Abstract
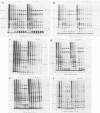
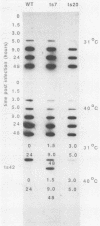
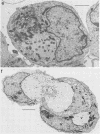
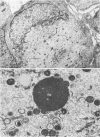
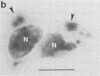
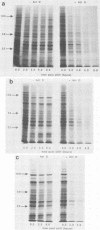
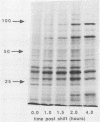
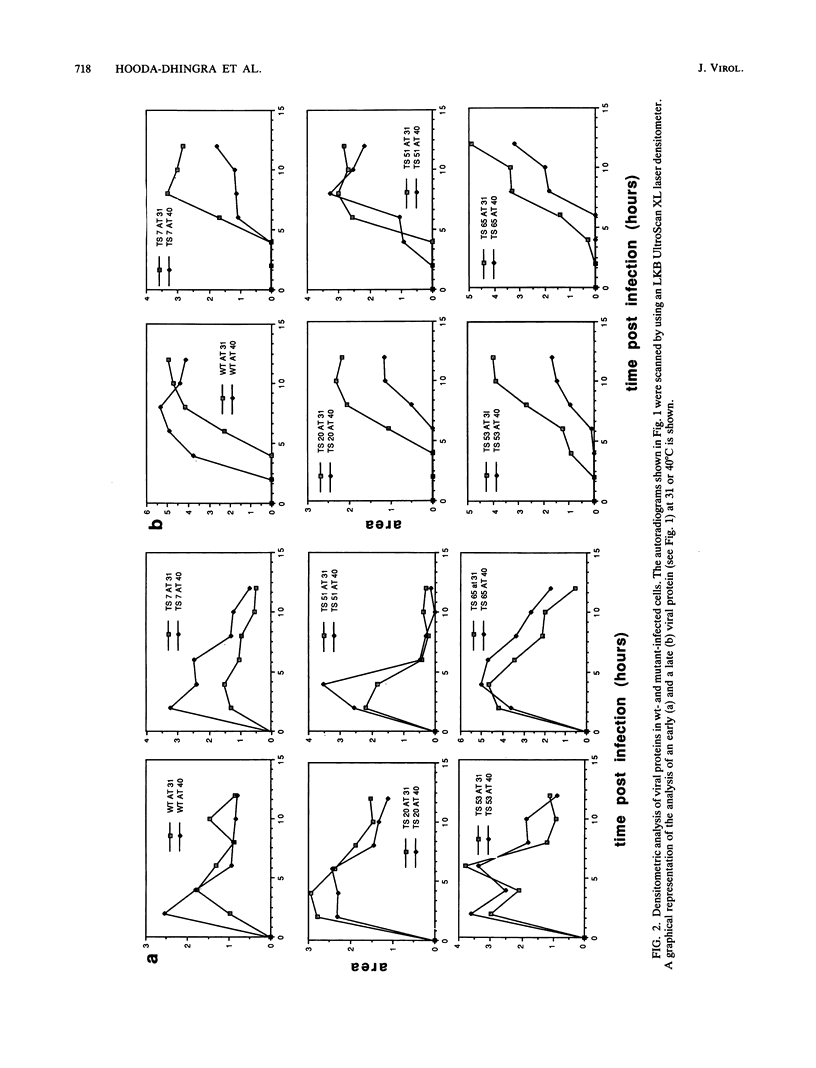
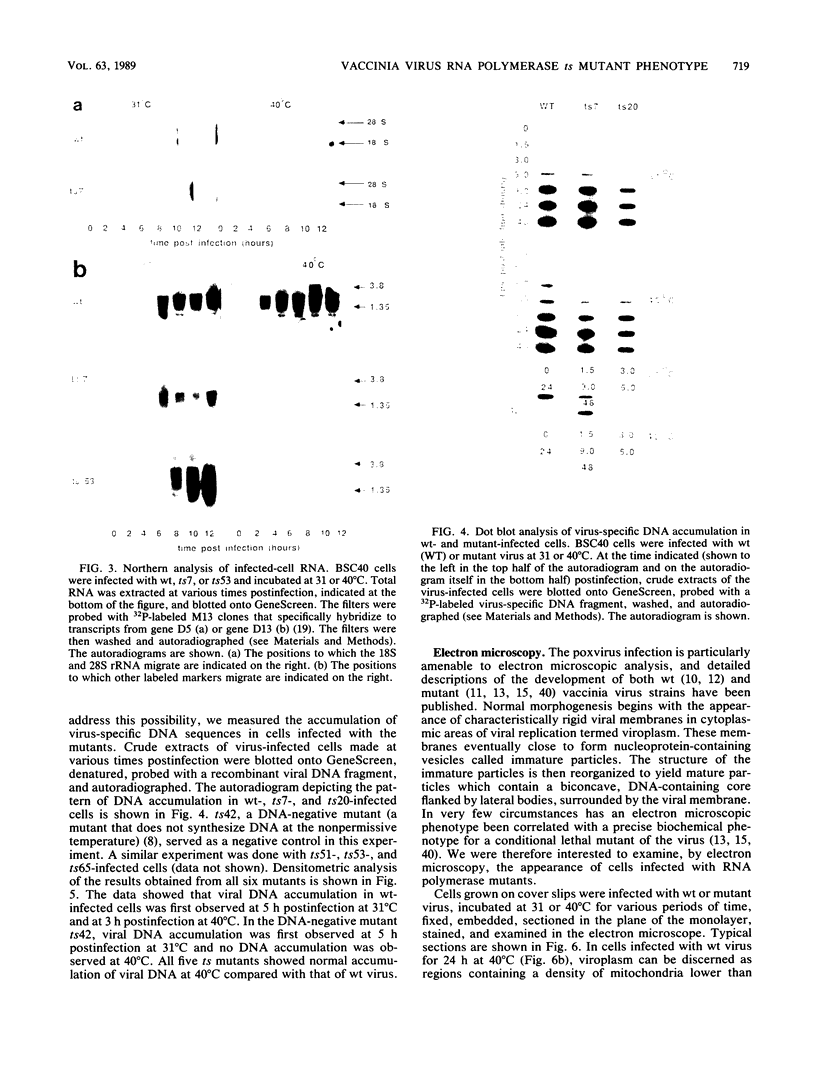
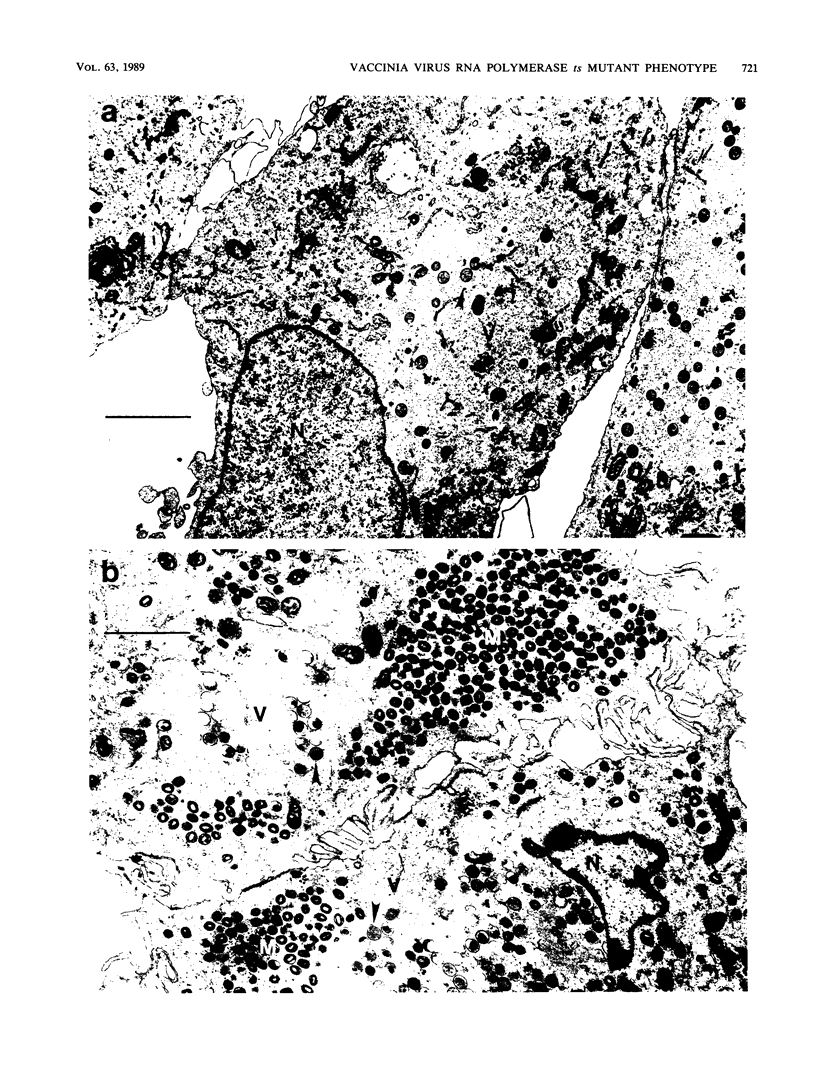
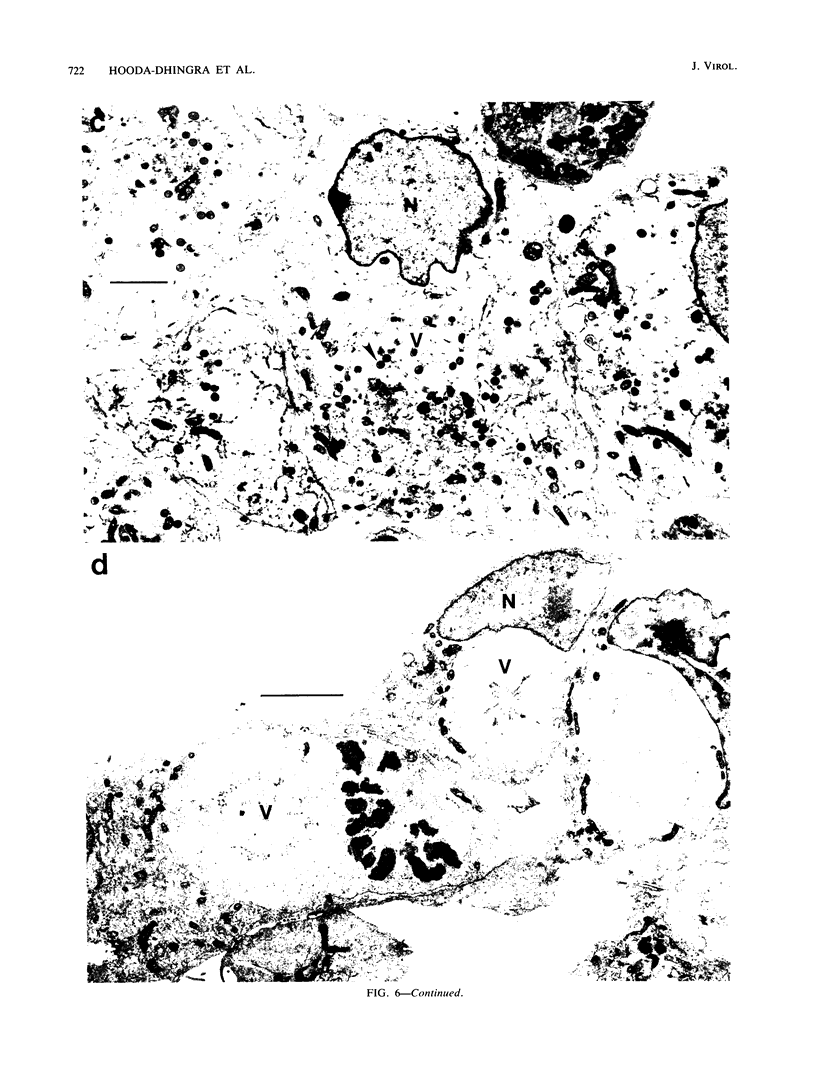
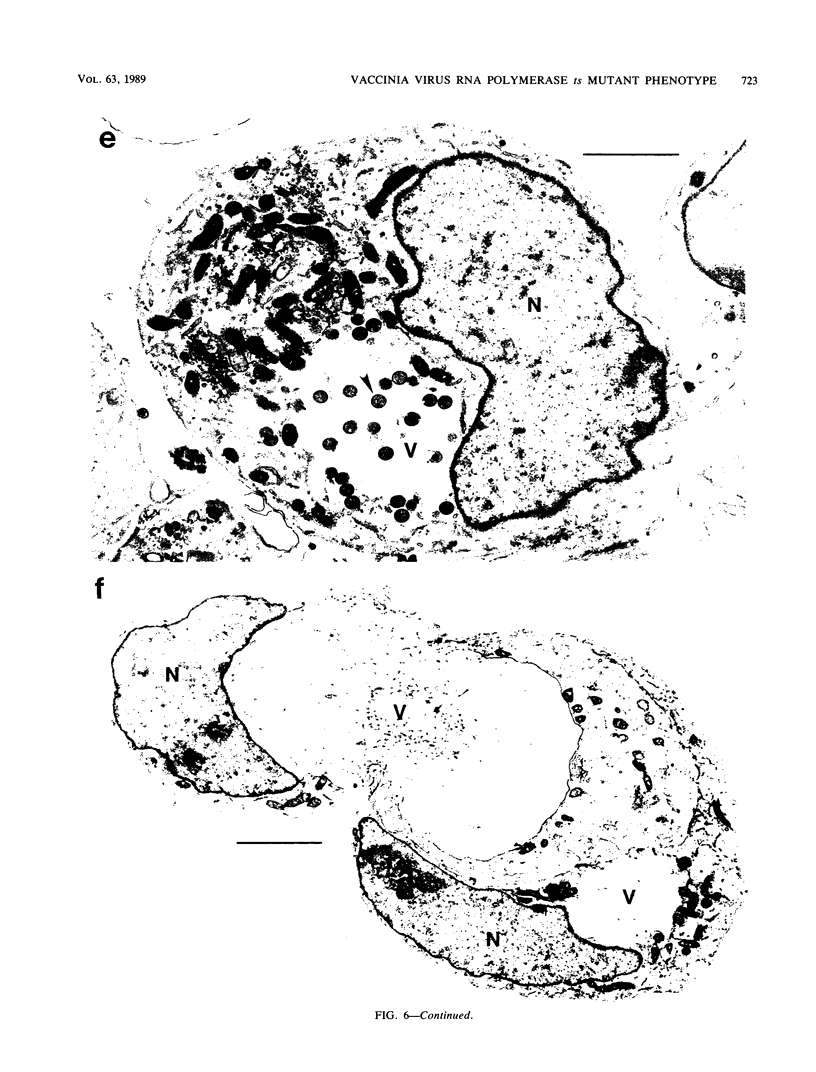
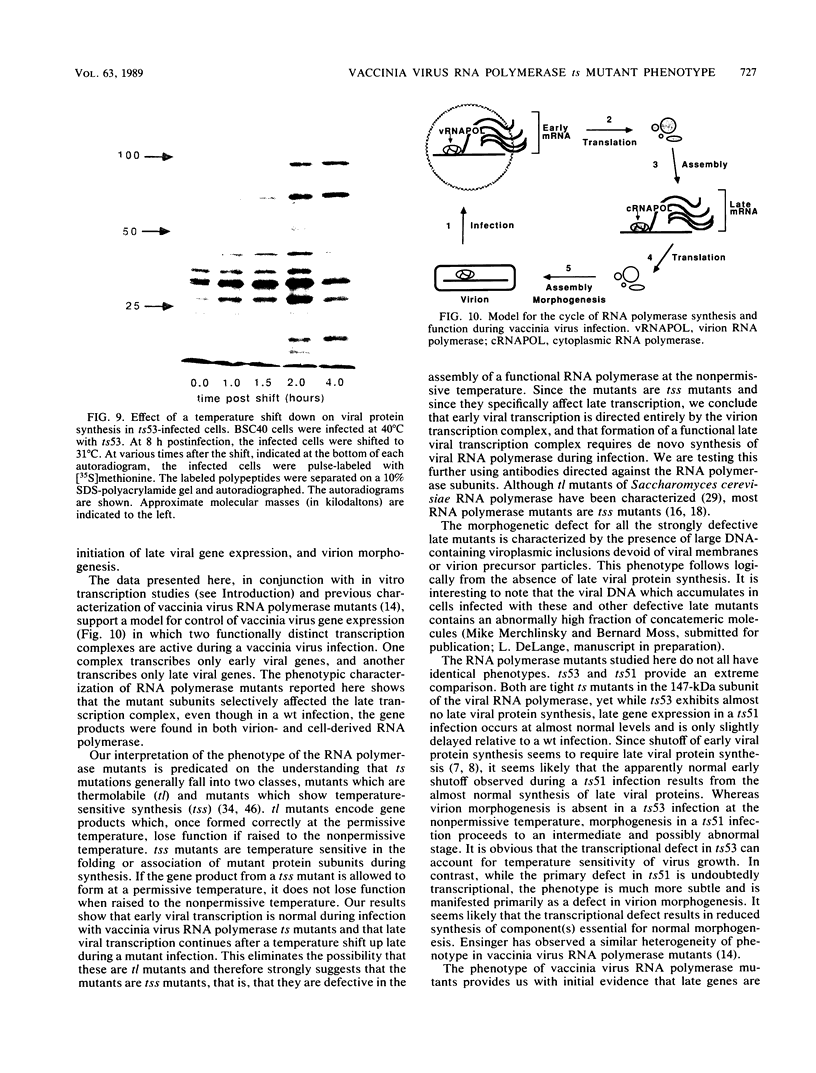
We have carried out detailed phenotypic characterization of five temperature-sensitive (ts) mutants of vaccinia virus, the ts lesions of which have previously been mapped to two different subunits of the viral RNA polymerase. We have also attempted to determine the mechanism of temperature sensitivity in these mutants. Phenotypic characterization of each of the mutants showed that at the nonpermissive temperature, all five mutants exhibited normal levels of early viral mRNA and protein synthesis, but for an extended period of time, all mutants accumulated normal levels of DNA in abnormally large pools in the cell cytoplasm; all mutants were defective in the synthesis of late viral mRNA and proteins and in viral morphogenesis. In an attempt to address the mechanism of temperature sensitivity in these mutants, we measured the effect of a temperature shift on the ability of the mutants to direct late viral protein synthesis. If infected cells were shifted down from a nonpermissive temperature late during infection, late