Abstract
After exposure to DNA-damaging agents, the p53 tumor suppressor protects against neoplastic transformation by inducing growth arrest and apoptosis. A series of investigations has also demonstrated that, in UV-exposed cells, p53 regulates the removal of DNA photoproducts from the genome overall (global nucleotide excision repair), but does not participate in an overlapping pathway that removes damage specifically from the transcribed strand of active genes (transcription-coupled nucleotide excision repair). Here, the highly sensitive ligation-mediated PCR was employed to quantify, at nucleotide resolution, the repair of UVB-induced cyclobutane pyrimidine dimers (CPDs) in genetically p53-deficient Li–Fraumeni skin fibroblasts, as well as in human lung fibroblasts expressing the human papillomavirus (HPV) E6 oncoprotein that functionally inactivates p53. Lung fibroblasts expressing the HPV E7 gene product, which similarly inactivates the retinoblastoma tumor-suppressor protein (pRb), were also investigated. pRb acts downstream of p53 to mediate G1 arrest, but has no demonstrated role in DNA repair. Relative to normal cells, HPV E6-expressing lung fibroblasts and Li–Fraumeni skin fibroblasts each manifested defective CPD repair along both the transcribed and nontranscribed strands of the p53 and/or c-jun loci. HPV E7-expressing lung fibroblasts also exhibited reduced CPD removal, but only along the nontranscribed strand. Our results provide striking evidence that transcription-coupled repair, in addition to global repair, are p53-dependent in UV-exposed human fibroblasts. Moreover, the observed DNA-repair defect in HPV E7-expressing cells reveals a function for this oncoprotein in HPV-mediated carcinogenesis, and may suggest a role for pRb in global nucleotide excision repair.
Inactivation of the p53 tumor-suppressor protein is a key event in carcinogenesis, since more than 50% of all human malignancies manifest p53 gene mutations (1), and individuals afflicted with Li–Fraumeni syndrome (LFS; a disease characterized by germ-line p53 alterations) are strongly predisposed to various cancers (2). In addition, the neoplastic potential of certain oncogenic viruses can be attributed, at least in part, to interactions of their associated viral gene products with p53. For example, the capacity of human papillomavirus (HPV) to promote epithelial malignancies is exerted largely through intracellular expression of the HPV E6 oncoprotein, which engenders functional inactivation of p53 by means of binding and accelerated proteasomal degradation (3, 4).
After exposure of cells to DNA-damaging agents, p53 triggers multiple protective pathways, i.e., through transactivation of genes and through protein–protein interactions, that regulate growth arrest and apoptosis (5, 6). Moreover, different lines of evidence (see below) demonstrate that p53 participates directly in nucleotide excision repair (NER), a major pathway for the removal of carcinogenic DNA damage inflicted by diverse mutagenic agents. The physiological importance of NER is exemplified by the autosomal recessive disease xeroderma pigmentosum (XP), in which affected individuals manifest defective removal of UV-induced DNA photoproducts and a concomitant 2000-fold increase in the frequency of sunlight-induced skin cancers (7). Two distinct, but overlapping, NER pathways have been identified: one for the removal of damage from the genome overall [global NER (GNER)], and another, which is mechanistically linked to transcription, that brings about rapid, preferential repair of adducts specifically on the transcribed strand (TS) of active genes [transcription-coupled NER (TCNER)] (8).
Early indications that p53-deficient cells might be defective in NER (9) led to studies revealing a potential role in this process for the p53 downstream effectors gadd45 (10) and p21waf1 (11), which sequester the DNA polymerase processivity factor PCNA (proliferating cell nuclear antigen) that is essential for NER (12). It was also shown that p53 binds RP-A (replication protein-A) (13), which participates in the initiation of the NER process (14). Moreover, Wang et al. (15) demonstrated that p53 interacts (i) with the XP-B and XP-D gene products, i.e., components of the TFIIH basal transcription/repair factor that participates in GNER as well as TCNER, and (ii) with the CS-B gene product (transcription-repair coupling factor) that is necessary for TCNER only. While a role for p53 in both GNER and TCNER was therefore indicated, this latter study could demonstrate an overall NER defect in p53-deficient cells only by employing a gene-specific DNA-repair assay that does not differentiate between GNER and TCNER. A series of succeeding investigations employing a strand-specific DNA-repair assay at the active dihydrofolate reductase locus in p53-deficient human skin fibroblasts, i.e., either HPV E6-expressing (16) or derived from LFS patients (17, 18) revealed strong evidence that p53 plays an essential role in GNER after treatment with UV, but does not modulate TCNER. In support of this conclusion, it was subsequently demonstrated that up-regulation of the p48 (XP complementation group E) gene product is p53-dependent and that human XP-E fibroblasts mutated at the p48 locus are deficient in GNER, but not in TCNER (19).
