in both vertebrates and invertebrates. One transforming growth factor-β-related gene, dbl-1, has been shown to regulate body length and male ray patterning in Caenorhabditis elegans. We screened arrayed cDNAs to identify downstream target genes for the DBL-1 signaling by using differential hybridization. C. elegans cDNAs representing 7,584 independent genes were arrayed on a nylon membrane at high density and hybridized with 33P-labeled DNA probes synthesized from the mRNAs of wild-type, dbl-1, sma-2, and lon-2 worms. Signals for all the spots representing hybridized DNA were quantified and compared among strains. The screening identified 22 and 2 clones, which were positively and negatively regulated, respectively, by the DBL-1 signal. Northern hybridization confirmed the expression profiles of most of the clones, indicating good reliability of the differential hybridization using arrayed cDNAs. In situ hybridization analysis revealed the spatial and temporal expression patterns of each clone and showed that at least four genes, including the gene for the type I receptor for DBL-1, sma-6, were transcriptionally regulated by the DBL-1 signal.
Members of the transforming growth factor-β (TGF-β) superfamily have critical roles in cell growth, differentiation, and embryogenesis (1). The bone morphogenetic proteins, a subfamily of the TGF-β superfamily, have conserved roles in vertebrates and insects to regulate the early dorsoventral patterning of embryos (2). The structure and function of type I and type II serine/threonine protein kinase receptors and Smad proteins, which are essential components in this signaling pathway, are also well conserved between species. Although much of the intracellular signaling mechanism has been elucidated, the whole spectrum of target genes responding to the signaling pathway has not. Thus, identification and functional analyses of the downstream genes regulated by TGF-β family members are necessary to understand the gene regulation underlying the animal body plan. Recently, a new TGF-β-related and bone morphogenetic protein-related ligand, DBL-1,§ was isolated in C. elegans and shown to play a role in the regulation of body length and male ray patterning (3, 4). The DBL-1 signal is transduced by type I (SMA-6) and type II (DAF-4) receptors and by Smad proteins (SMA-2, SMA-3, and SMA-4) (3–5). These elements of the signaling mechanism are conserved, allowing us to study the genetic cascade of the TGF-β signal by using C. elegans, which has many advantages as a model animal for genetic studies.
Recent advancements in genome sequencing and expressed sequence tag (EST) techniques allow a more comprehensive analysis of gene expression. DNA arrayed on a membrane filter or a glass slide has been used for large-scale quantification of gene expression (6, 7). For example, microarrays containing all the Saccharomyces cerevisiae genes were used to analyze changes in gene expression induced by a metabolic condition or the cell cycle (8, 9). This technique, using ESTs, is thought to be effective for identifying new genes that are differentially expressed under different conditions (10). Upon completion of the C. elegans genome project, approximately 19,000 ORFs were detected (11). ESTs representing about 10,000 independent genes are available (Y.K., unpublished data). Furthermore, a variety of mutants in which a single gene is affected are available in C. elegans. In this paper, we sought to identify genes that are transcriptionally regulated by DBL-1 signaling by differentially hybridizing arrayed cDNAs containing 7,584 ESTs with probes from wild-type and DBL-1-signaling mutants. Differentially expressed genes identified by this screening were further analyzed by Northern hybridization and whole-mount in situ hybridization to confirm their expression profiles. We identified at least four genes that are activated by DBL-1 signaling, demonstrating that screening with arrayed cDNA is an effective method for identifying target genes of a specific signaling pathway.
Materials and Methods
General Methods and Strains.
C. elegans strains were cultured as described by Brenner (12). For synchronous culture, worms were grown in a liquid medium as described previously (13). The following strains were used in this study: wild-type C. elegans Bristol strain (N2), dbl-1 (mk3), sma-2 (e502), and lon-2 (e678).
cDNA Array.
Preparation and characterization of the C. elegans cDNA clones will be published elsewhere (Y.K., unpublished work). Inserts of 7,584 independent cDNA clones were amplified with vector primers and arrayed on a nylon membrane filter (Hybond-N; Amersham Pharmacia) with a gridding robot (Bio Robotic, Cambridge, U.K.) at a high density (384 × 5 × 5 cDNA grids per 8 × 12 cm filter). The arrayed high-density filter was denatured with 0.5 M NaOH, neutralized with 0.5 M Tris⋅HCl (pH 7.0), and baked at 80°C for 2 h.
