Abstract
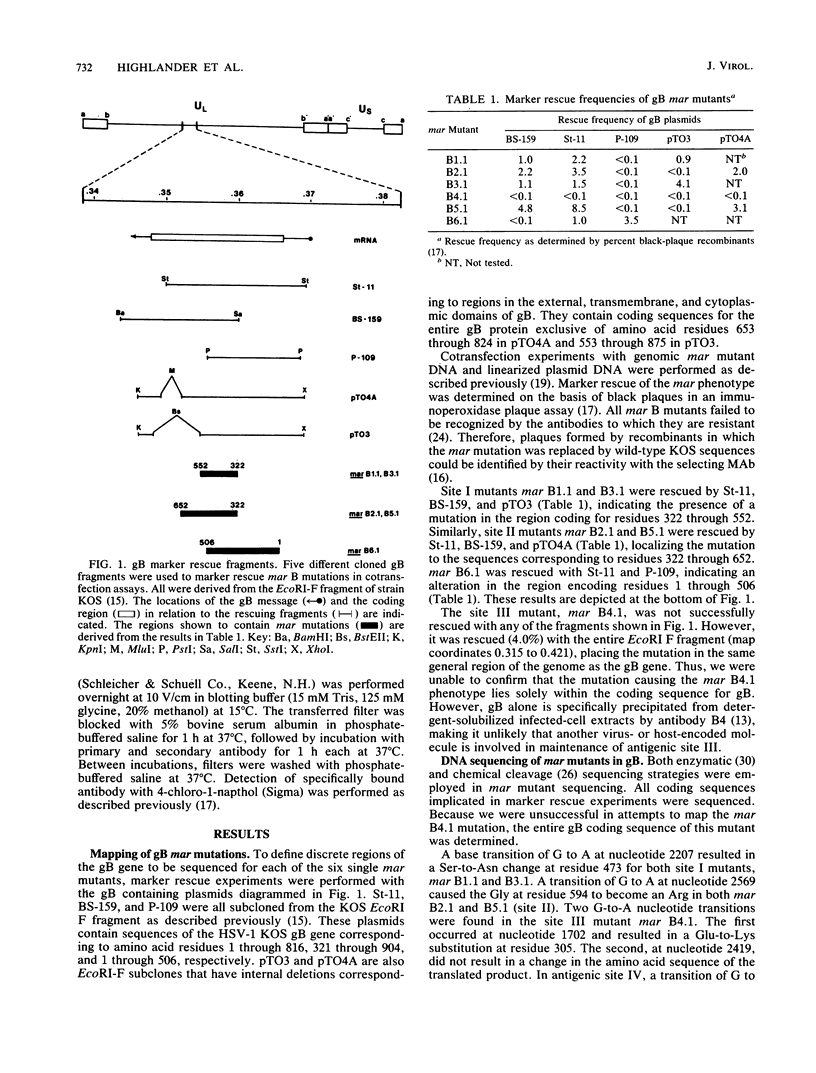
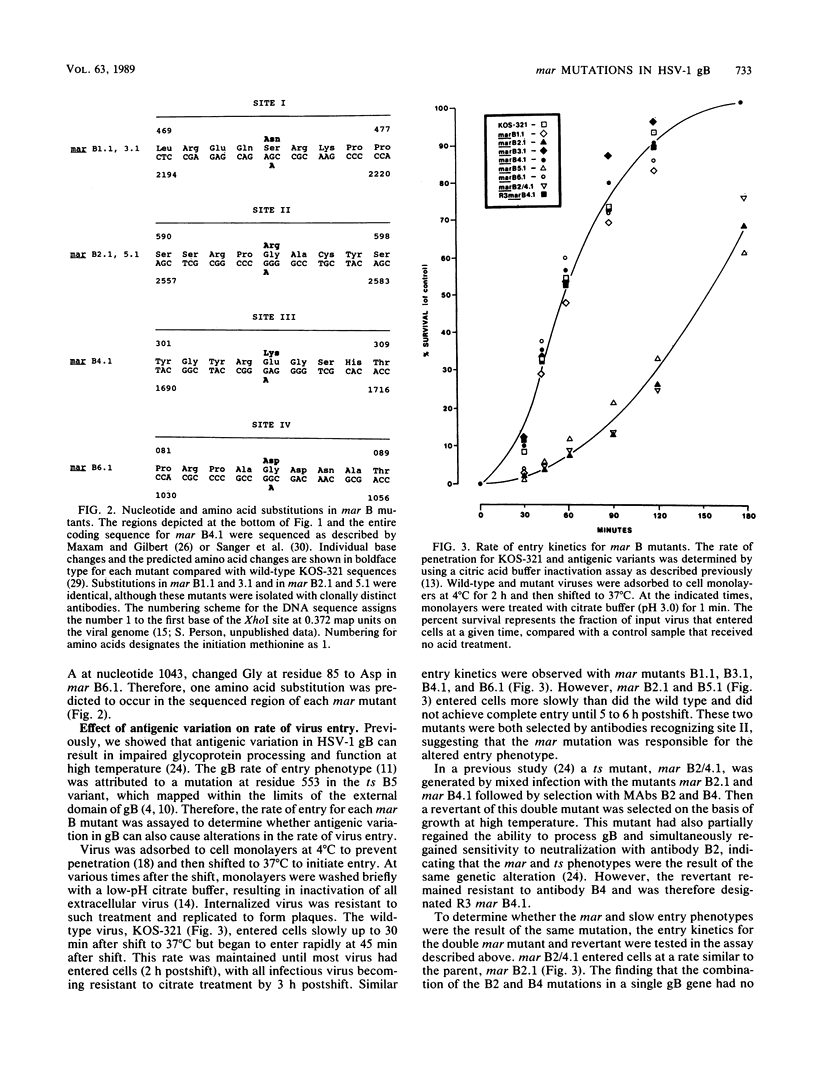
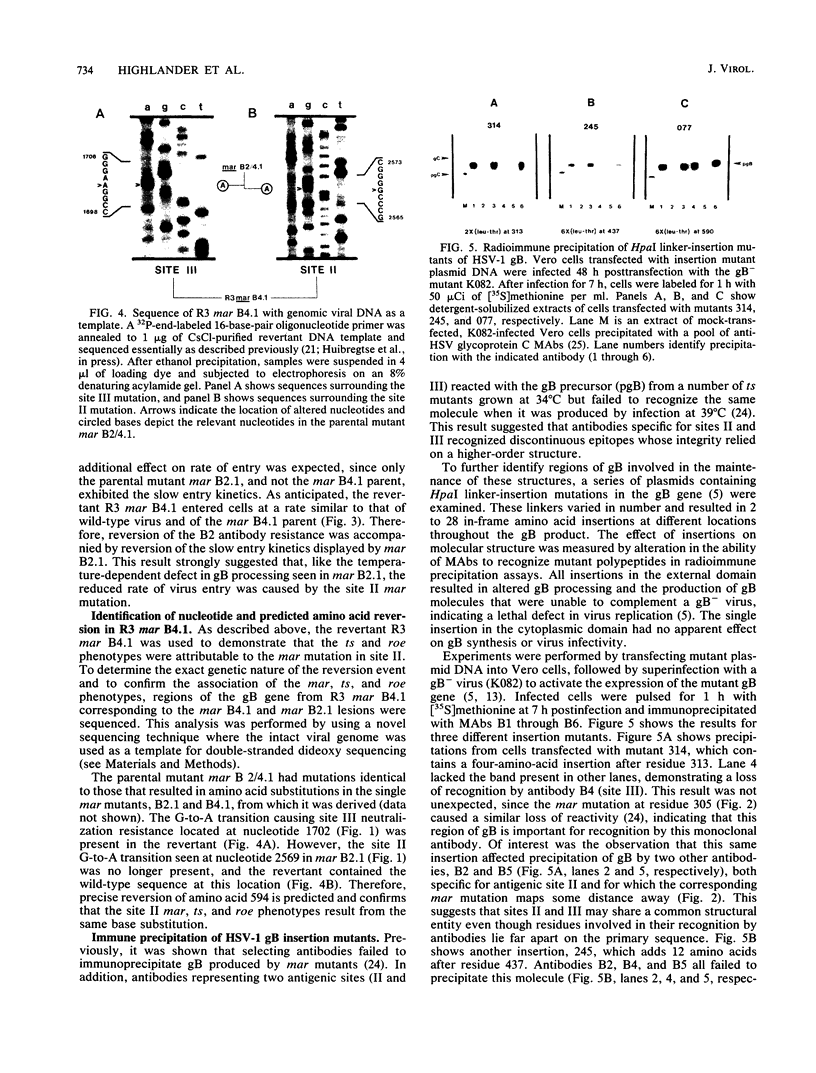
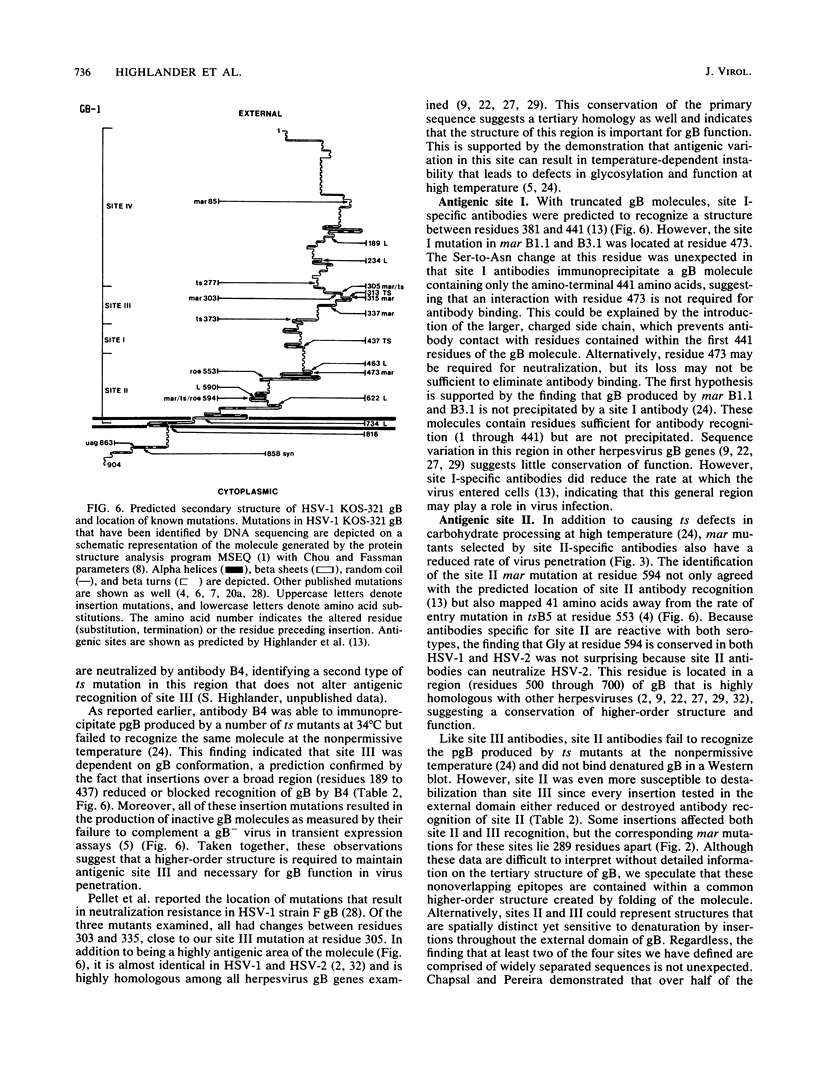
Analysis of six monoclonal antibody-resistant (mar) mutants in herpes simplex virus type 1 glycoprotein B identified two type-common (II and III) and two type-specific (I and IV) antigenic sites on this molecule. To derive additional information on the location of these sites, mar mutations were mapped and nucleotide alterations were identified by DNA sequencing. Each mutant carried a single amino acid substitution resulting from a G-to-A base transition. Alterations affecting antibody neutralization were identified at residues 473, 594, 305, and 85 for mutants in sites I through IV, respectively. Two clonally distinct site II antibodies each selected mar mutants (Gly to Arg at residue 594) that exhibited a reduction in the rate of entry (roe) into host cells. A site II mar revertant that regained sensitivity to neutralization by site II antibodies also showed normal entry kinetics. DNA sequencing of this virus identified a single base reversion of the site II mar mutation, resulting in restoration of the wild-type sequence (Arg to Gly). This finding demonstrated that the mar and roe phenotypes were the result of a single mutation. To further define structures that contributed to antibody recognition, monoclonal antibodies specific for all four sites were tested for their ability to immune precipitate a panel of linker-insertion mutant glycoprotein B molecules. Individual polypeptides that contained single insertions of 2 to 28 amino acids throughout the external domain were not recognized or were recognized poorly by antibodies specific for sites II and III, whereas no insertion affected antibody recognition of sites I and IV. mar mutations affecting either site II or III were previously shown to cause temperature-sensitive defects in glycoprotein B glycosylation, and variants altered in both these sites were temperature sensitive for virus production. Taken together, the data indicate that antigenic sites II and III are composed of higher-order structures whose integrity is linked with the ability of glycoprotein B to function in virus infectivity.