Abstract
Thymocytes and thymic dendritic cell (DC) lineages develop simultaneously and may originate from a common intrathymic progenitor. Mice deficient for two growth factor receptor molecules [c-kit and the common cytokine receptor γ chain (γc)] lack all thymocytes including T cell progenitors. Despite this lack of pro-T cells, thymic DC compartments were identified in c-kit−γc− mice. Thus, c-kit- and γc-mediated signals are not essential to generate thymic DCs. In addition, pro-T cells do not appear to be obligatory progenitors of thymic DCs, because DC development is dissociated from the generation of thymocytes in these mice. Thymic DCs in c-kit−γc− mice are phenotypically and functionally normal. In contrast to wild-type mice, however, thymic DCs in c-kit−γc− and, notably, in RAG-2-deficient mice are CD8αneg/low, indicating that CD8α expression on thymic DCs is not independent of thymocytes developing beyond the “RAG-block.”
Thymic dendritic cells (DCs) reside in the medulla and at the cortico-medullary junction, where they display antigenic peptides to T cells undergoing repertoire selection. In the adult thymus, DCs account for ≈0.1% of all hematopoietic cells. DCs are of bone marrow origin, and they are commonly assigned to the myeloid lineage. DCs can be generated in vitro from bone marrow or peripheral blood-derived progenitors under myeloid growth conditions, e.g., in the presence of granulocyte/monocyte colony-stimulating factor, tumor necrosis factor-α, or IL-3, indicating that at least some DCs are myeloid cells (reviewed in refs. 1–3). An additional view has been proposed, i.e., that subsets of DCs might be closely related to or descendants of the lymphoid rather than the myeloid lineage. In support of this idea, adoptively transferred intrathymic progenitors can generate donor-type thymic and splenic DCs. Moreover, T and DC lineages are turned over in parallel (“linked development”) (4, 5). Lymphoid progenitors, and mature T and DC lineages share several markers; for instance, both pro-T cells and thymic DCs express Sca-1, Sca-2, and, notably, c-kit. Like some T cells, subsets of thymic and splenic DCs express CD8α (reviewed in refs. 6 and 7). Thus, DC populations can be phenotypically heterogeneous, and DC subtypes may represent distinct lineages and/or functional stages.
Null mutations in the genes for both c-kit and the common cytokine receptor γ chain (γc) completely abrogate thymocyte development. In these mutants, thymocytes were undetectable by flow cytometry and by histology (8).
Here, we show that pro-T cell populations defined by expression of c-kit and CD25 are present only in single-mutant (c-kit−γc+ or c-kit+γc−) but are missing from double-mutant (c-kit−γc−) thymi. Given that T lineage cells were undetectable, we searched for other hematopoietic cells in c-kit−γc− thymi. Notably, thymic DCs were identified as the major and almost exclusive hematopoietic cell type present in these mutants. Here, these thymic DC populations are characterized in terms of cell number, phenotype, intrathymic location, and function.
Materials and Methods
Mice.
Mice lacking c-kit (9), γc (10), or c-kit + γc were genotyped as previously described (8). All experiments were performed on postnatal 5-day-old mice because c-kit− mice die ≈10 days after birth because of severe anemia (9).
mAbs and Flow Cytometry.
mAbs used were: HL3 (anti-CD11c), 53–6.7 (anti-CD8α), 3C7 (anti-CD25), R35–95 (rat IgG2a isotype control), and 104–2.1 (anti-CD45) (all from PharMingen); M1/70.15 (anti-Mac-1) and 5a-8 (anti-Thy-1) (both from Caltag, South San Francisco, CA); and ACK-4 (anti-c-kit) (11). Second-step reagent was streptavidin-APC (Molecular Probes). When required, propidium iodide was used at 5 μg/ml to gate out dead cells. Cells were stained and analyzed [FACStar (Becton Dickinson)] or sorted [FACSCalibur (Becton Dickinson)] as previously described (8).
Histological Analysis.
Immunohistology was performed as previously described (12). Primary mAbs used were MTS10 (undiluted supernatant) (13) and phycoerythrin-labeled HL3 (1:50 dilution). Second-step reagent was goat-anti-rat IgG (1:50 dilution) (Southern Biotechnology Associates).
Isolation of Thymic DCs.
Thymi were digested with collagenase dispase (Roche Molecular Biochemicals) (1 mg/ml)/DNase I (Sigma) (0.5 mg/ml) in PBS/5% FCS for a total of 60 min rotating at 37°C with the addition of fresh enzyme after 30 min. Cell surface expression of CD45 and CD11c was resistant to this treatment. For analysis of CD8α expression, thymus tissues were digested with collagenase D (Roche) (1 mg/ml) with DNase I. In contrast to collagenase dispase treatment, collagenase D digestion does not affect CD8α expression (data not shown).
To measure absolute numbers of DCs, each thymus was weighed and digested individually as described above, and CD45+CD11c+ cells were counted with a fluorescence-activated cell sorter (FACS, Becton Dickinson) by collecting data from 106 total cells or from the entire sample from each mouse (corresponding to all DCs from one thymus).
Mixed Leukocyte Reactions.