RNA Preparation and Differential Hybridization.
Total RNA was extracted from synchronously cultured larval stage 3 (L3) larvae with Trizol reagent (GIBCO/BRL), and poly(A)+ RNA was purified by using Oligotex-dT 30 (Roche Molecular Biochemicals). To remove oligo(dT) that might contaminate the poly(A)+ RNA preparation, 1 μg of poly(A)+ RNA was mixed with 1 μg of poly(dA), heated at 65°C for 10 min, then cooled as described by Nguyen et al. (14). For probe labeling, the oligo(dT)-depleted RNA was incubated with random hexamer, Superscript reverse transcriptase (GIBCO/BRL) and [33P]dCTP at 37°C for 2 h. Probes were purified with a G-50 spin column (Amersham Pharmacia). Two duplicate high-density filters were preincubated in a solution containing 4× SSC, 5× Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 0.5% SDS, and 100 μg/ml salmon sperm DNA at 65°C for 1 h, then incubated in the same solution containing a 33P-labeled probe at 65°C for 20 h. The hybridized filters were washed in a solution containing 0.1× SSC and 0.1% SDS at 60°C and exposed to a Fuji imaging plate for 24 h. The imaging plate was scanned in a Fuji BAS 2000 system. Detection and quantification of signals representing hybridized DNA was performed with hdg analyzer software (version 3.03; Genomic Solutions, Ann Arbor, MI). After quantification of the all spots, the intensities of corresponding spots from the two duplicate filters were averaged. The averaged intensities for dbl-1, sma-2, and lon-2 samples were normalized to the total-intensity intensities of the wild-type spots. The hybridized images and the relative intensities of all the spots will appear on a web site in The Nematode Expression Pattern DataBase (NEXTDB, http://watson.genes.nig.ac.jp/db/index.html).
Northern hybridization was performed by using 0.2 μg of the poly(A)+ RNA per lane as described previously (15). Probes for Northern hybridization were synthesized with cDNA inserts by using the Megaprime DNA labeling system (Amersham Pharmacia). The signals from hybridized DNA were detected and quantified with the Fuji BAS system. The intensities for each clone were normalized to the intensity of the CeIF probe (16).
In Situ Hybridization.
Whole-mount in situ hybridization was performed as described (17, 18) with some modifications. Collected worms were fixed in a solution of 3.7% formaldehyde/0.1 M Pipes buffer (pH 6.9)/1 mM MgSO4/1 mM EGTA for 15 min and stored in methanol at −20°C. The fixed worms were washed with TBST [10 mM Tris⋅HCl (pH 7.0)/150 mM NaCl/0.1% Triton X-100] and treated with 10% 2-mercaptoethanol in TBST for 10 min, 20 μg/ml proteinase K in 20 mM Tris⋅HCl (pH 7.5)/1 mM CaCl2 for 5 min, and then in the formaldehyde solution described above for 15 min. After being washed in TBST containing 100 μg/ml yeast RNA, the worms were treated with 100 mM triethanolamine (pH 8.0) for 5 min and 100 mM triethanolamine containing 0.5% acetic anhydride for 5 min. After washing with TBST containing yeast RNA, the worms were incubated in hybridization solution (5× SSC/0.1% Triton X-100/100 μg/ml salmon sperm DNA/100 μg/ml yeast RNA/50% formamide) at 60°C for 1 h, then incubated in the same solution containing digoxigenin-labeled RNA probes (Roche Molecular Biochemicals) for 16–24 h at 60°C. After hybridization, the worms were washed with TBST for 30 min at 37°C twice. The hybridized signals were detected by using alkaline-phosphatase-labeled anti-digoxigenin antibody and BM purple substrate (Roche Molecular Biochemicals).
Results and Discussion
Differential Hybridization with Arrayed cDNAs.