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bzik D. J., Debroy C., Fox B. A., Pederson N. E., Person S. The nucleotide sequence of the gB glycoprotein gene of HSV-2 and comparison with the corresponding gene of HSV-1. Virology. 1986 Dec;155(2):322–333. doi: 10.1016/0042-6822(86)90196-0. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984 Aug;137(1):185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Cai W. Z., Person S., DebRoy C., Gu B. H. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J Mol Biol. 1988 Jun 5;201(3):575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- Cai W. Z., Person S., Warner S. C., Zhou J. H., DeLuca N. A. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987 Mar;61(3):714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapsal J. M., Pereira L. Characterization of epitopes on native and denatured forms of herpes simplex virus glycoprotein B. Virology. 1988 Jun;164(2):427–434. doi: 10.1016/0042-6822(88)90556-9. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cranage M. P., Kouzarides T., Bankier A. T., Satchwell S., Weston K., Tomlinson P., Barrell B., Hart H., Bell S. E., Minson A. C. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986 Nov;5(11):3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca N., Bzik D. J., Bond V. C., Person S., Snipes W. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gb (VP7). Virology. 1982 Oct 30;122(2):411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- DeLuca N., Bzik D., Person S., Snipes W. Early events in herpes simplex virus type 1 infection: photosensitivity of fluorescein isothiocyanate-treated virions. Proc Natl Acad Sci U S A. 1981 Feb;78(2):912–916. doi: 10.1073/pnas.78.