Abstract
Inhibitory killer Ig-like receptors (KIR) at the surface of natural killer (NK) cells induced clustering of HLA-C at the contacting surface of target cells. In this manner, inhibitory immune synapses were formed as human NK cells surveyed target cells. At target/NK cell synapses, HLA-C/KIR distributed into rings around central patches of intercellular adhesion molecule-1/lymphocyte function-associated antigen-1, the opposite orientation to mature murine T cell-activating synapses. This organization of protein was stable for at least 20 min. Cells could support multiple synapses simultaneously, and clusters of HLA-C moved as NK cells crawled over target cells. Clustering required a divalent metal cation, explaining how metal chelators inhibit KIR function. Surprisingly, however, formation of inhibitory synapses was unaffected by ATP depletion and the cytoskeletal inhibitors, colchicine and cytochalsins B and D. Clearly, supramolecular organization within plasma membranes is critical for NK cell immunosurveillance.
Natural killer (NK) cells express a variety of inhibitory killer Ig-like receptors (KIR) that inhibit cytotoxicity upon recognition of class I MHC proteins (1). In this manner NK cells detect diseased cells through their loss of expression of self-MHC protein rather than by directly detecting foreign antigen, a conjecture known as the missing self-hypothesis (2). Inhibitory NK receptors containing two Ig domains, denoted KIR1 (or KIR2DL1) and KIR2 (or KIR2DL2), recognize the class I MHC proteins, HLA-Cw4 or -Cw6 and HLA-Cw3 or -Cw7, respectively (3, 4). Crystal structures of both class I MHC and KIR extracellular domains have been determined (5–9), and the appropriate binding sites have been mapped by site-directed mutagenesis (10–15). Although the binding kinetics between soluble KIR/MHC proteins, determined by surface plasmon resonance, are extremely fast (16–18), video microscopy of NK cell immunosurveillance shows that intercellular contacts last for minutes (not shown). Thus, we set out to delineate the molecular mechanisms of NK cell recognition that occur over this time frame.
An enhanced variant of green fluorescent protein (EGFP) (19), originally discovered and cloned from Aequorea victoria jellyfish (20, 21), was used to mark the location of HLA-C. Plasmids encoding EGFP attached to the intracellular C terminus of class I MHC protein were transfected into 721.221, a B cell line derived by mutagenesis that does not express class I MHC protein (22, 23). These transfectants then were incubated with various NK cell lines for 20 min at 37°C, after which time many NK cell/target cell conjugates were formed. Conjugates of living NK and target cells were imaged by laser-scanning confocal fluorescence microscopy. This methodology advances previous imaging of mouse T cell/target cell intercellular contacts that used paraformaldehyde-fixed cells (24) or live T cells interacting with MHC protein-rich lipid bilayers (25). Here, immune synapses are shown to exist at the contact between two living human cells.
Materials and Methods
Cell Lines and Transfectants.
Plasmids encoding EGFP attached to the C terminus of HLA-Cw3 or -Cw4 were prepared by PCR of the appropriate HLA-C allele to remove the stop codon and add an EcoRI restriction site at the 5′ end and a NotI site at the 3′ end. EGFP was prepared by PCR from the plasmid pEGFP (CLONTECH) to contain a NotI site at the 5′ end and a BamHI site at the 3′ end. These PCR products then were joined and amplified together by PCR by using primers at the 5′ end of HLA-C and the 3′ end of EGFP. The product was cloned first into pBABE and then into pcDNA3 (Invitrogen). Plasmids encoding other GFP-linked proteins were prepared by PCR of the appropriate HLA-C allele to remove the stop codon and cloned as KpnI/NotI fragments into the vector initially encoding HLA-Cw3-GFP. All primers were purchased from Life Technologies (Gaithersburg, MD) and all plasmid inserts were sequenced by the Core Facilities, Dana–Farber Cancer Institute (Boston, MA). 721.221 cells were transfected with 100 μg of linearized plasmids by electroporation, and peripheral blood NK cell lines were prepared from healthy donors as described (11). The correct molecular weight of GFP attached to HLA-C was checked by analyzing cell lysates by SDS/PAGE and Western blotting by using a mAb to GFP (CLONTECH).