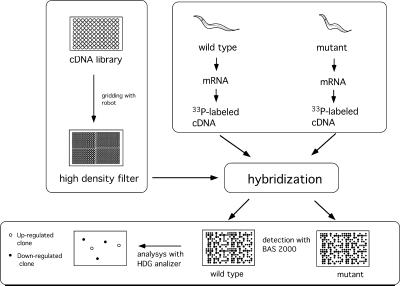
The strategy we used to isolate TGF-β-regulated genes is illustrated in Fig. 1. Probes were synthesized with RNA prepared from wild-type, dbl-1, sma-2, and lon-2 worms synchronously cultured and harvested at L3. The radioactivity was quantified as photostimulated luminescence (PSL) (Fuji BAS system) for each spot and ranged from 0 to >500 PSL. Spots with intensities higher than 10 PSL in at least one of the strains were chosen for further analysis, because an intensity lower than 10 PSL was sometimes not reproducible in duplicate filters, and because background signals in a control experiment with unrelated DNAs showed signals with intensities under 10 PSL (data not shown). The intensities of 3,390 spots were chosen and compared between the wild-type probes and those of the mutants. Table 1 shows the numbers of clones whose intensities differed by more than 2-fold in the mutants. As shown in Table 1, 88 and 70 clones showed increased signals in dbl-1 and sma-2 worms, respectively, and 169 and 61 clones showed decreased signals in these same mutants. We then chose clones whose intensities increased or decreased in both dbl-1 and sma-2 worms, ensuring that the genes were indeed regulated by the DBL-1 pathway; clones whose intensities increased or decreased in only one of the two mutants were not likely to be real targets of the DBL-1 signal. In addition, slight differences in the growth rate or nutritive conditions between strains could have caused different developmental and/or physiological states, even if the worms were synchronously cultured in the same manner. Differential hybridization with probes prepared from such worms might result in the identification of genes unrelated to the DBL-1 signal. To minimize such false positives, we used three strains with mutations in different DBL-1 signaling components. DBL-1 and SMA-2 have been shown to be involved in the same signaling pathway (3, 4), and mutants in the genes that encode them have very similar phenotypes, suggesting that the expression of the DBL-1 target genes is regulated in the same manner in these mutants. On the other hand, lon-2 has an opposite phenotype to dbl-1 and sma-2. Therefore, real target genes of the DBL-1 signal should be repressed in both dbl-1 and sma-2, but not in lon-2 worms, or activated in both dbl-1 and sma-2, but not in lon-2 worms. By these criteria, we identified 2 candidate clones as genes that are negatively regulated and 22 candidate clones as genes that are positively regulated by the DBL-1 signal (Table 1).
Figure 1.
Scheme of the differential hybridization technique with arrayed cDNA. Amplified cDNAs were arrayed on a nylon membrane to make a high-density filter. The filter was then hybridized with probes prepared from wild-type and mutant worms. Hybridized signals were detected by using the Fuji BAS system and quantified with hdg analyzer software (Genomic Solutions). Comparing the intensities between wild-type and mutant hybridizations identified differentially expressed genes.
Table 1.
Differentially expressed genes identified with arrayed cDNAs
| Clones | Genes
|
|||
|---|---|---|---|---|
| dbl-1 | sma-2 | Both dbl-1 and sma-2 | Both dbl-1 and sma-2, but not lon-2 | |
| Up-regulated | 88 | 70 | 3 | 2 |
| Down-regulated | 169 | 61 | 31 | 22 |
Number of clones whose expression was up-regulated or down-regulated in the mutants is indicated.
We used a conventional filter-hybridization technique to screen cDNA arrays, whereas others have used a microarray method with fluorescent dyes (7–10). The microarray technique has the advantage that two or more fluorescent dyes can be used for probe labeling, which allows multiprobe labeling in a single hybridization step and eliminates the false signals that may result from inconsistencies in the amount of DNA spotted onto different arrays. To minimize such false signals, we used two duplicate filters for each probe. Even though the microarray technique has some advantages, the conventional hybridization does not require a laser scanner for detection and may thus be available to more laboratories.
Expression Analysis by Northern Hybridization.