protein synthesis was initiated after a lag period of 1 to 2 h. If infected cells were shifted up from a permissive temperature early during infection, late protein synthesis continued to be defective. If infected cells were shifted up to the nonpermissive temperature after late protein synthesis had commenced, late protein synthesis was maintained at the nonpermissive temperature at the level observed when the temperature was shifted up. We interpret these results to mean that once a functional RNA polymerase has been assembled at the permissive temperature during a mutant infection, it remains functional at the nonpermissive temperature, but that the ts mutants are defective in the assembly of a newly synthesized RNA polymerase at the nonpermissive temperature. This interpretation implies that the virion RNA polymerase is responsible for early viral transcription and that a newly synthesized RNA polymerase transcribes late viral genes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Moss B. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem. 1980 May 10;255(9):4372–4380. [PubMed] [Google Scholar]
- Bertholet C., Stocco P., Van Meir E., Wittek R. Functional analysis of the 5' flanking sequence of a vaccinia virus late gene. EMBO J. 1986 Aug;5(8):1951–1957. doi: 10.1002/j.1460-2075.1986.tb04449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet C., Van Meir E., ten Heggeler-Bordier B., Wittek R. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell. 1987 Jul 17;50(2):153–162. doi: 10.1016/0092-8674(87)90211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci U S A. 1986 May;83(10):3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Sedimentation of an RNA polymerase complex from vaccinia virus that specifically initiates and terminates transcription. Mol Cell Biol. 1987 Jan;7(1):7–14. doi: 10.1128/mcb.7.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran M. A., Puckett C., Moss B. In vitro mutagenesis of the promoter region for a vaccinia virus gene: evidence for tandem early and late regulatory signals. J Virol. 1985 Apr;54(1):30–37. doi: 10.1128/jvi.54.1.30-37.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Coupar B. E., Boyle D. B., Both G. W. Effect of in vitro mutations in a vaccinia virus early promoter region monitored by herpes simplex virus thymidine kinase expression in recombinant vaccinia virus. J Gen Virol. 1987 Sep;68(Pt 9):2299–2309. doi: 10.1099/0022-1317-68-9-2299. [DOI] [PubMed] [Google Scholar]
- DALES S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J Cell Biol. 1963 Jul;18:51–72. doi: 10.1083/jcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S., Milovanovitch V., Pogo B. G., Weintraub S. B., Huima T., Wilton S., McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978 Feb;84(2):403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- Dales S., Mosbach E. H. Vaccinia as a model for membrane biogenesis. Virology. 1968 Aug;35(4):564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- Drillien R., Tripier F., Koehren F., Kirn A. A temperature-sensitive mutant of vaccinia virus defective in an early stage of morphogenesis. Virology. 1977 Jun 15;79(2):369–380. doi: 10.1016/0042-6822(77)90364-6. [DOI] [PubMed] [Google Scholar]