Laser-Scanning Confocal Microscopy.
NK cells and target cells (3 × 106 of each) were mixed together in 6 ml of medium, briefly centrifuged at 1,500 rpm to bring cells into contact, and incubated for 20 min at 37°C in 5% CO2. Cells then were resuspended in ≈50 μl of tissue culture medium and ≈7 μl sealed between a glass slide and 0.17-mm coverslip. Slides were imaged immediately by laser-scanning confocal microscopy (LSM 410; Zeiss) by using a ×100 objective. For counting the percentage of clustering, slides were scanned by Nomarski imaging and cells that clearly were conjugated were examined for their fluorescence distribution. Results in all figures and Tables are representative of at least three independent experiments.
ATP Depletion and Inhibition of Actin- or Tubulin-Mediated Cytoskeletal Movement.
When required, both NK cells and target cells were preincubated with additional reagents for the times indicated. For d-glucose, 2-deoxyglucose, and antimycin-A, cells were first spun down and resuspended in glucose-free RPMI 1640 medium (Life Technologies). After such preincubation, cells were mixed together, centrifuged briefly, and incubated for a further 20 min, again in the presence of the additional reagents. Stock solutions of reagents (all purchased from Sigma unless stated) were used as follows: 500 mM 1,10-phenanthroline or 1,7-phenanthroline (Aldrich) in ethanol, 9.5 mM NNN′N′-tetrakis-(2-pyridylmethyl)ethylenediamine in ethanol, 18.5 mM antimycin-A in DMSO, 125 mM rotenone in chloroform, 270 μM 2,4-dinitrophenol in chloroform, 3 M sodium azide in aqueous solution, 50 mM d-glucose or 2-deoxyglucose in glucose-free RPMI 1640, 4 mM cytochalasin B or D in DMSO, and 25 mM colchicine in DMSO. ATP depletion was measured in parallel to the imaging by assaying luciferase activity in the presence of cell lysates according to the manufacturer's instructions (ATPLite-M; Packard).
Staining Cells with mAb.
EB6 and HP3E4, mAbs to KIR1, were purchased from Immunotech or were a kind gift from M. Lopez-Botet (Hospital Universitario de la Princesa, Madrid, Spain). IgG1 mAb EB6, HA58, and HI111 (PharMingen) against KIR1, intercellular adhesion molecule-1 (ICAM-1) (CD54), and lymphocyte function-associated antigen-1 (LFA-1) (CD11a), respectively, were labeled with Alexa 568 according to the manufacturer's instructions (Alexa 568 Protein Labeling Kit; Molecular Probes). Labeled mAb EB6 (50 μg/ml) was used to stain YTS/KIR1 and 221/Cw6-GFP, preincubated together for 20 min at 37°C in 5% CO2, according to the manufacturer's instructions (Cytofix/Cytoperm kit; PharMingen). Labeled mAb HA58 (1 μg/ml) was used to stain 1.5 × 106/ml 221/Cw4-GFP cells before incubation with YTS/KIR1. Labeled mAb HI111 (1 μg/ml) was used to label 1.5 × 106/ml YTS/KIR1 cells before incubation with 221/Cw6-GFP.
KIR-Ig Coated Beads.
KIR1-Ig was covalently coupled to 10-μm carboxylated microspheres (Polybead; Polysciences) by using carbodiimide (Carbodiimide kit; Polysciences). Coated beads (2 × 107) were incubated with 5 × 106 221, 221/Cw3-GFP, or 221/Cw4-GFP cells in 5 ml of media for 20 min at 37°C and prepared for microscopy.
Results
KIR Induce Clustering of HLA-C.