Cell sorter-purified C3H lymph node T cells (2 × 105; CD4+Thy-1+ and CD4−Thy-1+ corresponding to CD4+CD8− and CD4− CD8+ T cells) were cultured in 200-μl round-bottom wells with graded numbers of cell sorter-purified thymic DCs (CD45+CD11c+ cells). After 6 days of culture, 1 μCi of [3H]thymidine (Amersham Pharmacia; 1 Ci = 37 GBq) per well was added, and after 12 hours, 3H incorporation was measured in a β-counter.
Results
Pro-Thymocytes Are Undetectable in c-kit and γc Double-Mutant Mice.
Thymocyte numbers are reduced ≈30-fold in γc− and ≈5-fold in c-kit− single-mutant mice, but are undetectable by cell counting or histology in c-kit and γc double-mutant mice (8). Because the developmental block found in c-kit−γc− mice clearly precedes the appearance of T cell receptor (TCR) Vβ(Dβ)Jβ, VγJγ, or Vδ(Dδ)Jδ rearrangements (14), the earliest thymocyte compartments were analyzed. The CD3−CD4−CD8− thymocytes can be subdivided based on differential expression of CD44 and CD25 (15). Together with c-kit (16), the earliest stage can be defined by a c-kit+CD44+CD25− phenotype. Acquisition of CD25 defines the next stage (c-kit+CD44+CD25+). If c-kit is omitted from the analysis, the CD44+CD25− population also includes other hematopoietic cells such as B cells and myeloid cells. In c-kitW/W mice, c-kit is not expressed on the cell surface because of a deletion of the transmembrane region (17).
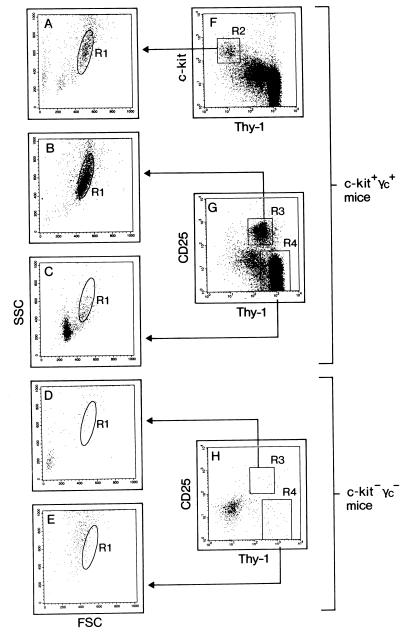
Wild-type and mutant thymi were analyzed for the presence of progenitors defined by forward scatter (FSC)/side scatter (SSC) analysis and expression of Thy-1, CD25, and, when possible, c-kit (Fig. 1). Most c-kit+ thymocytes were Thy-1low [Fig. 1F, region 2 (R2)], and “backgating” of c-kit+Thy-1low identified a distinct cell population by FSC/SSC analysis (Fig. 1A, R1). Thy-1 expression is up-regulated in CD25+ thymocytes (Fig. 1G, R3), and this population falls into the same FSC/SSC gate as c-kit+ thymocytes (Fig. 1B, R1). In contrast, more mature Thy-1+CD25− thymocytes (Fig. 1G, R4) are small lymphoid cells, most of which fall out of the progenitor size gate (Fig. 1C). c-kit+ thymocytes were also found in single-mutant mice lacking only γc, and CD25+ thymocytes were present in c-kit−γc+ mice (data not shown). All of these populations satisfied the scatter criteria for wild-type progenitors (data not shown). In marked contrast, CD25+ thymocytes were absent from double-mutant thymi (Fig. 1 D and H), and cells apparently staining for Thy-1 (Fig. 1H, R4) neither fell into the progenitor gate nor overlapped with the pattern found for small lymphoid cells (Fig. 1 C vs. E). We cannot directly demonstrate that c-kit+CD44+CD25− progenitors are lacking. However, the fact that cells with progenitor scatter properties and phenotype (lack of Thy-1) were undetectable suggests that pro-T cells are, indeed, absent.
Figure 1.
Pro-T cells are undetectable in mice lacking c-kit and γc. Wild-type (c-kit+γc+) (A–C, F, and G) and c-kit−γc− (D, E, and H) thymic cells were analyzed for expression of Thy-1 vs. c-kit (F), or Thy-1 vs. CD25 (G and H). In wild-type mice, c-kit+Thy-1low (R2) and CD25+Thy-1+ (R3) pro-thymocytes were further defined by FSC/SSC gate R1 (A and B). More mature thymocytes (R4) are outside of R1 (C). Pro-thymocytes defined by these criteria are lacking in c-kit−γc− mice (D, E, and H). Cells in the lower left area (dot plot in H) are thymic epithelial cells. The absence of c-kit+ progenitors cannot be demonstrated directly in c-kit-deficient mice because of lack of c-kit surface expression.
Thus, by histology, cell counting, and flow cytometry, we concluded that the thymus in c-kit−γc− mice is devoid of lymphoid cells. In previous analyses, however, clonal rearrangements at the TCR-β, TCR-γ, and TCR-δ loci were detectable in some double-mutant mice (14). Because we failed to detect RAG mRNA in c-kit−γc− thymi (data not shown), it is not clear whether these rare TCR recombination events reflect true intrathymic or, alternatively, extrathymic rearrangements. At such low levels, TCR rearrangements have also been identified previously in extrathymic sites such as fetal blood, fetal liver, and fetal gut (reviewed in ref. 14). Collectively, despite the presence of single TCR rearrangements, all evidence points to the absence of the earliest intrathymic progenitors in c-kit−γc− mice.