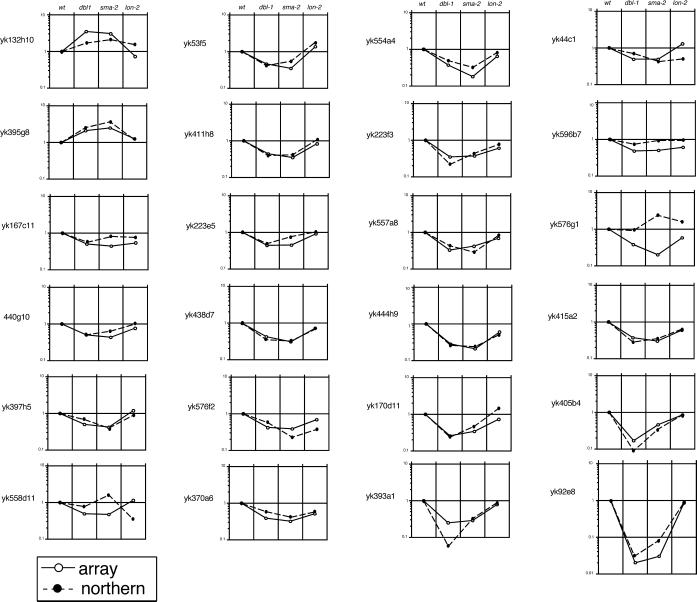
Northern hybridization was used to confirm the expression profiles of the 2 up-regulated and 22 down-regulated clones identified by differential hybridization as described above. The signal representing hybridized DNA was quantified by using the Fuji BAS system and compared with the results obtained from differential hybridization using the cDNA array (Fig. 2; Table 2). There were 21 of 24 clones that showed similar expression profiles in the differential hybridization and the Northern hybridization. Three clones described below were excluded from the candidate DBL-1 target genes because their expression profiles were inconsistent between the two methods. Two clones, yk558d11 and yk576g1, showed quite different profiles, and yk596b7 showed almost the same expression level in all the mutants (Fig. 2). These inconsistent results may have been caused by the crosshybridization of labeled probe to unrelated sequences. In a preliminary experiment in which the washing stringency for the differential hybridization was lower than that described in Materials and Methods, much more inconsistent results were obtained and were thought to be caused by crosshybridization. With the stringent washing conditions used in this work, about 90% of randomly selected clones showed expression profiles similar in the two methods (data not shown). These results indicate the reliability of the differential hybridization with arrayed cDNA to screen for a set of target genes, but also suggest that the precise expression profile should be determined by combining the results from more than two different methods before target genes can be confirmed.
Figure 2.
Comparison of gene-expression profiles obtained from differential hybridization using a cDNA array and Northern hybridization. Quantified intensities obtained from the two different methods were plotted relative to the wild-type level. The vertical line shows relative intensity. Open and filled circles show results from the differential hybridization and Northern hybridization, respectively.
Table 2.
Characterization of differentially expressed genes
| Clone name | Fold increase by array* (dbl-1, sma-2, lon-2) | Fold increase by Northern blot (dbl-1, sma-2, lon-2) | Expression domain (in situ hybridization) | Gene | Identification or similarity |
|---|---|---|---|---|---|
| yk132h10 | 3.48, 3.04, 0.73 | 1.69, 2.08, 1.52 | Head | T01A4.1 | Similarity to natriuretic peptide receptors |
| yk395g8 | 2.08, 2.48, 1.25 | 2.51, 3.61, 1.17 | Hypodermis | D1014.5 | Weak similarity to Plasmodium merozoite surface protein 1 |
| yk167c11 | 0.50, 0.44, 0.54 | 0.57, 0.82, 0.77 | Intestine | C46H11.2 | Similarity to flavin-containing monooxygenases |
| yk440g10 | 0.50, 0.43, 0.76 | 0.50, 0.64, 0.99 | Head and intestine | K05G3.3 | Similarity to carbonic anhydrase |
| yk397h5 | 0.50, 0.42, 1.19 | 0.68, 0.40, 0.89 | Intestine | R09H10.5 | Similarity to epidermal growth factor domains |
| yk558d11 | 0.49, 0.47, 1.14 | 0.77, 1.60, 0.34 | No signal | T10C6.5 | Similarity to Schizosaccharomyces pombe hypothetical protein |
| yk44c1 | 0.49, 0.49, 1.29 | 0.72, 0.93, 0.93 | Intestine | K10C2.1 | Similarity to human lysosomal protective protein |