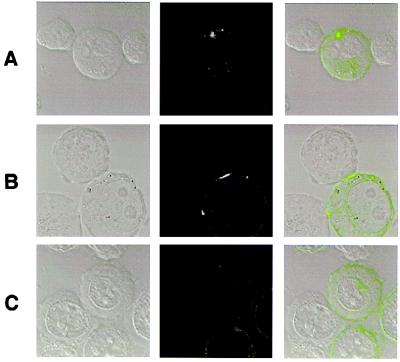
A peripheral blood NK line, 484, expressing both KIR1 and KIR2 by flow cytometry, was incubated with the 721.221 transfectants. Clusters of GFP-linked HLA-Cw3 (Fig. 1A; Table 1) or HLA-Cw4 (Table 1) were clearly seen in about 70% of cell conjugates. A single NK cell was able to simultaneously cause clustering of MHC protein on several target cells, and one target cell could simultaneously present clusters of MHC protein at multiple NK cell contacts (Fig. 1A). Also, one NK cell may simultaneously bind cells expressing different MHC alleles and form an MHC cluster with none, one, or many (not shown). This may provide the molecular basis for the fact that NK cells are unable to be inhibited “in trans” (3), i.e., the inhibitory effect of KIR may be local within the cell, only at the synapse.
Figure 1.
KIR induce clustering of HLA-C. Various transfectants of 721.221 were imaged when conjugated to peripheral blood NK cells or YTS/KIR1. The first five rows show the Nomarski image of a cell conjugate, the corresponding fluorescence, and an overlay of the fluorescence (colored green for GFP or red for Alexa 594) and Nomarski fields. The target cells are those highlighted by green or red fluorescence. Rows show conjugates of peripheral blood NK line 484 (small cells) and 221/Cw3-GFP (larger cells) (A), YTS/KIR1 and 221/Cw3-GFP (B), YTS/KIR1 and 221/Cw4-GFP (C), YTS/KIR1 and 221/Cw4-GFP in the presence of 50 μg/ml mAb EB6 (anti-KIR1) (D), 221-Cw6, labeled with fluorescent β2m, and YTS/KIR1 (E). (F) YTS/KIR1 and 221/Cw6-GFP, preincubated together, then fixed and permeabilized, stained with 50 μg/ml Alexa-568-conjugated mAb EB6 (red), the corresponding GFP fluorescence (green), and an overlay of the fluorescence fields over the Nomarski image in which colocalized green and red staining is colored yellow. (G) 221/Cw4-GFP conjugated to a peripheral blood NK cell after 0, 10, and 20 min.
Table 1.
KIR induce clustering of HLA-C
| Cell conjugates (target cell + effector cell) | n, number of conjugates imaged | % clustering |
|---|---|---|
| 221/Cw3-GFP + NK line 484 (KIR1+, KIR2+) | 52 | 73 |
| 221/Cw4-GFP + NK line 484 (KIR1+, KIR2+) | 11 | 72 |
| 221/Cw3-GFP + YTS/KIR1 | 102 | 5 |
| 221/Cw4-GFP + YTS/KIR1 | 335 | 71 |
| 221/Cw6-GFP + YTS/KIR1 | 294 | 74 |
| 221/Cw6-GFP + YTS/KIR1 + mAb EB6 | 103 | 7 |
| 221/Cw6-GFP + YTS/KIR1 + control IgG1 mAb | 63 | 76 |
| 221/Cw6-TMG-GFP + YTS/KIR1 | 115 | 70 |
| 221/Cw6-C321Y-GFP + YTS/KIR1 | 94 | 70 |
Various transfectants of 721.221 were imaged when conjugated to peripheral blood NK cells or YTS/KIR1 as listed. The percentage of cell conjugates for which immune synapses formed at the intercellular contact is shown. Where stated, mAbs were added at 50 μg/ml.
To determine whether the clustering of class I MHC protein was orchestrated specifically by NK receptors, YTS, an NK tumor line that expresses no known KIR, was transfected to express NK receptors by retroviral infection (26). YTS expressing KIR1 (YTS/KIR1) triggered intercellular clustering of GFP-linked HLA-Cw4 or HLA-Cw6, but not HLA-Cw3 (Fig. 1 B and C; Table 1). The diameter of HLA-C clusters ranged from 3 to 6 μm, approximately half the diameter of the intercellular contact. Blocking KIR1 with mAb EB6 abrogated clustering of HLA-Cw6 (Fig. 1D; Table 1). Thus, the clustering of distinct class I MHC alleles is controlled by the specificity of the NK receptors expressed.