Thymic DCs Constitute the Major Hematopoietic Component in c-kit−γc−, but Not in c-kit or γc Single Mutants.
Given that pro-thymocytes were absent, we considered the possibility that the thymus in c-kit−γc− mice was not colonized by hematopoietic cells at all. To search for other hematopoietic cells, thymic tissue was collagenase digested to release all possible cellular subsets, and the resulting cell suspension was analyzed for the presence of CD45-expressing cells. Because DCs are functionally important hematopoietic cells in the thymus, analysis for expression of the integrin CD11c, within the thymus a specific DC marker (18), was included. After collagenase digestion, three populations could be distinguished using CD45 and CD11c as markers: (i) very large CD45−CD11c− cells (nonhematopoietic stromal cells), (ii) small and large CD45+CD11c− thymocytes, and (iii) very large CD45+CD11c+ cells (thymic DCs). This staining yields rather homogenous populations because cells, overlapping by size, differ in CD45 and CD11c expression, and CD45+ cells fall into small CD11c− vs. large CD11c+ cells (data not shown).
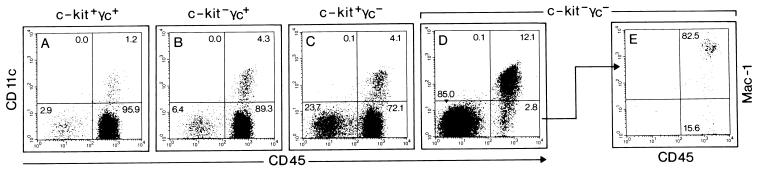
Thymi isolated from 5-day-old wild-type and mutant mice were compared by using these markers (Fig. 2). In the wild-type thymus, stromal cells (CD45−CD11c−) and DCs (CD45+CD11c+) represented ≈3% and ≈1%, respectively, of all cells; the remaining cells (>95%) were CD45+CD11c− thymocytes (Fig. 2A). The proportion of stroma and DC vs. thymocytes increased as thymocyte cellularity decreased in mice lacking either c-kit (Fig. 2B), or γc (Fig. 2C) alone. In all of these strains, >70% of all cells were thymocytes.
Figure 2.
Thymic epithelium and DCs are strongly enriched in c-kit−γc− mice. Wild-type (A), c-kit−γc+ (B), c-kit+γc− (C), and c-kit−γc− (D) postnatal-day-5 thymi were analyzed for cells expressing CD45 vs. CD11c (A–D), or CD45 vs. Mac-1 (E) to distinguish thymic stroma (CD45−CD11c−) from thymocytes (CD45+CD11c−) and DCs (CD45+CD11c+). Thymocytes are the major cell population in wild-type (A), single c-kit (B), and single γc (C) mutants. CD45−CD11c− stromal cells and CD45+CD11c+ DCs are strongly enriched in c-kit−γc− mice (D) (stroma: ≈85% in c-kit−γc− vs. ≈3% in wild-type mice; DC: ≈12% in c-kit−γc− vs. ≈1% in wild-type mice). In double-mutants, cells gated as CD45+CD11c− are mostly monocytes (CD45+Mac-1+) (E).
In c-kit−γc− mice, the stroma component was, as expected, greatly enriched (85% in c-kit−γc− mice vs. 3% in wild-type mice). It is interesting that thymic DCs, defined by their CD45+CD11c+ phenotype, were the second most prominent population (Fig. 2D). Thymic DCs were enriched ≈10-fold in c-kit−γc− when compared with wild-type mice [12.1% (c-kit−γc−) vs. 1.2% (wild type)]; absolute numbers of thymic DCs are discussed below). The vast majority (>80%) of the small fraction (<3%) of the remaining hematopoietic cells (CD45+CD11c− phenotype) (Fig. 2D) could be assigned to the monocytic/macrophage lineage as shown by staining for Mac-1 (Fig. 2E). Collectively, these data demonstrate that DCs are the major hematopoietic cell type in the c-kit−γc− thymus.
Correlation of Thymus Mass and Thymic DC Numbers in Wild-Type, c-kit, γc, and RAG-2 Mutant Mice.