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- HOLMES I. H., WATSON D. H. AN ELECTRON MICROSCOPE STUDY OF THE ATTACHMENT AND PENETRATION OF HERPES VIRUS IN BHK21 CELLS. Virology. 1963 Sep;21:112–123. doi: 10.1016/0042-6822(63)90309-x. [DOI] [PubMed] [Google Scholar]
- HUANG A. S., WAGNER R. R. PENETRATION OF HERPES SIMPLEX VIRUS INTO HUMAN EPIDERMOID CELLS. Proc Soc Exp Biol Med. 1964 Aug-Sep;116:863–869. doi: 10.3181/00379727-116-29392. [DOI] [PubMed] [Google Scholar]
- Highlander S. L., Cai W. H., Person S., Levine M., Glorioso J. C. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol. 1988 Jun;62(6):1881–1888. doi: 10.1128/jvi.62.6.1881-1888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander S. L., Sutherland S. L., Gage P. J., Johnson D. C., Levine M., Glorioso J. C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987 Nov;61(11):3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland T. C., Marlin S. D., Levine M., Glorioso J. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J Virol. 1983 Feb;45(2):672–682. doi: 10.1128/jvi.45.2.672-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland T. C., Sandri-Goldin R. M., Holland L. E., Marlin S. D., Levine M., Glorioso J. C. Physical mapping of the mutation in an antigenic variant of herpes simplex virus type 1 by use of an immunoreactive plaque assay. J Virol. 1983 May;46(2):649–652. doi: 10.1128/jvi.46.2.649-652.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homa F. L., Otal T. M., Glorioso J. C., Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases -34 to +124 relative to the 5' terminus of the mRNA. Mol Cell Biol. 1986 Nov;6(11):3652–3666. doi: 10.1128/mcb.6.11.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Direct identification of small sequence changes in chromosomal DNA. Gene. 1986;44(1):151–158. doi: 10.1016/0378-1119(86)90056-9. [DOI] [PubMed] [Google Scholar]