YTS/KIR1 was able to cause clustering of GFP-linked HLA-Cw6 in which the transmembrane sequence was replaced with that of HLA-G (221/Cw6-TMG-GFP, Table 1), which lacks a critical transmembrane cysteine. Thus, although recognition of HLA-Cw6 by a subset of NK cells is dependent on a cysteine within the transmembrane sequence (Cys309) (27), clustering of HLA-Cw6 by KIR1 is independent of this substitution. Similarly, YTS/KIR1 efficiently clustered HLA-Cw6 in which the cysteine residue in the cytoplasmic tail unique to HLA-C (Cys321) was replaced with tyrosine, the corresponding residue in HLA-A or -B (221/Cw6-C321Y-GFP, Table 1).
A mutant of β2-microglobulin (β2m) was prepared that could be labeled with a single red fluorophore (Alexa 594 maleimide; Molecular Probes) at a site distal to its binding site with the class I MHC heavy chain (28–30). Fluorescent images of Alexa 594-conjugated β2m exogenously exchanged with its natural counterpart were dim compared with those of GFP-linked HLA-C because only low amounts of β2m readily exchange at the cell surface. Nevertheless, clusters of Alexa 594-conjugated β2m were clear at intercellular contacts between 721.221/Cw6 and YTS/KIR1 (Fig. 1E). This novel and minimally invasive method proves that clustering of HLA-C at the target/NK cell interface is independent of GFP attachment and other potential sources of artifact such as mAb cross-linking. Clusters of class I MHC were also evident by labeling with fluorescein-conjugated mAb G46–2.6 (PharMingen), which recognizes HLA-C but does not efficiently block its interaction with KIR (data not shown).
Because mAb EB6 blocks the clustering of HLA-C by KIR, it could not be used to reveal the location of KIR at the interface of live cell conjugates. Instead, cells were incubated together to allow synapse formation and subsequently were fixed with formaldehyde before staining with fluorophore-conjugated anti-KIR mAb EB6 in the presence of a mild detergent (see Materials and Methods). This methodology allowed clustering of KIR1 to be detected on the NK cell (Fig. 1F, red field). Superposition of the red and green fluorophores gave a yellow band at the interface, clearly demonstrating the overlap of clustered KIR and HLA-C protein at the intercellular contact (Fig. 1F).
Clusters of class I MHC protein followed the intercellular contact as NK cells moved about the surface of the target cell (Fig. 1G). Such extensive immunosurveillance by NK cells may be required when surveying cells that express class I MHC protein in patches at the cell surface or at a pole.
Supramolecular Organization of the Human NK Cell Immune Synapse.
Images within the plane of intercellular contact were taken by motorized stepping of the objective lens at 0.05-μm intervals while the illuminating laser scanned a plane bisecting the flattened intercellular contact. This method showed a uniform distribution of GFP-linked class I MHC protein at the edge of an unconjugated cell (not shown) or at the contact between 221/Cw4-GFP and untransfected YTS (Fig. 2A). However, a ring of class I MHC protein was clearly visible in intercellular contacts between 221/Cw4-GFP and YTS/KIR1 (Fig. 2A). A red membrane dye, DiD (Molecular Probes), showed a uniform distribution at the interface, demonstrating that the apparent ring of class I MHC protein was not caused by the shape of the plasma membrane at the contact (Fig. 2A). Using Alexa 568-conjugated mAb HA58, ICAM-1 (CD54) was found to concentrate at the center of the class I MHC ring (Fig. 2A). mAb HI111, which binds LFA-1 but does not block its interaction with its ligand ICAM-1, also moves to a central patch within the ring of MHC protein (Fig. 2B). mAb EB6, used to stain KIR1 in fixed and permeabilized cell conjugates, reveals that KIR organize into a ring-shaped structure opposite the MHC protein at the intercellular synapse. Thus, at the synapse, the distributions of MHC and ICAM-1 proteins on the target cell correspond exactly to their receptors, KIR1 and LFA-1, on the NK cell side of the interface.