In a manner analogous to HPV E6, intracellular expression of the HPV E7 oncogene product plays a major role in the molecular development of HPV-positive malignancies by binding and functionally inactivating (also through accelerated proteasomal degradation) the retinoblastoma tumor suppressor (pRb) (20). This latter protein, which is frequently altered in a wide variety of sporadic tumor types (21), acts downstream of p53, as well as through p53-independent pathways, to regulate G1 arrest in cells exposed to DNA-damaging agents (22). Preliminary investigations suggesting a role for pRb in DNA repair (e.g., ref. 23) have not been adequately substantiated or refuted to date. However, it should be noted that HPV E7 interacts with a considerable number of proteins aside from pRb, some of which may conceivably be involved in DNA repair (see Discussion).
Here, we have employed the ligation-mediated PCR (LMPCR) to investigate, at nucleotide resolution, the repair of UVB-induced cyclobutane pyrimidine dimers (CPDs) in genetically p53-deficient LFS human skin fibroblasts, as well as in primary human lung fibroblasts expressing either the HPV E6 or HPV E7 oncoprotein. Introduction of these oncoproteins into cultured cells has been widely used as a model to dissect the phenotypic consequences of p53 or pRb inactivation, respectively. Our primary goal was to reassess the function of p53 in NER because, notwithstanding the investigations cited above showing p53 dependence for GNER but not TCNER, other studies have reported data consistent with a role for this protein in both NER pathways (reviewed in ref. 24). We were also interested in addressing the possibility that the HPV E7 oncoprotein modulates DNA repair in human cells. Our data provide compelling evidence that p53 participates in TCNER as well as GNER, in disagreement with the prevailing notion that this protein regulates only GNER. In addition, we show that HPV E7-expressing fibroblasts are deficient in GNER, but not TCNER, revealing a role for this oncoprotein in the pathogenesis of HPV-positive tumors.
Materials and Methods
Cell Strains.
Normal human foreskin fibroblasts, and the spontaneously immortalized (i.e., post-crisis; approximately 200 population doublings) LFS skin fibroblast strain LF041 (a gift of M. Tainsky, University of Texas M. D. Anderson Cancer Center, Houston, TX) were maintained in DMEM supplemented with 10% FBS plus antibiotics. LF041 cells have lost one p53 allele and carry a frameshift mutation at codon 184 in the remaining copy. The normal primary diploid lung fibroblast strain LF-1 (no relation with Li–Fraumeni; kindly provided by John Sedivy, Brown University, Providence, RI) and its derivatives were grown in Ham's F-10 medium containing 15% FBS plus antibiotics. Low-passage LF-1 cells were infected as previously described (25) with a replication-defective retrovirus (LXSN) expressing G418 resistance, and the HPV E6, HPV E7, or both oncoproteins derived from the high-risk HPV type 16. Briefly, culture medium was harvested from a confluent LXSN murine producer cell line (obtained from American Type Culture Collection), and passed through a 0.22-μm filter. Fibroblasts at 50–70% confluence on 60-mm dishes were incubated for 2 hr with 1 ml of this viral supernatant containing 8 μg/ml Polybrene, followed by aspiration of the viral supernatant and addition of normal growth medium. After a 2-day phenotypic expression period, fibroblasts expressing the empty vector, or ones expressing HPV E6 and/or HPV E7, were selected in growth medium containing 200 mg/ml G418 (GIBCO/BRL).
UV Irradiation.
Cells were grown to confluence on 150-mm Petri dishes and irradiated with UVB (290–320 nm) at room temperature after replacing the medium with 0.9% NaCl. The UVB source consisted of two tubes (FS20T12/UVB/BP, Phillips) delivering a fluence of 7.45 J/m2·s that was filtered through a screen of cellulose acetate (Kodacel TA-407, clear 0.015-inch; Eastman Kodak). All cells were irradiated with 1 kJ/m2 of UVB and allowed to repair for various times in freshly prepared culture medium. Cells were harvested, nuclear DNA was extracted and quantified, and the global CPD frequency was determined in T4-endonuclease V-digested total genomic DNA by alkaline gel electrophoresis (26, 27).
LMPCR.