- Ensinger M. J. Phenotypic characterization of temperature-sensitive mutants of vaccinia virus with mutations in a 135,000-Mr subunit of the virion-associated DNA-dependent RNA polymerase. J Virol. 1987 Jun;61(6):1842–1850. doi: 10.1128/jvi.61.6.1842-1850.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essani K., Dugre R., Dales S. Biogenesis of vaccinia: involvement of spicules of the envelope during virion assembly examined by means of conditional lethal mutants and serology. Virology. 1982 Apr 30;118(2):279–292. doi: 10.1016/0042-6822(82)90347-6. [DOI] [PubMed] [Google Scholar]
- Gross G. C., Fields D. A., Bautz E. K. Temperature-sensitive mutants of Escherichia coli with defects in the assembly of RNA polymerase in vitro. Eur J Biochem. 1977 Dec 1;81(2):333–338. doi: 10.1111/j.1432-1033.1977.tb11956.x. [DOI] [PubMed] [Google Scholar]
- Hänggi M., Bannwarth W., Stunnenberg H. G. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J. 1986 May;5(5):1071–1076. doi: 10.1002/j.1460-2075.1986.tb04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A., Shimamoto N., Aiba H., Kawakami K., Nashimoto H., Tsugawa A., Uchida H. Temperature-sensitive mutations in the alpha subunit gene of Escherichia coli RNA polymerase. J Mol Biol. 1980 Feb 25;137(2):137–150. doi: 10.1016/0022-2836(80)90321-6. [DOI] [PubMed] [Google Scholar]
- Lee-Chen G. J., Bourgeois N., Davidson K., Condit R. C., Niles E. G. Structure of the transcription initiation and termination sequences of seven early genes in the vaccinia virus HindIII D fragment. Virology. 1988 Mar;163(1):64–79. doi: 10.1016/0042-6822(88)90234-6. [DOI] [PubMed] [Google Scholar]