Figure 2.
Supramolecular organization of the human NK cell immune synapse. Images within the plane of the intercellular contact were taken by laser-scanning confocal microscopy. (A) Distribution of GFP-linked HLA-Cw4 at the contact between 221/Cw4-GFP untransfected both YTS and YTS/KIR1, as well as the intercellular distribution of DiD (a membrane dye) and a mAb to ICAM-1 at the contact between 221/Cw4-GFP and YTS/KIR1. Also shown is the HLA-Cw4/GFP (green) and ICAM-1 (red) distribution overlaid. (B) Intercellular distribution of mAb to LFA-1 or KIR1 on the NK cell (red) in relation to the HLA-C/GFP (green) on the target cell. Superposition of the red and green fluorescence, i.e., where KIR1 and HLA-C staining overlap, is colored yellow. (C) Distribution of HLA-Cw4/GFP and ICAM-1 over 22 min. The diameter of the MHC ring corresponds to the width of MHC clusters at intercellular contacts in Fig. 1.
Strikingly, this intercellular arrangement of class I MHC protein/KIR protein and ICAM-1/LFA-1 is inverted compared with the distribution of class II MHC protein and ICAM-1 at the mature mouse T cell/target cell interface (24). Instead, this distribution of class I MHC protein and ICAM-1 is like the transient arrangement seen at the nascent contact between a mouse T cell and lipid bilayer, lasting up to 5 min (25). Here, the arrangement of MHC protein and ICAM-1 at the contact between live human peripheral blood NK cells and B cell transfectants was stable for at least 22 min (Fig. 2C). After 22 min, the fluorescence from both GFP and Alexa 594 was bleached significantly. It will be interesting to determine whether the NK-activating receptor(s) and their ligand(s) colocalize in the ring with KIR1 and HLA-C or in the center with LFA-1 and ICAM-1 under different conditions, when the components of the NK cell lysis receptor/ligand system(s) have been fully identified.
Formation of the Human NK Cell Immune Synapse Requires a Divalent Metal Cation.
Addition of the metal ion chelator 1,10-phenanthroline, but not its isomer, 1,7-phenanthroline, abrogated the clustering of HLA-C at the NK cell/target cell synapse. Inclusion of an excess of Zn2+ or Cu2+, but not Mg2+ ions, restored MHC clustering in the presence of 1,10-phenanthroline (Fig. 3; Table 2). Addition of another chelator with similarly high affinity for Zn2+ or Cu2+, NNN′N′-tetrakis-(2-pyridylmethyl)ethylenediamine, also abrogated MHC clustering (Table 2). Clearly, a divalent transition metal cation, presumably Zn2+ physiologically, is required for clustering of HLA-C by KIR.
Figure 3.
Clustering of HLA-C by YTS/KIR1 is controlled by metal ions. Rows show conjugates of YTS/KIR1 and 221/Cw4-GFP in the presence of 1 mM 1,10-phenanthroline (A), 1 mM 1,10-phenanthroline and 3 mM ZnCl2 (B), and 1 mM 1,10-phenanthroline and 3 mM MgSO4 (C). For each condition, the Nomarski image of a cell conjugate, the corresponding fluorescence, and an overlay of the fluorescence and Nomarski fields are shown.
Table 2.
Clustering of Cw4/GFP by YTS/KIR1 is controlled by metal ions
| Chelator added | Salt added, 3 mM | n, number of conjugates imaged | % clustering |
|---|---|---|---|
| 1 mM 1,10-phenanthroline | None | 180 | 23 |
| 1 mM 1,10-phenanthroline | ZnCl2 | 144 | 64 |
| 1 mM 1,10-phenanthroline | MgSO4 | 20 | 20 |
| 1 mM 1,10-phenanthroline | CuSO4 | 18 | 67 |
| 1 mM 1,7-phenanthroline | None | 17 | 59 |
| 70 μm TPEN | None | 24 | 20 |
Listed is the number of 221/Cw4-GFP cells with a cluster of HLA-C at the intercellular contact with YTS/KIR1 in the presence of metal chelators and salts. TPEN, N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine.