The LMPCR protocol has been described previously in detail (26, 28). Briefly, after irradiation and incubation of cells, genomic DNA was isolated and digested with T4 endonuclease V to efficiently incise the DNA at CPD sites. The resulting 5′-pyrimidine overhangs were then removed by photoreactivation, using Escherichia coli photolyase to generate ligatable ends. A gene-specific oligonucleotide was annealed downstream of the break site, and the set of genomic cleavage products was extended using Sequenase (United States Biochemical). An asymmetric double-stranded linker was ligated to the phosphate groups at the fragment termini, providing a common sequence on the 5′ end of all fragments. The longer oligonucleotide of this same linker, in conjunction with another gene-specific primer, was used in a PCR to amplify the cleavage products of interest. These products were subjected to electrophoresis on 8% polyacrylamide gels alongside a Maxam and Gilbert sequencing ladder, transferred to nylon membranes, hybridized to a 32P-labeled gene-specific probe, and visualized by autoradiography. Each experimental condition was assayed in duplicate. A screening sequencing gel was run using a portion of the DNA to ensure that there was no significant variation between samples. The two samples were then pooled on a combined gel, and the resulting autoradiogram was analyzed with a Fuji BAS 1000 phosphorimager (Fuji Medical Systems, Stanford, CT). Each band represents a nucleotide position where a break was induced by CPD cleavage, and the intensity of the band reflects the number of DNA molecules with ligatable ends terminating at that position. To assess proficiency in GNER and TCNER for the various strains investigated in this study, relative repair rates (reflected in Table 1) were determined along (i) the TS of the p53 gene (exon 7; nucleotides 14,030–14,080), (ii) the TS of the c-jun gene (exon 1; nucleotides +30 to +117), (iii) the TS of the c-jun promoter (immediately downstream of the transcription start site; nucleotides −40 to −10), and/or (iv) the nontranscribed strand (NTS) of the p53 gene (exons 5 and 8; nucleotides 13,095–13,166 and 14,510–14,575, respectively). The LMPCR primer sets used in each case have been presented previously (29, 30).
Table 1.
Relative delay in nucleotide excision repair for LF041 skin fibroblasts, and for lung fibroblasts expressing the HPV E6, HPV E7, or both oncoproteins
| Fibroblast type | Strand | Average 50% repair time, hr* | Fold delay in repair | No. and location of sites analyzed |
|---|---|---|---|---|
| Normal skin | TS | 3 (1, 8) | 1 | 16 (exon 1 c-jun); 5 (exon 7 p53) |
| NTS | 10 (4, 16) | 1 | 10 (exon 5 p53) | |
| LF041 skin | TS | 14 (4, 28) | 6.1 ± 2.8 | 16 (exon 1 c-jun); 5 (exon 7 p53) |
| NTS | 28 (16, 32) | 3.1 ± 1.1 | 10 (exon 5 p53) | |
| LXSN lung | TS | 3.5 (1, 8) | 1 | 9 (c-jun promoter); 4 (exon 7 p53) |
| NTS | 6 (2, 16) | 1 | 7 (exon 5 p53); 9 (exon 8 p53) | |
| E6 lung | TS | 12 (6, 24) | 3.8 ± 1.4 | 9 (c-jun promoter); 4 (exon 7 p53) |
| NTS | 18 (8, 24) | 4.1 ± 1.3 | 7 (exon 5 p53); 9 (exon 8 p53) | |
| E7 lung | TS | 4 (1, 8) | 1.5 ± 0.8 | 9 (c-jun promoter); 4 (exon 7 p53) |
| NTS | 16 (4, 32) | 3.6 ± 1.9 | 5 (exon 5 p53); 5 (exon 8 p53) | |
| E6E7 lung | TS | 13 (4, 28) | 4.1 ± 0.9 | 9 (c-jun promoter); 4 (exon 7 p53) |
| NTS | 17 (6, 32) | 4.1 ± 1.3 | 5 (exon 5 p53); 5 (exon 8 p53) |
Each value reflects the average time required to achieve 50% repair, and was extrapolated from repair-rate determinations for at least 10 different dipyrimidine sites pooled from different gels. For example (as indicated by the first entry in the column at the far right), the average 50% repair time for the TS strand of normal skin fibroblasts was calculated by pooling repair-rate calculations from 21 sites, i.e., 16 from exon 1 of c-jun and 5 from exon 7 of p53. The values in parentheses represent the slowest and fastest 50% repair times, to indicate the site-to-site variation in repair rates.
Results and Discussion
LMPCR is an extremely sensitive genomic-sequencing method because it utilizes a single-sided exponential PCR amplification step. LMPCR can be employed to assess DNA-repair rates in living cells, and allows quantification, at nucleotide resolution in single-copy genes, of any DNA adduct that can be revealed as a ligatable strand break. UV-induced CPDs are easily amenable to LMPCR analysis, as these photoproducts can be efficiently cleaved with extremely high specificity by the enzyme T4-endonuclease V (31), followed by digestion with DNA photolyase to create ligatable 5′ ends at the termini of incised strands (28). We therefore utilized LMPCR to investigate the repair of UVB-induced CPDs in human fibroblasts differing in p53 status.