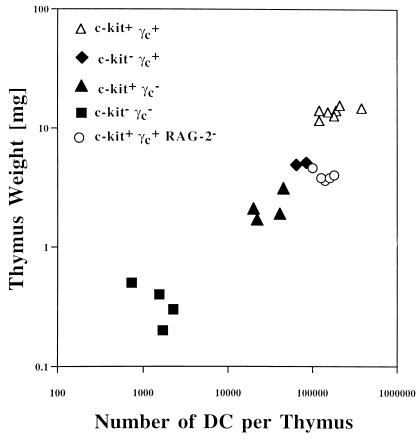
The absolute number of thymic DCs per thymus is expected to be proportional to the thymus mass, provided that mutations in the genes for c-kit, γc, or both do not affect thymic DC development. Thymus mass and number of thymic DCs were measured in c-kit and γc single and double mutants and, for comparison, in age-matched (postnatal day 5) RAG-2−/− mice (Fig. 3). Thymi were digested, and numbers of DCs were determined as shown in Fig. 2. Lack of c-kit or γc or both receptors causes a roughly parallel decline of both thymus weight and DC cell number. Such a correlation was not observed for RAG-2−/− thymi, which, at this stage in development, harbor approximately normal numbers of thymic DCs although the weight is reduced approximately threefold. In adult RAG-1−/− mice, however, a massive drop (≈10-fold) in absolute thymic DC numbers has been reported (7). Collectively, DC numbers depend primarily on thymus size in young c-kit and γc single and double mutants and in adult RAG mutants.
Figure 3.
Correlation of thymus mass and absolute numbers of DCs per thymus in wild-type (▵), c-kit−γc+ (⧫), c-kit+γc− (▴), c-kit−γc− (■), and c-kit+γc+RAG-2− (○) mutants. Postnatal-day-5 thymi from 22 mice of the indicated genotypes were individually analyzed for weight in milligrams and for the total number of DCs that were quantified by FACS as shown in Fig. 2. For c-kit and γc mutants, the number of DCs is roughly proportional to the thymus mass, rather than genotype. For RAG-2 mutants, the weight is reduced approximately threefold whereas the number of DCs is comparable with wild-type mice.
Failure to Properly Organize Thymic Epithelium into Cortical and Medullary Zones Alters Thymic DC Compartmentalization.
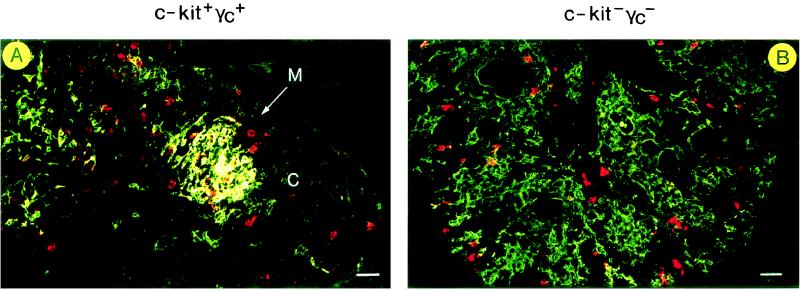
Thymic DCs are normally located in the medulla and at the cortico-medullary junction. The alymphoid thymus in the c-kit−γc− mouse lacks a regular medulla/cortex organization (14; see also below). The intrathymic location of DCs in double-mutant mice was examined histologically (Fig. 4). Staining for MTS10, a marker of medullary epithelium (13), showed a well-developed medullary zone in the postnatal-day-5, wild-type thymus (Fig. 4A, MTS10 staining in green). MTS10+ medullary epithelium was surrounded by MTS10− cortex. Staining for CD11c identified thymic DCs (Fig. 4A, CD11c staining in red), many but not all of which resided in close proximity to MTS10+ medullary epithelium.
Figure 4.
Localization of DCs in wild-type (A) and c-kit−γc− (B) thymi. Sections were analyzed by immunofluorescence for expression of the medullary marker MTS10 (13) (green) vs. the DC marker CD11c (red). An MTS10+ medullary zone [“M” (see arrow)] is visible (A). Many, but not all, of the thymic DCs are localized in the medullary area, and DCs are rarely found in the cortex (C). In B, MTS10+ epithelium fails to form medullary areas, and single or small clusters of CD11c+ thymic DCs are distributed throughout the thymus. (Bars = 30 μm.)
In the c-kit−γc− thymus, the patterns of both MTS10 and CD11c stainings were altered (Fig. 4B). First, MTS10+ medullary epithelium failed to form medullary clusters. Second, single or small clusters of CD11c+ thymic DCs were distributed throughout the thymus. Thus, the distribution of DCs in the c-kit−γc− thymus does not resemble medulla-associated DC clusters in the wild-type thymus.
Thymic DCs in c-kit−γc− Mice Include Both Myeloid and Lymphoid DC Phenotypes.