Previously, removal of the metal ion-binding site or addition of 1,10-phenanthroline was shown not to effect the binding of KIR to HLA-C, yet did impair signal transduction (31, 32). Now, these results can be explained. Metal ion chelation is required for MHC clustering by KIR, and such clustering is required for the initiation of signal transduction by KIR. Because MHC clustering is an integral part of T cell recognition (24, 25, 33, 34), NK cells could kill targets containing a viral product that interfered with MHC clustering.
Formation of the Human NK Cell Immune Synapse Requires Neither ATP nor Actin- or Tubulin-Mediated Cytoskeletal Movement.
Treatment with standard metabolic inhibitors, namely, azide, 2-deoxyglucose, antimycin-A, rotenone, and 2,4-dinitrophenol, depleted the intracellular ATP in NK cell/target cell mixtures between 43% and 97% (Table 3). The ATP-dependent process of internalizing fluorophore-conjugated transferrin was inhibited by azide (not shown). However, none of these poisons affected the number of immune synapses formed (Fig. 4; Table 3). Furthermore, neither cytochalasin B, cytochalasin D, nor colchicine had any effect on the number of synapses formed. Cytochalasins B and D affect actin filaments and colchicine inhibits tubulin polymerization. Thus, neither ATP nor tubulin- or actin-mediated cytoskeletal movement is necessary for clustering of HLA-C by KIR. It is possible that ATP or tubulin- or actin-mediated cytoskeletal movement is required for the MHC/ICAM-1 distribution to invert, as seen at the mature T cell immune synapse (24, 25). It is also possible that the number of cell conjugates formed, or the absolute amount of protein clustered, may be affected by metabolic or cytoskeletal inhibitors.
Table 3.
Neither ATP nor tubulin- or actin-mediated cytoskeletal movement is required for HLA-C clustering by KIR
| Compound added | Concentration | Time of incubation, hr | n, number of conjugates imaged | % clustering | % ATP depletion |
|---|---|---|---|---|---|
| d-glucose | 50 mM | 2 | 153 | 72 | 0 |
| 2-Deoxyglucose | 50 mM | 2 | 182 | 72 | 89 |
| Azide | 50 mM | 2 | 137 | 79 | 94 |
| Antimycin-A | 13 μM | 1 | 175 | 66 | 97 |
| 2,4-Dinitrophenol | 450 μM | 1 | 88 | 75 | 44 |
| Rotenone | 25 μM | 1 | 111 | 68 | 43 |
| Colchicine | 10 μM | 1 | 42 | 78 | N/A |
| Cytochalasin B | 10 μM | 1 | 83 | 75 | N/A |
| Cytochalasin D | 10 μM | 1 | 183 | 66 | N/A |
Table shows the number of 221/Cw6-GFP cells with a cluster of GFP fluorescence at the contact with YTS/KIR1 in the presence of metabolic or cytoskeletal inhibitors. The extent of ATP depletion is shown for each metabolic inhibitor. By calibrating the luciferase assay with known ATP concentrations, the total amount of ATP in 50 × 103 of untreated NK and target cells (25 × 103 of each) was about 2 × 10−10 moles. The time of incubation indicated refers to the time that each cells type was preincubated before the cells were mixed together. Cells then were incubated together for a further 20 min in the presence of the compounds. “% clustering” refers to the number of target cells that have a cluster of HLA-C at an NK cell contact. These data do not preclude the possibility that the number of cell conjugates formed or the quantity of clustered protein is affected by the compounds used. N/A, not applicable.
Figure 4.