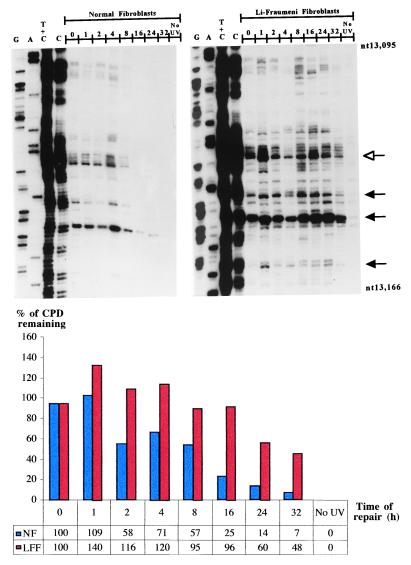
Fig. 1 shows a representative autoradiogram and accompanying histogram that reflect CPD removal along the NTS of the p53 tumor-suppressor gene at different time points up to 32 hr postirradiation in LF041- vs. normal-skin fibroblasts. It should be emphasized that the LF041 strain employed here is the same as that used in previous investigations to demonstrate p53 dependence for GNER, but not for TCNER (18). As expected, our data revealed that most of the CPDs were removed from the NTS strand of the p53 gene in wild-type skin fibroblasts by 24 hr postirradiation, whereas LF041 cells manifested a significant impairment in this respect, i.e., quantification by phosphorimager and averaging of repair rates for 10 different dipyrimidine sites revealed that there was a delay, relative to normal cells, of approximately 3-fold in the time needed to achieve 50% repair (Table 1).
Figure 1.
Repair of UVB-induced CPDs at nucleotide resolution along the NTS of the human p53 tumor-suppressor gene (exon 5, nucleotides 13,095–13,166) in normal human skin fibroblasts (Left) and LF041 skin fibroblasts (Right). The first four lanes from the left on each autoradiogram show LMPCR of DNA treated in standard Maxam–Gilbert cleavage reactions. The following eight lanes show LMPCR of DNA isolated from UVB-irradiated cells that have undergone repair for the indicated times (hr). The far right lane shows LMPCR of unirradiated DNA followed by T4 endonuclease V/photolyase digestion. The shaded arrows indicate dipyrimidine sites quantified with a Fuji BAS 1000 phosphorimager, equipped with the MacBAS v2.5 program. The open arrow indicates a representative dipyrimidine site (i.e., CC site indicated in bold, located within 5′-CCCCCG-3′, codon 151–152) for which the relative repair rate has been graphically illustrated below the autoradiograms. NF, normal fibroblasts; LFF, LF fibroblasts.
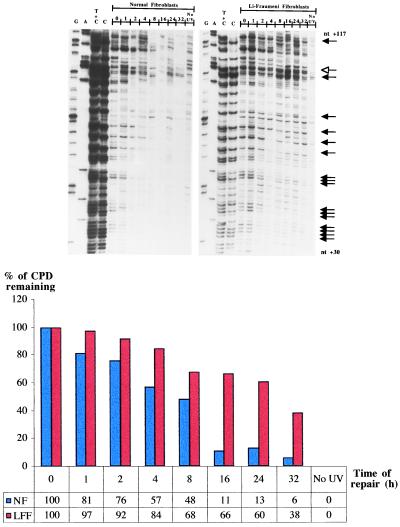
Exon 1 of the c-jun oncogene as well as exon 7 of the p53 gene were then employed as targets to assess CPD removal along the TS of active sequences in LF041 vs. normal fibroblasts. Along the portion of the c-jun gene chosen for analysis (nucleotides +30 to +117), both the TS and NTS strands are repaired at a relatively high rate (32). Visual inspection of the representative autoradiogram in Fig. 2, and the accompanying histogram, show that in wild-type cells most of the CPDs were rapidly removed from the TS of c-jun, i.e., the average time required to achieve 50% repair was approximately 3 hr (Table 1). Strikingly, however, LF041 fibroblasts were profoundly impaired in CPD removal along the TS of c-jun, manifesting an average 50% repair time of 14 hr, i.e., a delay of up to 6-fold compared with wild-type cells (Table 1).
Figure 2.
Repair of UVB-induced CPDs at individual nucleotide positions along the TS of the human c-jun protooncogene (exon 1; nucleotides +30 to +117). The lane designations, arrow indications, and the histogram below the autoradiograms are as for Fig. 1. The representative dipyrimidine site depicted in the histogram is located within 5′-TACTGC-3′ (nucleotides +92 to +97).