| yk596b7 | 0.48, 0.50, 0.61 | 0.74, 0.93, 0.93 | Germ cell | Unidentified | ND |
| yk53f5 | 0.46, 0.35, 1.36 | 0.41, 0.55, 1.79 | Head, posterior, and intestine | C32D5.2 | Serine/threonine kinase SMA-6 |
| yk411h8 | 0.44, 0.35, 0.83 | 0.40, 0.41, 1.08 | Intestine | C17G10.5 | Similarity to Entamoeba histolytica N-acetylmuraminidase |
| yk223e5 | 0.44, 0.45, 0.92 | 0.49, 0.77, 1.03 | No signal | W09D12.1 | Similarity to human collagenase |
| yk438d7 | 0.42, 0.31, 0.72 | 0.35, 0.32, 0.68 | Intestine | F56D6.2 | Similarity to lectin C type domains |
| yk576f2 | 0.42, 0.39, 0.69 | 0.57, 0.23, 0.37 | Intestine | Unidentified | ND |
| yk370a6 | 0.39, 0.32, 0.51 | 0.60, 0.42, 0.60 | Intestine | F35C5.9 | Similarity to lectin C type domains |
| yk576g1 | 0.38, 0.20, 0.58 | 0.91, 2.40. 1.60 | Germ cell | Unidentified | ND |
| yk415a1 | 0.37, 0.30, 0.60 | 0.28, 0.35, 0.64 | Intestine | ZK1320.3 | No similarity |
| yk554a4 | 0.37, 0.18, 0.65 | 0.49, 0.32, 0.82 | Intestine | F35C5.6 | Similarity to lectin C type domains |
| yk223f3 | 0.35, 0.37. 0.61 | 0.22, 0.44, 0.76 | Hypodermis | F46F2.3 | No similarity |
| yk557a8 | 0.33, 0.42, 0.69 | 0.43, 0.29, 0.82 | Intestine | F35C5.8 | Similarity to von Willebrand factor type A domains |
| yk444h9 | 0.28, 0.21, 0.60 | 0.26, 0.24, 0.51 | No signal | F35C5.8† | ND |
| yk170d11 | 0.26, 0.34, 0.73 | 0.24, 0.47, 1.46 | Intestine | F09C8.1 | Similarity to phospholipase precursor |
| yk393a1 | 0.25, 0.29, 0.79 | 0.06, 0.34, 0.93 | Intestine | K01A2.3 | No similarity |
| yk405b4 | 0.17, 0.46, 0.86 | 0.09, 0.35, 0.85 | Head, intestine, and vulva | F42A8.1 | Weak similarity to Drosophila probable epidermal cell surface receptor |
| yk92e8 | 0.02, 0.03, 0.84 | 0.03, 0.08, 0.84 | Intestine and vulva | C08E3.13 | Weak similarity to Mycobacterium tuberculosis hypothetical protein |
Sequence and other information about each cDNA clone will be available at http://www.ddbj.nig.ac.jp/htmls/c-elegans/html/CE_INDEX.html. ND, not determined.
Differential hybridization by using cDNA array.
† yk444h9 is encoded by predicted introns of F35C5.8 gene.
Expression Analysis by Whole-Mount in Situ Hybridization.
We used three mutants for the differential hybridization to minimize false signals. However, this choice was not sufficient to exclude all the false positives. Temporal and spatial expression analysis at every developmental stage in each strain is necessary to determine the precise expression profile and, thus, to identify real target genes of the DBL-1 signal. Neither the differential hybridization nor Northern hybridization is suitable for this purpose. Therefore, we used whole-mount in situ hybridization to determine the temporal and spatial expression patterns of the candidate genes identified by the differential hybridization and Northern hybridization.
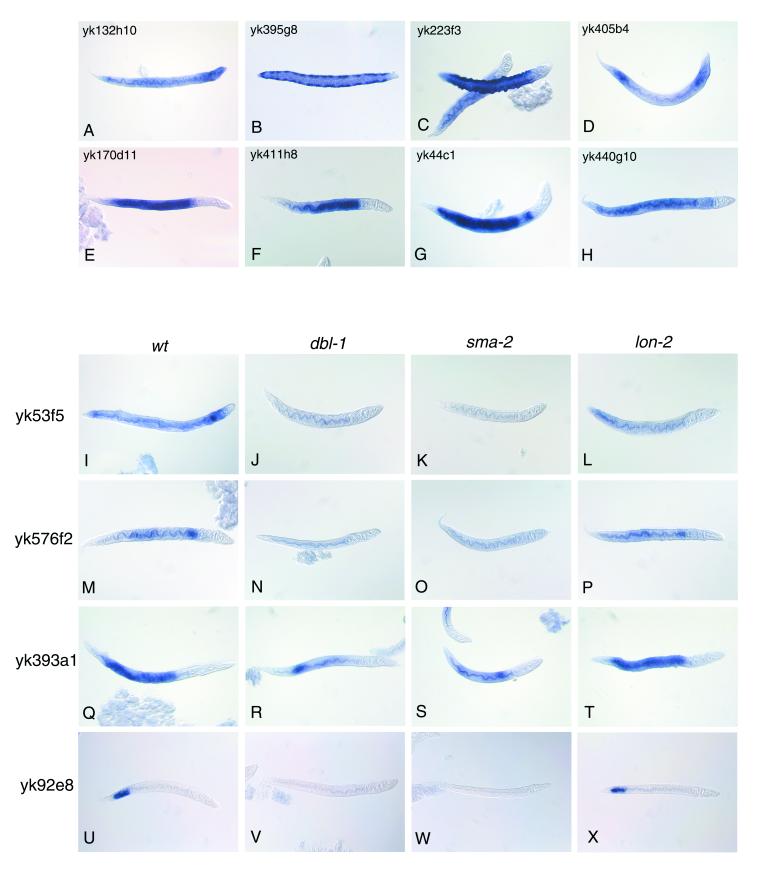
First, wild-type worms at various stages were used for whole-mount in situ hybridization analysis to obtain the expression profile for each clone, and then dbl-1, sma-2, and lon-2 worms were used to determine whether the expression pattern of each clone differed among the mutants. The signal for yk132h10, one of the up-regulated clones, was detected in the whole body with an intense signal in the head region from the L3 to adulthood (Fig. 3A). Another up-regulated clone, yk395g8, was expressed as two lateral rows in L3 and L4 larvae, suggesting a specific expression in the seam hypodermis (Fig. 3B). Changes in the expression pattern or intensity of these clones were not observed in dbl-1, sma-2, or lon-2 worms (data not shown).