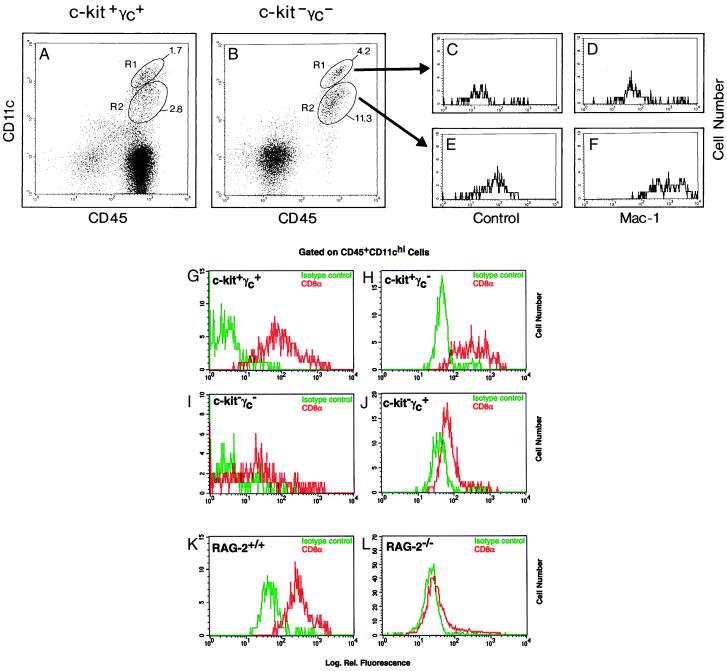
Based on marker expression and putative developmental origin, DCs are frequently divided into CD11c+Mac-1+CD8α− myeloid and CD11c+Mac-1−/lowCD8α+ lymphoid DC phenotypes (6, 7). Because absolute numbers of thymic DCs were much reduced in c-kit−γc− mice (Fig. 3), we next determined whether both myeloid and lymphoid DC phenotypes were present in c-kit−γc− mice. Thymic DCs were analyzed for expression of CD45, CD11c, and either Mac-1 (CD11b) or CD8α (Fig. 5). As shown before in Fig. 2, c-kit−γc− (Fig. 5B) but not wild-type mice (Fig. 5A) lacked CD45+CD11c− thymocytes. Thymic DCs could be separated into CD11chigh and CD11clow populations, and this separation was evident in both wild-type (Fig. 5A) and double-mutant mice (Fig. 5B). Further analysis showed that the CD45+CD11chigh cells (Fig. 5B, R1) were Mac-1−/low (Fig. 5C vs. D), whereas CD45+CD11clow cells (Fig. 5B, R2) were Mac-1+ (Fig. 5E vs. F). Thus, based on Mac-1 expression, CD11chigh DCs have a lymphoid and CD11clow DCs have a myeloid DC phenotype. Of note, DCs satisfying these criteria were found in both wild-type and c-kit−γc− thymi. The ratio of CD11chigh (lymphoid) to CD11clow (myeloid) DCs was roughly comparable between DCs from wild-type and c-kit−γc− thymi [experiment 1 (shown in Fig. 5): 1 to 1.6 (CD11chigh to CD11clow) for wild type and 1 to 2.7 (CD11chigh to CD11clow) for c-kit−γc− thymi; experiment 2: 1 to 1.8 (CD11chigh to CD11clow) for wild-type, and 1 to 2.5 (CD11chigh to CD11clow) for c-kit−γc− thymi] (data not shown).
Figure 5.
Both myeloid and lymphoid DC phenotypes (see text) are generated in the absence of pro-thymocytes. Wild-type (A, G, and K), c-kit−γc− (B–F and I), c-kit+γc− (H), c-kit−γc+ (J), RAG-2+/+ (K), and RAG-2−/− (L) mice were analyzed for expression of CD45 and CD11c (A and B), or CD45, CD11c and Mac-1 (B–F), or CD45, CD11c, and CD8α (G–L). CD45+CD11c+ DC can be separated into CD11chigh and CD11clow populations (A and B) which, purified by cell sorting, were restained with isotype controls (C and E) or with anti-Mac-1 (D and F). CD11chigh DC are Mac-1−/low [mean fluorescences are 38 (C) vs. 201 (D)], whereas CD11clow DCs are Mac-1+ [mean fluorescences are 88 (E) vs. 1297 (F)]. CD8α expression on CD11chigh DCs was compared between wild-type (G), c-kit+γc− (H), c-kit−γc+ (J), and c-kit−γc− (I) mice and between RAG-2+/+ (K) vs. RAG-2−/− (L) mice (isotype control in green; CD8α staining in red). CD8α expression was negative/low in DCs from c-kit−γc− thymi and from RAG-2−/− thymi when compared with DCs from wild-type thymi. Mean fluorescences and peak channels were 5 and 1 [isotype (G)], 163 and 63 [anti-CD8α (G)]; 57 and 46 [isotype (H)], 445 and 289 [anti-CD8α (H)]; 9 and 1 [isotype control (I)], 67 and 1 [anti-CD8α (I)], 48 and 31 [isotype control (J)], 84 and 64 [anti-CD8α (J)], 57 and 39 [isotype control (K)]; 355 and 228 [anti-CD8α (K)]; 23 and 19 [isotype control (L)], and 51 and 23 [anti-CD8α (L)].
Next, CD8α expression, a marker reported to allow discrimination of lymphoid from myeloid DC phenotypes (7), was analyzed on lymphoid (CD45+CD11chigh) DCs from wild-type and double-mutant thymi. As shown in Fig. 5, wild-type CD45+CD11chigh DCs were clearly CD8α+ (Fig. 5G). In contrast, CD45+CD11chigh DCs from double-mutant mice were CD8αneg/low (Fig. 5I). Such loss of CD8α on lymphoid thymic DCs was not observed in γc (Fig. 5H) or c-kit (Fig. 5J) single-mutant mice although the level of CD8α+ expression was reduced in the latter strain. To analyze whether lack of CD8α expression on thymic DC was associated with the absence of more mature thymocytes (which may provide CD8 protein for DC pickup), we next compared CD8α expression on CD45+CD11chigh thymic DC from postnatal-day-5 RAG-2+/+ vs. RAG-2−/− mice. Although wild-type DCs were again CD8α+ (Fig. 5K), surprisingly, thymic DCs from RAG-2−/− mice were CD8αneg/low (Fig. 5L).