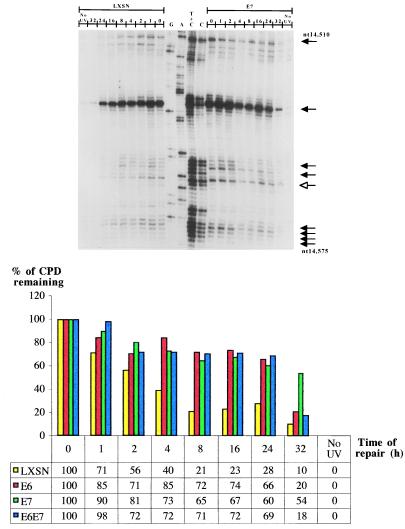
Because the immortalized LF041 and normal primary skin fibroblasts were not isogenic, and to elucidate the role of HPV in tumorigenesis, we investigated repair at different sites along the TS vs. the NTS of the p53 gene in HPV E6-expressing primary lung fibroblasts (which are functionally compromised for p53 protein, but which retain a normal p53 genotype). We also studied repair rates along the TS of the c-jun promoter region (located downstream of the transcription start site; nucleotides −40 to −10), which is very rapidly repaired by TCNER (32). HPV E6-expressing cells manifested extremely low basal levels of p53 protein on Western blots, which did not increase after treatment with ionizing radiation or UVB, unlike the wild-type strain, where p53 protein was strongly up-regulated after DNA damage (data not shown). In agreement with results for genetically p53-deficient LF041 fibroblasts, HPV E6-expressing lung fibroblasts presented, relative to the normal isogenic counterpart, (i) an approximately 4-fold slower rate of repair along the NTS of the p53 gene, and (ii) a similar 4-fold delay in repair along the TS of p53 and of the c-jun promoter region (autoradiograms not shown, Table 1; compare histograms in Figs. 3 and 4).
Figure 3.
Repair of UVB-induced CPDs at nucleotide resolution along the NTS of the human p53 gene (exon 8, nucleotides 14,510–14,575). The lane and arrow designations are as for Figs. 1 and 2, except note that the autoradiograms are presented in an inverse orientation. The accompanying histogram shows repair rates at one representative CC dipyrimidine site quantified by phosphorimager (5′-CACCAC-3′, codon 296–297) for each of LXSN-, HPV E6-, HPV E7-, and HPV E6/E7-expressing lung fibroblasts.
Figure 4.
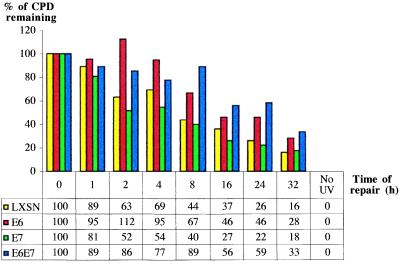
Graphical representation of UVB-induced CPD-repair rates at a representative CC dipyrimidine site (5′-CCTCCG-3′, exon 7, codon 248) on the TS of the human p53 gene for LXSN-, HPV E6-, HPV E7-, and HPV E6/E7-expressing lung fibroblasts.
Taken together, these data demonstrate that p53 deficiency, whether genetically or functionally imposed, engenders a profound defect in TCNER as well as in GNER, as measured along two different target genes (p53 and c-jun), and in cells derived from different tissues (skin and lung). This result is consistent with studies supporting a role for p53 in TCNER, based on, e.g., host-cell reactivation of a reporter gene carried on an adenovirus vector in p53-proficient vs. -deficient cell lines (33, 34). Remarkably, however, our results are in complete disagreement with the persuasive studies cited earlier showing that p53 modulates only GNER, which used a strand-specific DNA-repair assay. While this Southern blot-based assay is certainly capable of detecting differences in TCNER vs. GNER, we emphasize that the LMPCR technique employed here is PCR-based, and, therefore, much more sensitive. In addition, LMPCR measures repair at nucleotide resolution, i.e., at many different sites simultaneously, compared with only one overall measurement at the gene level in the previous studies. Formal explanations for the discrepancy could also be related to the different genetic targets employed (dihydrofolate reductase gene vs. p53 and c-jun) or to the different UV wavelengths used to irradiate cells, i.e., 254-nm UV in the previous studies vs. polychromatic UVB here, as these wavelengths exhibit dissimilar properties with respect to both genotoxicity and patterns of gene activation (35). However, it should be noted that the doses of 254-nm UV and UVB employed in the respective studies, i.e., 10 J/m2 of 254-nm UV compared with 1 kJ/m2 of UVB, would be expected, according to predictions based on UV action spectra in human fibroblasts (36), to induce similar yields of total CPD in the genome. Finally, in complete contrast with our own findings, Wani et al. (37) have recently presented LMPCR data that indicate defective GNER, but normal TCNER, in the same LFS skin fibroblast strain, and using one of the the same target genes (i.e., p53), employed here. Aside from emphasizing that we have presented a much more extensive set of LMPCR results relative to the latter study, we are unable to provide an explanation for this discrepancy.