- Lee-Chen G. J., Niles E. G. Map positions of the 5' ends of eight mRNAs synthesized from the late genes in the vaccinia virus HindIII D fragment. Virology. 1988 Mar;163(1):80–92. doi: 10.1016/0042-6822(88)90235-8. [DOI] [PubMed] [Google Scholar]
- Lee-Chen G. J., Niles E. G. Transcription and translation mapping of the 13 genes in the vaccinia virus HindIII D fragment. Virology. 1988 Mar;163(1):52–63. doi: 10.1016/0042-6822(88)90233-4. [DOI] [PubMed] [Google Scholar]
- Mahr A., Roberts B. E. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984 Feb;49(2):510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. Osmium ferricyanide fixation improves microfilament preservation and membrane visualization in a variety of animal cell types. J Ultrastruct Res. 1984 Feb;86(2):107–118. doi: 10.1016/s0022-5320(84)80051-9. [DOI] [PubMed] [Google Scholar]
- Miner J. N., Weinrich S. L., Hruby D. E. Molecular dissection of cis-acting regulatory elements from 5'-proximal regions of a vaccinia virus late gene cluster. J Virol. 1988 Jan;62(1):297–304. doi: 10.1128/jvi.62.1.297-304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K., Moyer R. W. Detection of a subunit of cellular Pol II within highly purified preparations of RNA polymerase isolated from rabbit poxvirus virions. Cell. 1986 Feb 28;44(4):587–596. doi: 10.1016/0092-8674(86)90268-0. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Condit R. C., Caro P., Davidson K., Matusick L., Seto J. Nucleotide sequence and genetic map of the 16-kb vaccinia virus HindIII D fragment. Virology. 1986 Aug;153(1):96–112. doi: 10.1016/0042-6822(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K. I., Joklik W. K. Hybridization and sedimentation studies on "early" and "late" vaccinia messenger RNA. J Mol Biol. 1967 Aug 14;27(3):395–419. doi: 10.1016/0022-2836(67)90047-2. [DOI] [PubMed] [Google Scholar]
- Pacha R. F., Condit R. C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985 Nov;56(2):395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmann G., Yuen L., Moss B. Transcription of vaccinia virus early genes by enzymes isolated from vaccinia virions terminates downstream of a regulatory sequence. Cell. 1986 Sep 26;46(7):1029–1035. doi: 10.1016/0092-8674(86)90702-6. [DOI] [PubMed] [Google Scholar]
- SADLER J. R., NOVICK A. THE PROPERTIES OF REPRESSOR AND THE KINETICS OF ITS ACTION. J Mol Biol. 1965 Jun;12:305–327. doi: 10.1016/s0022-2836(65)80255-8. [DOI] [PubMed] [Google Scholar]
- Schwer B., Stunnenberg H. G. Vaccinia virus late transcripts generated in vitro have a poly(A) head. EMBO J. 1988 Apr;7(4):1183–1190. doi: 10.1002/j.1460-2075.1988.tb02929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Visca P., Vos J. C., Stunnenberg H. G. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5' poly(A) leader. Cell. 1987 Jul 17;50(2):163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebring E. D., Salzman N. P. Metabolic properties of early and late vaccinia virus messenger ribonucleic acid. J Virol. 1967 Jun;1(3):550–558. doi: 10.1128/jvi.1.3.550-558.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S., Broyles S. S., Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987 Sep 5;262(25):12372–12380. [PubMed] [Google Scholar]
- Silver M., Dales S. Biogenesis of vaccina: interrelationship between post-translational cleavage, virus assembly, and maturation. Virology. 1982 Mar;117(2):341–356. doi: 10.1016/0042-6822(82)90474-3. [DOI] [PubMed] [Google Scholar]
- Spencer E., Shuman S., Hurwitz J. Purification and properties of vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1980 Jun 10;255(11):5388–5395. [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Thompson C. L., Hooda-Dhingra U., Condit R. C. Fine structure mapping of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J Virol. 1989 Feb;63(2):705–713. doi: 10.1128/jvi.63.2.705-713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Determination of the promoter region of an early vaccinia virus gene encoding thymidine kinase. Virology. 1987 May;158(1):206–210. doi: 10.1016/0042-6822(87)90254-6. [DOI] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Determination of the transcriptional regulatory region of a vaccinia virus late gene. J Virol. 1987 Jan;61(1):75–80. doi: 10.1128/jvi.61.1.75-80.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. H., King J. Surface amino acids as sites of temperature-sensitive folding mutations in the P22 tailspike protein. J Biol Chem. 1988 Jan 25;263(3):1424–1431. [PubMed] [Google Scholar]
- Yuen L., Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]