Collectively, thymic DCs from c-kit−γc− mice include both lymphoid and myeloid DCs, with regard to their CD11chighMac-1−/low and CD11clowMac-1+ phenotypes, respectively. However, in c-kit−γc− mice, but notably also in RAG-2−/− mice, DCs with a lymphoid phenotype were CD8αlow/neg when compared with the corresponding population from wild-type mice.
Thymic DCs from c-kit−γc− Mice Are Potent Stimulators of an Allogeneic Mixed-Leukocyte Reaction.
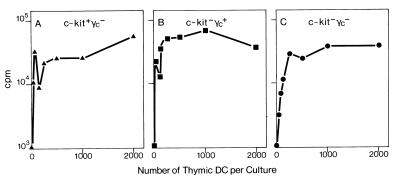
Thymic DCs from c-kit−γc− mice appeared normal in relative numbers, subset composition, and, with the exception of CD8α expression, cell surface phenotype. Therefore, we next determined whether DCs from double-mutant mice were also functionally indistinguishable from wild-type DCs. Characteristically, DCs are very potent stimulators of mixed-leukocyte reactions (19). c-kit and γc double-mutant mice were generated from crosses of mouse strains WB (H-2j), C57BL/6, 129/ola (both H-2b), and BALB/c (H-2d). Responder lymph node T cells were isolated from C3H mice (H-2k). Thus, mixed-leukocyte reactions were set up across full H-2 mismatches (H-2b, H-2d, or H-2j anti H-2k) (Fig. 6). DCs from wild-type (data not shown) or c-kit+γc− (Fig. 6A) or c-kit−γc+ mice (Fig. 6B) were potent stimulators. C-kit−γc− thymic DCs behaved indistinguishably in this assay, i.e., at ≥250 DCs per culture, maximum plateau level T cell proliferation was induced (Fig. 6C). Thus, thymic DCs from c-kit−γc− mice were as competent at mixed-leukocyte reaction induction as DCs from wild-type or single-mutant mice.
Figure 6.
DCs from c-kit−γc− thymi are potent stimulators of a mixed leukocyte reaction. DCs from c-kit+γc− (A), c-kit−γc+ (B), or c-kit−γc− (C) thymi were cultured at indicated cell numbers with responder T cells. Mixed leukocyte reactions were set up across full H-2 mismatches (H-2b, H-2d, or H-2j anti-H-2k) (see text).
Discussion
Our data support four main conclusions: (i) thymic DCs can develop independently of c-kit- and γc-mediated signals, (ii) development of thymic DCs can be dissociated from the generation of thymocytes and their progenitors, (iii) CD8α expression on thymic lymphoid DCs is not independent of thymocytes developing beyond the RAG-block, and (iv) the distribution of DCs in the c-kit−γc− thymus, which lacks proper epithelial organization, is altered.
Thymic DCs Can Develop in the Absence of c-kit- and γc-Mediated Signals.
Because of the absence of thymocytes (see below), relative numbers of thymic DCs (and thymic epithelium) were significantly increased in single and, more dramatically, in double mutants (Fig. 2). However, the ratio of stroma to DCs was variable: In wild-type controls and in c-kit single mutants, the stroma:DC ratio was ≈2, but this ratio was elevated to 6–7 in γc single or double mutants (Fig. 2). Although all mutants were clearly permissive for thymic DC development, this altered stroma:DC ratio in γc mutants could imply that DC development is not completely unaffected by lack of γc. Comparison of absolute numbers of DCs to thymus mass, however, suggests that DC numbers depend on the thymus mass rather than the genotype in these mutant strains (Fig. 3). At this stage in ontogeny (postnatal-day 5), such correlation was not observed for the RAG-2−/− thymus, which showed an approximately threefold reduction in thymus weight but no decrease in total DC numbers. However, in adult RAG-1−/− mice, a 10-fold reduction in DC numbers has been reported (7). The basis for this delayed effect of thymus size on DC numbers in RAG-deficient mice when compared with the growth factor mutants remains to be determined.
The finding that thymic DC development can proceed in the absence of either c-kit- or γc-mediated signals is noteworthy because candidate progenitors for thymic DCs (see below) such as circulating hematopoietic stem cells or the most immature intrathymic progenitors express both c-kit and γc (reviewed in ref. 20). Moreover, thymic progenitors with DC potential are responsive in vitro to cytokine cocktails containing c-kit ligand and IL-7 in addition to IL-1β, IL-3, IL-4, tumor necrosis factor, and granulocyte/monocyte colony-stimulating factor. Omission of c-kit ligand reduces DC colony formation from thymic progenitors by ≈50%. If granulocyte/monocyte colony-stimulating factor is omitted from the cytokine cocktail, DC development becomes IL-7 dependent in vitro (21). In line with these studies, roles for IL-2 (22) or IL-7 (23) in DC development have been proposed. Based on these reports and on the notion that even mature thymic DCs express c-kit (24, 25), c-kit- or γc-mediated signals in thymic-DC development had to be considered. Our data show that these growth factor receptors are not critical for thymic DC development in vivo. In contrast to c-kit, the flk-2/flt-3 tyrosine kinase can affect DC development because injection of flk-2/flt-3 ligand into mice leads to greatly increased DC numbers (26). It is not known whether DCs are affected in flk-2/flt-3−/− mice (27).