To explore the possibility that HPV E7 contributes to carcinogenesis by interfering with DNA repair, primary lung fibroblasts expressing this oncoprotein were investigated for rates of CPD removal along the NTS strand of the p53 gene, and along the TS strands of p53 and the c-jun promoter. Levels of pRb were barely detectable by Western blotting in HPV E7-expressing lung fibroblasts, and were at drastically reduced levels relative to wild-type lung fibroblasts (data not shown). An autoradiogram reflecting CPD-repair rates along the NTS of the p53 gene in HPV E7-expressing- vs. wild type-lung fibroblasts is presented in Fig. 3. The accompanying histogram reflects quantification of one representative dipyrimidine site (TT, exon 8 of p53, codon 286) for LXSN-, HPVE6-, E7-, and E6/E7-expressing cells. Averaging of repair rates for at least 10 different sites in each case revealed that HPV E7-expressing cells exhibit an approximate 3.6-fold delay, relative to cells expressing an empty LXSN vector, in the time required to achieve 50% repair along the NTS of p53 (Table 1). However, HPV E7 expression did not significantly affect CPD repair along the TS of either the p53 gene or the c-jun promoter (Fig. 4, and Table 1). Cells coexpressing HPV E6 and HPV E7 exhibited a very similar NER deficiency compared with cells expressing HPV E6 alone (Table1), suggesting that these oncoproteins interfere in the same pathway leading to the global removal of UV-induced CPDs.
While exogenous expression of HPV E6 and HPV E7 has been widely used in cultured cells to inactivate p53 and pRb, respectively, such experiments must be interpreted with caution. Indeed, these viral oncoproteins each interact with a variety of gene products that may contribute to carcinogenesis in a p53/pRb-independent manner (38), e.g., HPV E6-mediated degradation of c-myc (39), activation of telomerase by HPV E6 and HPV E7 (40), and activation of cyclins A and E by HPV E7 (41). Nonetheless, we have presented evidence that HPV E6-mediated functional inactivation of p53 in lung fibroblasts might fully account for the impairment in GNER and TCNER observed in these cells, because essentially the same repair-deficient phenotype was also revealed in genetically p53-deficient LF041 skin fibroblasts.
On the other hand, the precise molecular basis for the observed GNER defect in HPV E7-expressing cells remains to be determined. Because HPV E7 binds and inactivates pRb, as well as the functionally related “pocket proteins” p107 and p130 (42, 43), it is tempting to speculate that pRb (and/or pRb family members) plays a role in GNER. This might occur in a number of ways. For example, both pRb and HPV E7 are known to interact with histone deacetylases (44, 45), and, furthermore, HPV E7 has been shown to associate with the nuclear matrix (46). This raises the interesting speculation that HPV E7-mediated modification of chromatin structure, possibly by means of pRb binding, leads to transcriptional repression of DNA-repair genes, and/or to alterations in repair-protein accessibility to damaged DNA, which may significantly compromise GNER (47). In addition, pRb is known to bind and possibly modulate the activity of topoisomerase II α protein (48), which is clearly involved in DNA replication and recombination (49), and evidently in DNA repair (50, 51). Finally, pRb binds and stimulates the ubiquitous transcription factor AP-1, and HPV E7 abolishes this interaction (52). This abolition may certainly have important consequences with respect to the current findings, because cellular signaling mediated by AP-1 has a major impact on the DNA damage response, i.e., by modulating apoptosis, cell cycle arrest, and possibly DNA repair (53).
The situation is further complicated because HPV E7 influences proteins, other than pRb, that could affect DNA-repair rates. HPV E7 expression leads to elevated basal p53 protein expression (54), as well as up-regulation of the cyclin-dependent kinase inhibitor p21waf1 independently of p53 (55, 56). This may have consequences for CPD removal because p21waf1 may negatively regulate NER (57), although this protein has also been shown to stimulate NER in cultured cells (11). Finally, HPV E7 also interacts with TATA box-binding protein (58, 59), thereby suggesting that various genes involved in GNER might be transcriptionally modulated in HPV E7-expressing cells.