Figure 3.
Whole-mount in situ hybridization showing the expression pattern of DBL-1-regulated genes. Wild-type (A–I, M, Q, and U), dbl-1 (J, N, R, and V), sma-2 (K, O, S, and W), and lon-2 (L, P, T, and X) larvae were hybridized with digoxigenin-labeled RNA probes to localize the expression of each cDNA clone. The names of the cDNA clones used for probe synthesis are indicated above (A–H) and to the left (I–X) of the figures.
Twelve of the down-regulated clones showed specific signals only in the intestine from L1 to adulthood (Fig. 3 E, F, M, and Q). Of these clones, signals were ubiquitous throughout the whole intestine for eight clones (yk170d11, yk167c11, yk397h5, yk438d7, yk415a1, yk554a4, yk557a8, and yk393a1) (Fig. 3E and data not shown). An anterior to posterior gradient of the intestinal signal was observed for two of the clones (yk411h8 and yk370a6) (Fig. 3F and data not shown). Intestine-specific expression with a narrow gap near the pharynx was observed for two clones (yk44c1 and yk576f2) (Fig. 3G and data not shown). Another down-regulated clone, yk92e8, showed specific expression in the posterior intestine (Fig. 3U) of L1 larvae to young adults and in the vulva (data not shown) of young adults. The signal for yk405b4 was found in the head, posterior gut, and hypodermis from L3 to adulthood (Fig. 3D) and in the vulva from L4 to adulthood (data not shown). The signal for yk53f5 was detected in the head, posterior regions, and intestine from L2 to young adulthood. The signal for yk440g10 was observed in the whole intestine and head region, including neurons, of L3 larvae to adults (Fig. 3H). The signal for yk223f3 was found in the hypodermis of L3 and L4 larvae (Fig. 3C).
Different expression patterns between the wild-type and the mutant worms were observed for yk53f5, yk576f2, yk393a1, and yk92e8, whose expression was decreased in both dbl-1 and sma-2 but not in lon-2 worms (Fig. 3 I–X). yk53f5 encodes SMA-6, which is thought to be a type 1 receptor for the DBL-1 ligand. It is conceivable that a ligand regulates the expression of genes encoding its own signaling components. Three intestine-specific genes were down-regulated in dbl-1 and sma-2 worms. The most marked case was that of yk92e8 expression. The expression in the posterior intestine found in wild-type and lon-2 worms was missing in most dbl-1 and sma-2 worms, but in a few cases, a very weak signal was seen in the same region of dbl-1 or sma-2 worms. The yk92e8 signal in the vulva of young adults was seen in all the mutants, suggesting that its expression in the vulva was not regulated by the DBL-1 signal. In sum, we found that the expressions of four clones were decreased in both dbl-1 and sma-2 worms. However, we have no evidence to exclude other candidate clones from being DBL-1-regulated genes based on this analysis, because it is impossible to quantify expression levels accuratly by whole-mount in situ hybridization.