Thymic DCs and Thymocytes Can Be Developmentally Dissociated.
DC and T cell lineages are turned over in parallel (24), and no mouse mutant was known in which DC but not T cell development was permissive. Mice expressing a dominant negative Ikaros protein (Ikaros C-terminal−/− mice) lacked both thymocytes and DCs. In contrast, true Ikaros-null mice, in which only postnatal but not prenatal thymocyte development was permissive, showed DC development, albeit at reduced levels (28). Thus, thymocyte development per se might be required for DC development. Data from c-kit−γc− mice now reveal that this is not the case. A similar conclusion was derived from other T cell-deficient mice in which thymocyte development is blocked at later stages when compared with c-kit−γc− mice (7).
If the absence of a normal pro-T cell compartment does not preclude the generation of thymic DC populations, what is the developmental origin of thymic DC populations? In addition to hematopoietic stem cells, early intrathymic progenitors contain potential for thymic DCs (4, 5). One CD4low progenitor generated ≈500 thymocytes. In marked contrast, approximately two CD4low progenitors were required to give rise to one DC (4, 5, 24). Thus, the CD4low progenitor population generates ≈1000-fold more T lineage cells than DC progeny. Because intrathymic transfer is a very efficient method of thymus seeding (29), these quantitative analyses of precursor-product ratios could reflect the in vivo potential of the CD4low population.
Each thymus rudiment in c-kit−γc− mice harbored ≈1 × 103-2 × 103 DCs (Fig. 3). Considering the numbers discussed above, one should have expected to find on the order of 2 × 103–4 × 103 pro-T cells per thymus. Because 1 × 103 to 2 × 103 DCs were readily detectable (Fig. 2), it is unlikely that a comparable number of progenitors escaped our analyses. However, such cells were not identified. Hence, the generation of both myeloid and lymphoid DC populations, each with normal cellularity compared with thymus mass, was not affected by the absence of pro-T cell compartments. We cannot rule out that very few as yet undetected progenitors are present in the double-mutant thymus. If such progenitors gave rise to thymic DCs, they would have done so very efficiently, unlike the known CD4low cells. Clearly, on a per-cell basis, pro-T cells are not essential to generate thymic DCs. Such a T-cell-independent, nonlymphoid progenitor pathway could also produce thymic DCs in normal mice.
CD8α Expression on Thymic Lymphoid DCs Is Not Independent of Thymocyte Development.
C-kit−γc− lymphoid DCs were CD8αneg/low. In γc single mutants, lymphoid DCs were CD8α+, whereas CD8α expression was reduced in c-kit single mutants. In addition, we also tested the possibility that absence of more mature, i.e., CD8-expressing thymocytes might play a role for CD8α expression on thymic DCs. As proposed previously for CD4 or Thy-1 (5), DCs may acquire surface expression by passive pickup from surrounding thymocytes. The reduction in CD8α expression by DCs may reflect the difference between authentic CD8α expression (seen in c-kit−γc−) and endogenous plus passive CD8α expression (seen in the wild type). This idea is strongly supported by the fact that thymic DCs are CD8αneg/low in RAG-2−/− mice. In these mutants, all potential DC progenitor populations are expected to be present in normal numbers (30). Lack of CD8α expression on thymic DCs in RAG-2−/− mice most likely indicates the absence of the CD8 marker on DCs rather than absence of the entire lymphoid DC compartment. In contrast to our findings, Shortman et al. have reported that >90% of thymic DCs from adult RAG-1−/− mice are CD8α+ (7). It remains to be determined whether these discrepancies are due to the age of the RAG-deficient mice analyzed (postnatal vs. adult). Of note, DCs generated in vitro from thymic progenitors (under conditions not permissive for thymocyte development) satisfied the criteria for lymphoid DCs, but lacked CD8α (21). By combining in vivo (RAG-2−/−) and in vitro evidence, CD8α is not an inevitable marker of the lymphoid origin of thymic DCs.
DCs Were Distributed Throughout the c-kit−γc− Thymus Without Major Cluster Formation.
Although no mechanisms are known to account for the normal tissue distribution of thymic DCs predominantly in the medulla and at the cortico-medullary junction, our data indicate that thymic DC development can proceed independently of the physiological intrathymic location of DCs. Thymic DCs do not appear to undergo extensive local proliferation (which should have been evident as DC foci), suggesting that thymic DCs are generated from circulating DC progenitors that colonize the thymus stroma without further cell division. Future work must focus on further delineation of hematopoietic pathways leading to DC development.
Acknowledgments
We thank Drs. H. J. Fehling, S. Gilfillan, A. Lanzavecchia, and K. Shortman for discussions; M. Dessing and A. Pickert for cell sorting; Dr. R. Boyd for antibodies; B. Aschwanden for animal husbandry; B. Kugelberg for help with histology; H. Stahlberger for artwork; and B. Pfeiffer and H. Spalinger for photography. The Basel Institute for Immunology was founded and is supported by F. Hoffmann La Roche, Basel.