We have presented strong evidence that human cells, either genotypically or functionally compromised for p53 function, manifest defective GNER and TCNER, whereas cells functionally compromised for pRb function are defective only in GNER. Taken together, these findings have clear implications for the mechanism of p53-regulated tumor suppression and also shed light on the molecular pathogenesis of HPV-mediated carcinogenesis. The high-risk HPVs play a major role in the etiology of anogenital carcinomas, especially cervical cancer, and to a lesser (but significant) extent, of other epithelial malignancies, including solar UV-associated nonmelanoma skin cancer (60, 61). Given the critical importance of NER in the prevention of carcinogenesis, our results provide impetus for further investigations (e.g., LMPCR studies on cells derived from pRb family knock-out mice) to identify the precise mechanism underlying the observed GNER defect in UV-exposed, HPV E7-expressing human fibroblasts.
Acknowledgments
We thank Mrs. Nathalie Bastien and Isabelle Paradis for technical assistance, and Drs. R. Stephen Lloyd and Aziz Sancar for kindly supplying T4 endonuclease V and UV photolyase, respectively. This work was supported by grants (held separately by E.A.D. and R.D.) from the National Cancer Institute of Canada (with funds from the Canadian Cancer Society), and from the Medical Research Council of Canada. E.A.D. is a scholar of the Medical Research Council of Canada, and R.D. is a scholar of the Cancer Research Society Inc./Medical Research Council of Canada. C.B. is supported by a graduate student fellowship from the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche (Quebec).
Abbreviations
- LFS
Li–Fraumeni syndrome
- HPV
human papillomavirus
- NER
nucleotide excision repair
- XP
xeroderma pigmentosum
- GNER
global NER
- TS
transcribed strand
- TCNER
transcription-coupled NER
- pRb
retinoblastoma protein
- LMPCR
ligation-mediated PCR
- CPD
cyclobutane pyrimidine dimer
- NTS
nontranscribed strand
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harris C C. Environ Health Perspect. 1996;104:435–439. doi: 10.1289/ehp.96104s3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malkin D. Biochim Biophys Acta. 1994;1198:197–213. doi: 10.1016/0304-419x(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 3.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann H, Degenkolbe R, Bernard H U, O'Connor M J. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Giaccia A J, Kastan M B. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer K H, Lee M M, Scotto J. Carcinogenesis. 1984;5:511–514. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- 8.de Laat W L, Jaspers N G, Hoeijmakers J H. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 9.Smith M L, Chen I T, Zhan Q, O'Connor P M, Fornace A J., Jr Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 10.Smith M L, Chen I T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O'Connor P M, Fornace A J., Jr Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 11.Sheikh M S, Chen Y Q, Smith M L, Fornace A J., Jr Oncogene. 1997;14:1875–1882. doi: 10.1038/sj.onc.1201004. [DOI] [PubMed] [Google Scholar]
- 12.Shivji K K, Kenny M K, Wood R D. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 13.Dutta A, Ruppert J M, Aster J C, Winchester E. Nature (London) 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 14.Coverley D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Nature (London) 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 15.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 16.Ford J M, Baron E L, Hanawalt P C. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 17.Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 18.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang B J, Ford J M, Hanawalt P C, Chu G. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyer S N, Wazer D E, Band V. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 21.Bookstein R, Lee W H. Crit Rev Oncog. 1991;2:211–227. [PubMed] [Google Scholar]
- 22.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 23.Weichselbaum R R, Nove J, Little J B. Proc Natl Acad Sci USA. 1978;75:3962–3964. doi: 10.1073/pnas.75.8.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKay B C, Ljungman M, Rainbow A J. Carcinogenesis. 1999;20:1389–1396. doi: 10.1093/carcin/20.8.1389. [DOI] [PubMed] [Google Scholar]