DBL-1 Signaling Regulates Intestine-Specific Genes.
We identified at least 4 and probably 12 additional genes whose expression in the intestine was regulated by DBL-1 signaling. dbl-1 is expressed mainly in the nervous system (3, 4), but low-level expression of a dbl-1∷gfp fusion gene is observed in the intestine (K.M. and N.U., unpublished data). The type I receptor, SMA-6, was expressed in the intestine (Fig. 3 I and L; ref. 5). These observations suggest that these genes are directly regulated by DBL-1 signaling in the intestine. Further investigation of the regulatory regions of these genes is required to confirm this hypothesis.
The expression patterns of these genes in the intestine were classified into four categories: (i) ubiquitous expression throughout the whole intestine; (ii) graded expression from the anterior to posterior end; (iii) expression with a gap near the pharynx; and (iv) posterior expression. These results showed that gene expression in the intestine varied along the anterior–posterior axis as reported previously (19). Functional analysis of the gene products expressed in specific regions of the intestine may reveal specialized roles of these regions.
Conclusions
Similarities between known proteins and the deduced products encoded by the genes differentially expressed in dbl-1 and sma-2 worms are shown in Table 2. We presently have no information about whether any of the genes in Table 2 besides sma-6 regulate the body length of C. elegans. Such information will be revealed by the functional analysis of DBL-1-regulated genes, for example, by gene disruption. Of course, genes with critical roles in body-length regulation might have been missed in our screening, because cDNAs representing all of the C. elegans genes were not available at this time. Also, some genes might have escaped detection because of their low expression level. Continued improvement of the technique and the acquisition of cDNAs covering all of the C. elegans genes will address these problems. The present work showed that analysis with arrayed DNA is effective for identifying target genes that are directly or indirectly regulated by a certain signal. We believe that this kind of approach will facilitate understanding of the whole gene network that regulates specific biological phenomena, not only for C. elegans, but also for any species, including vertebrates.
Acknowledgments
We thank Dr. A. Fujiyama for the gridding robot and the technicians in the Y.K. lab for producing the cDNA arrays. This work was supported by the Research for the Future program of the Japan Society for the Promotion of Science (to N.U.), and Grants-in-Aid for Priority Research Area from the Ministry of Education, Science, Culture, and Sports (to Y.K.). Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, National Center for Research Resources.
Abbreviations
- TGF-β
transforming growth factor-β
- L1–L4
larval stages 1–4
- PSL
photostimulated luminescence
Footnotes
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Hogan B L. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki Y, Yandell M D, Roy P J, Krishna S, Savage-Dunn C, Ross R M, Padgett R W, Wood W B. Development (Cambridge, UK) 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 4.Morita K, Chow K L, Ueno N. Development (Cambridge, UK) 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- 5.Krishna S, Maduzia L L, Padgett R W. Development (Cambridge, UK) 1999;126:251–260. doi: 10.1242/dev.126.2.251. [DOI] [PubMed] [Google Scholar]
- 6.Zhao N, Hashida H, Takahashi N, Misumi Y, Sakaki Y. Gene. 1995;156:207–213. doi: 10.1016/0378-1119(95)00023-y. [DOI] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 11.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis J A, Fleming J T. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. San Diego: Academic; 1997. pp. 4–29. [Google Scholar]
- 14.Nguyen C, Rocha D, Granjeaud S, Baldit M, Bernard K, Naquet P, Jordan B R. Genomics. 1995;29:207–216. doi: 10.1006/geno.1995.1233. [DOI] [PubMed] [Google Scholar]
- 15.Mochii M, Agata K, Kobayashi H, Yamamoto T S, Eguchi G. Cell Differ. 1988;24:67–74. doi: 10.1016/0045-6039(88)90087-5. [DOI] [PubMed] [Google Scholar]
- 16.Roussell D L, Bennett K. Nucleic Acids Res. 1992;20:3783. doi: 10.1093/nar/20.14.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitani S, Du H, Hall D H, Driscoll M, Chalfie M. Development (Cambridge, UK) 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. J Biol Chem. 1998;273:2192–2198. doi: 10.1074/jbc.273.4.2192. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder D F, McGhee J D. Development (Cambridge, UK) 1998;125:4877–4887. doi: 10.1242/dev.125.24.4877. [DOI] [PubMed] [Google Scholar]