Abbreviations
- DC
dendritic cell
- γc
common cytokine receptor γ chain
- TCR
T cell receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Peters J H, Gieseler R, Thiele B, Steinbach F. Immunol Today. 1996;17:273–278. doi: 10.1016/0167-5699(96)80544-5. [DOI] [PubMed] [Google Scholar]
- 2.Cella M, Sallusto F, Lanzavecchia A. Curr Opin Immunol. 1997;9:10–16. doi: 10.1016/s0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- 3.Steinman R M, Inaba K. J Leukocyte Biol. 1999;66:205–208. doi: 10.1002/jlb.66.2.205. [DOI] [PubMed] [Google Scholar]
- 4.Ardavin C, Wu L, Li C-L, Shortman K. Nature (London) 1993;362:761–763. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Li C L, Shortman K. J Exp Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardavin C. Immunol Today. 1997;18:350–361. doi: 10.1016/s0167-5699(97)01090-6. [DOI] [PubMed] [Google Scholar]
- 7.Shortman K, Vremec D, Corcoran L M, Georgopoulos K, Lucas K, Wu L. Immunol Rev. 1998;165:39–46. doi: 10.1111/j.1600-065x.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 8.Rodewald H-R, Ogawa M, Haller C, Waskow W, DiSanto J P. Immunity. 1997;6:265–272. doi: 10.1016/s1074-7613(00)80329-5. [DOI] [PubMed] [Google Scholar]
- 9.Russell E. Adv Genet. 1979;20:357–459. [PubMed] [Google Scholar]
- 10.DiSanto J P, Müller W, Guy-Grand D, Fischer A, Rajewsky K. Proc Natl Acad Sci USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S-I, Kunisada T, Sudo T, Kina T, Nakauchi H, Nishikawa S-I. J Exp Med. 1991;174:63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brocker T. J Exp Med. 1997;186:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd R L, Tucek C L, Godfrey D I, Izon D J, Wilson T J, Davidson N J, Bean A G, Ladyman H M, Ritter M A, Hugo P. Immunol Today. 1993;14:445–459. doi: 10.1016/0167-5699(93)90248-J. [DOI] [PubMed] [Google Scholar]
- 14.Rodewald H-R, Fehling H J. Adv Immunol. 1998;69:1–112. doi: 10.1016/s0065-2776(08)60606-9. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey D I, Kennedy J, Suda T, Zlotnik A. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 16.Godfrey D I, Zlotnik A, Suda T. J Immunol. 1992;149:2281–2285. [PubMed] [Google Scholar]
- 17.Hayashi S, Kunisada T, Ogawa M, Yamaguchi K, Nishikawa S. Nucleic Acids Res. 1991;19:1267–1271. doi: 10.1093/nar/19.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metlay J P, Witmer-Pack M D, Agger R, Crowley M T, Lawless D, Steinman R M. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crowley M, Inaba K, Witmer-Pack M, Steinman R M. Cell Immunol. 1989;118:108–125. doi: 10.1016/0008-8749(89)90361-4. [DOI] [PubMed] [Google Scholar]
- 20.DiSanto J P, Rodewald H-R. Curr Opin Immunol. 1998;10:196–207. doi: 10.1016/s0952-7915(98)80249-5. [DOI] [PubMed] [Google Scholar]
- 21.Saunders D, Lucas K, Ismaili J, Wu L, Maraskovsky E, Dunn A, Shortman K. J Exp Med. 1996;184:2185–2196. doi: 10.1084/jem.184.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bykovskaja S N, Buffo M J, Bunker M, Zhang H, Majors A, Herbert M, Lokshin A, Levitt M L, Jaja A, Scalise D, Kosiban D, Evans C, Marks S, Shogan J. J Leukocyte Biol. 1998;63:620–630. doi: 10.1002/jlb.63.5.620. [DOI] [PubMed] [Google Scholar]
- 23.Varas A, Vicente A, Sacedon R, Zapata A G. Blood. 1998;92:93–100. [PubMed] [Google Scholar]
- 24.Wu L, Vremec D, Ardavin C, Winkel K, Suss G, Georgiou H, Maraskovsky E, Cook W, Shortman K. Eur J Immunol. 1995;25:418–425. doi: 10.1002/eji.1830250217. [DOI] [PubMed] [Google Scholar]
- 25.Vremec D, Shortman K. J Immunol. 1997;159:565–573. [PubMed] [Google Scholar]
- 26.Maraskovsky E, Brasel K, Teepe M, Roux E R, Lyman S D, Shortman K, McKenna H J. J Exp Med. 1996;184:1953–1962. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackarehtschian K, Hardin J D, Moore K A, Boast S, Goff S P, Lemischka I R. Immunity. 1995;3:147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Nichogiannopoulou A, Shortman K, Georgopoulos K. Immunity. 1997;7:483–492. doi: 10.1016/s1074-7613(00)80370-2. [DOI] [PubMed] [Google Scholar]
- 29.de Vries P, Brasel K A, McKenna H J, Williams D E, Watson J D. J Exp Med. 1992;176:1503–1509. doi: 10.1084/jem.176.6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]