The number of NK cell/target cell immune synapses is unaffected by the presence of metabolic or cytoskeletal inhibitors. The first six rows show conjugates of YTS/KIR1 and 221/Cw6-GFP in the presence of 50 mM d-glucose (A), 50 mM azide (B), 13 μM antimycin-A (C), 50 mM 2-deoxyglucose (D), 50 mM 2-deoxyglucose (E), and 10 μM cytochalasin D (NK cell adopts a cone shape in the presence of cytochalasin D) (F). (G) KIR-Ig-coated beads attached to 221/Cw4-GFP. Each row, except E, shows the Nomarski image of a cell conjugate, the corresponding fluorescence, and an overlay of the fluorescence and Nomarski fields. E shows the Nomarski image of a cell conjugate, the corresponding fluorescence image, and the image of the cluster within the plane of the interface (in green box), demonstrating that the ring-like organization of HLA-C also is retained in the absence of ATP.
Beads (10 μm) Coated with KIR-Ig Cannot Trigger Clustering of HLA-C.
Beads (10 μm) were coated with a chimeric protein consisting of the extracellular portion of KIR1 attached to the Fc portion of IgG Ab (35). By flow cytometry, such beads stained with mAb HP3E4 (anti-KIR1) about two orders of magnitude greater than YTS/KIR1 (not shown). However, although these beads efficiently attached themselves to 221/Cw4-GFP (and not to 221/Cw3-GFP or untransfected 221), they did not facilitate clustering of HLA-C (Fig. 4G). Thus, although ATP and tubulin- or actin-mediated cytoskeletal movement are not required, a solid-phase support for KIR alone is not sufficient for HLA-C clustering. Thus, consistent with the superposition of proteins at the synapse (Fig. 2B), the NK cell immune synapse probably is formed, at least in part, by passive diffusion of both MHC and KIR proteins within the plasma cell membrane.
Discussion
This demonstration of an inhibitory synapse between living cells firmly establishes immune synapses as broadly significant in molecular recognition between cell surfaces. That neither ATP nor tubulin- or actin-mediated cytoskeletal movement is required for formation of the NK cell immune synapse suggests that organization of membrane microdomains, protein/lipid interactions, and receptor oligomerization are critical determinants of intercellular recognition, at least for NK cells. Pharmaceutical intervention of specific intercellular clustering may be of use therapeutically, e.g., to enhance NK killing of HIV-infected or tumor cells by inhibition of HLA-C clustering. Rapid assays of intercellular protein clustering, perhaps by using spectroscopic techniques (e.g., refs. 29 and 36), must be developed before large chemical libraries can be screened.
For understanding NK cell immunosurveillance, it remains to be determined how formation of the NK cell immune synapse described here intersects with known biochemical signal transduction pathways (37). Once again, as a professor of physiology in Göttingen wrote 165 years ago, “the great improvement in microscopes and the growing interest of younger observers will shortly throw new light on this subject” (38).
Acknowledgments
We thank Guido Guidotti, Christoph Wülfing, Brent Stockwell, Pratap Malik, and Jonathan Boyson for discussion and Dave Smith and Robert Erskine for technical assistance. This work is supported by National Institutes of Health Grant CA-47554 (J.L.S.) and postdoctoral fellowships from The Irvington Institute (D.M.D.) and The Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship, Grant No. DRG 1454 (O.M.).