- 25.Foster S A, Galloway D A. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 26.Drouin R, Rodriguez H, Holmquist G P, Akman S A. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 211–225. [Google Scholar]
- 27.Drouin R, Gao S, Holmquist G P. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 37–43. [Google Scholar]
- 28.Tornaletti S, Pfeifer G P. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 199–209. [Google Scholar]
- 29.Rozek D, Pfeifer G P. Mol Cell Biol. 1993;13:5490–5499. doi: 10.1128/mcb.13.9.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornaletti S, Rozek D, Pfeifer G P. Oncogene. 1993;8:2051–2057. [PubMed] [Google Scholar]
- 31.Gordon L K, Haseltine W A. Radiat Res. 1982;89:99–112. [PubMed] [Google Scholar]
- 32.Tu Y, Tornaletti S, Pfeifer G P. EMBO J. 1996;15:675–683. [PMC free article] [PubMed] [Google Scholar]
- 33.McKay B C, Winrow C, Rainbow A J. Photochem Photobiol. 1997;66:659–664. doi: 10.1111/j.1751-1097.1997.tb03203.x. [DOI] [PubMed] [Google Scholar]
- 34.McKay B C, Francis M A, Rainbow A J. Carcinogenesis. 1997;18:245–249. doi: 10.1093/carcin/18.2.245. [DOI] [PubMed] [Google Scholar]
- 35.Tyrrell R M. BioEssays. 1996;18:139–148. doi: 10.1002/bies.950180210. [DOI] [PubMed] [Google Scholar]
- 36.Rosenstein B S, Mitchell D L. Photochem Photobiol. 1989;45:775–780. doi: 10.1111/j.1751-1097.1987.tb07881.x. [DOI] [PubMed] [Google Scholar]
- 37.Wani M A, Zhu Q Z, El-Mahdy M, Wani A A. Carcinogenesis. 1999;20:765–772. doi: 10.1093/carcin/20.5.765. [DOI] [PubMed] [Google Scholar]
- 38.Garbe J, Wong M, Wigington D, Yaswen P, Stampfer M R. Oncogene. 1999;18:2169–2180. doi: 10.1038/sj.onc.1202523. [DOI] [PubMed] [Google Scholar]
- 39.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias K E, Schwartz A L, Kahana C, Ciechanover A. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greider C W. Trends Genet. 1999;15:109–112. doi: 10.1016/s0168-9525(98)01681-3. [DOI] [PubMed] [Google Scholar]
- 41.McIntyre M C, Ruesch M N, Laimins L A. Virology. 1996;215:73–82. doi: 10.1006/viro.1996.0008. [DOI] [PubMed] [Google Scholar]
- 42.Jones D L, Munger K. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu T S, Ferril S C, Snider A M, Barbosa M S. Int J Oncol. 1995;6:167–174. doi: 10.3892/ijo.6.1.167. [DOI] [PubMed] [Google Scholar]
- 44.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 45.Brehm A, Nielsen S J, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenfield I, Nickerson J, Penman S, Stanley M. Proc Natl Acad Sci USA. 1991;88:11217–11221. doi: 10.1073/pnas.88.24.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smerdon M J, Conconi A. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 48.Bhat U G, Raychaudhuri P, Beck W T. Proc Natl Acad Sci USA. 1999;96:7859–7864. doi: 10.1073/pnas.96.14.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 50.Dresler S L, Robinson-Hill R M. Carcinogenesis. 1987;8:813–817. doi: 10.1093/carcin/8.6.813. [DOI] [PubMed] [Google Scholar]
- 51.Stevnsner T, Bohr V A. Carcinogenesis. 1993;14:1841–5180. doi: 10.1093/carcin/14.9.1841. [DOI] [PubMed] [Google Scholar]
- 52.Nead M A, Baglia L A, Antinore M J, Ludlow J W, McCance D J. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bender K, Blattner C, Knebel A, Iordanov M, Herrlich P, Rahmsdorf H J. J Photochem Photobiol B. 1997;37:1–17. doi: 10.1016/s1011-1344(96)07459-3. [DOI] [PubMed] [Google Scholar]
- 54.Jones D L, Thompson D A, Munger K. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 55.Jones D L, Thompson D A, Suh-Burgmann E, Grace M, Munger K. Virology. 1999;258:406–414. doi: 10.1006/viro.1999.9733. [DOI] [PubMed] [Google Scholar]
- 56.Jian Y, Schmidt-Grimminger D C, Chien W M, Wu X, Broker T R, Chow L T. Oncogene. 1998;17:2027–2038. doi: 10.1038/sj.onc.1202142. [DOI] [PubMed] [Google Scholar]
- 57.Cooper M P, Balajee A S, Bohr V A. Mol Biol Cell. 1999;10:2119–2129. doi: 10.1091/mbc.10.7.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massimi P, Pim D, Banks L. J Gen Virol. 1997;78:2607–2613. doi: 10.1099/0022-1317-78-10-2607. [DOI] [PubMed] [Google Scholar]
- 59.Phillips A C, Vousden K H. J Gen Virol. 1997;78:905–909. doi: 10.1099/0022-1317-78-4-905. [DOI] [PubMed] [Google Scholar]
- 60.zur Hausen H. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 61.Shamanin V, zur Hausen H, Lavergne D, Proby C M, Leigh I M, Neumann C, Hamm H, Goos M, Haustein U F, Jung E G, et al. J Natl Cancer Inst. 1996;88:802–811. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]