Abbreviations
- KIR
inhibitory killer Ig-like receptors
- NK
natural killer
- β2m
β2-microglobulin
- GFP
green fluorescent protein
- ICAM-1
intercellular adhesion molecule-1
- LFA-1
lymphocyte function-associated antigen-1
- EGFP
enhanced variant of GFP
References
- 1.Long E O. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren H-G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Colonna M, Brooks E G, Falco M, Ferrera G B, Strominger J L. Science. 1993;260:1121–1124. doi: 10.1126/science.8493555. [DOI] [PubMed] [Google Scholar]
- 4.Colonna M, Borsellino G, Falco M, Ferrera G B, Strominger J L. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 6.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 7.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;390:315. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 8.Fan Q R, Wiley D C. J Exp Med. 1999;190:113–123. doi: 10.1084/jem.190.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maenaka K, Juji T, Stuart D I, Jones E Y. Structure. 1999;7:391–398. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- 10.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, Conte R, Di Donato C, Parham P, Moretta L. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandelboim O, Reyburn H T, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger J L. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelboim O, Reyburn H T, Sheu E G, Vales-Gomez M, Davis D M, Pazmany L, Strominger J L. Immunity. 1997;6:341–350. doi: 10.1016/s1074-7613(00)80336-2. [DOI] [PubMed] [Google Scholar]
- 13.Winter C C, Long E O. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 14.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 15.Winter C C, Gumperz J E, Parham P, Long E O, Wagtmann N. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 16.Vales-Gomez M, Reyburn H T, Mandelboim M, Strominger J L. Immunity. 1998;9:337–344. doi: 10.1016/s1074-7613(00)80616-0. [DOI] [PubMed] [Google Scholar]
- 17.Vales-Gomez M, Reyburn H T, Mandelboim M, Strominger J L. Immunity. 1998;9:892. doi: 10.1016/s1074-7613(00)80616-0. [DOI] [PubMed] [Google Scholar]
- 18.Vales-Gomez M, Reyburn H T, Erskine R A, Strominger J L. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kain S R. In: Fluorescent and Luminescent Probes for Biological Activity. 2nd Ed. Mason W T, editor. New York: Academic; 1999. pp. 284–292. [Google Scholar]
- 20.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 21.Chalfie M, Kain S, editors. Green Fluorescent Protein: Properties, Applications and Protocols. New York: Wiley; 1998. [Google Scholar]
- 22.Shimizu Y, Geraghty D E, Koller B H, Orr H T, DeMars R. Proc Natl Acad Sci USA. 1988;85:227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu Y, DeMars R. J Immunol. 1989;142:3320–3228. [PubMed] [Google Scholar]
- 24.Monks C R, Freiberg B A, Kupfer H, Sciaky N, Kupfer A. Nature (London) 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 25.Grakoui A, Bromley S K, Sumen C, Davis M M, Shaw A S, Allen P M, Dustin M L. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 26.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 27.Davis D M, Mandelboim O, Luque I, Baba E, Boyson J, Strominger J L. J Exp Med. 1999;189:1265–1274. doi: 10.1084/jem.189.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis D M, Reyburn H T, Pazmany L, Chiu I, Mandelboim O, Strominger J L. Eur J Immunol. 1997;27:2714–2719. doi: 10.1002/eji.1830271035. [DOI] [PubMed] [Google Scholar]
- 29.Gakamsky D M, Davis D M, Haas E, Strominger J L, Pecht I. Biophys J. 1999;76:1552–1560. doi: 10.1016/S0006-3495(99)77314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gakamsky D M, Boyd L M, Margulies D H, Davis D M, Strominger J L, Pecht I. Biochemistry. 1999;38:12165–12173. doi: 10.1021/bi9905821. [DOI] [PubMed] [Google Scholar]
- 31.Rajagopalan S, Long E O. J Immunol. 1998;161:1299–1305. [PubMed] [Google Scholar]
- 32.Rajagopalan S, Winter C C, Wagtmann N, Long E O. J Immunol. 1995;155:4143–4146. [PubMed] [Google Scholar]
- 33.Wülfing C W, Davis M M. Science. 1998;282:2266–2269. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- 34.Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. Science. 1999;283:680–682. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
- 35.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 36.De Angelis D A, Miesenbock G, Zemelman B V, Rothman J E. Proc Natl Acad Sci USA. 1998;95:12312–12316. doi: 10.1073/pnas.95.21.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolland S, Ravetch J V. Adv Immunol. 1999;72:149–177. doi: 10.1016/s0065-2776(08)60019-x. [DOI] [PubMed] [Google Scholar]
- 38.Wagner R. Lehrbuch der Vergleichenden Anatomie. Leipzig, Germany: Voss; 1834–1835. , quoted in Harris, H. (1999) The Birth of the Cell (Yale Univ. Press, New Haven, CT). [Google Scholar]