- Keller P. M., Davison A. J., Lowe R. S., Bennett C. D., Ellis R. W. Identification and structure of the gene encoding gpII, a major glycoprotein of varicella-zoster virus. Virology. 1986 Jul 15;152(1):181–191. doi: 10.1016/0042-6822(86)90383-1. [DOI] [PubMed] [Google Scholar]
- Little S. P., Jofre J. T., Courtney R. J., Schaffer P. A. A virion-associated glycoprotein essential for infectivity of herpes simplex virus type 1. Virology. 1981 Nov;115(1):149–160. doi: 10.1016/0042-6822(81)90097-0. [DOI] [PubMed] [Google Scholar]
- Marlin S. D., Highlander S. L., Holland T. C., Levine M., Glorioso J. C. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J Virol. 1986 Jul;59(1):142–153. doi: 10.1128/jvi.59.1.142-153.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin S. D., Holland T. C., Levine M., Glorioso J. C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985 Jan;53(1):128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Biggin M. D., Barrell B., Roizman B. Epstein-Barr virus genome may encode a protein showing significant amino acid and predicted secondary structure homology with glycoprotein B of herpes simplex virus 1. J Virol. 1985 Dec;56(3):807–813. doi: 10.1128/jvi.56.3.807-813.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Kousoulas K. G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985 Jan;53(1):243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Dorney D. J., Wathen M. W., Whealy M. E., Gold C., Watson R. J., Holland L. E., Weed S. D., Levine M., Glorioso J. C. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987 Sep;61(9):2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento M., Haffey M., Spear P. G. Membrane proteins specified by herpes simplex viruses. III. Role of glycoprotein VP7(B2) in virion infectivity. J Virol. 1979 Mar;29(3):1149–1158. doi: 10.1128/jvi.29.3.1149-1158.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuve L. L., Brown-Shimer S., Pachl C., Najarian R., Dina D., Burke R. L. Structure and expression of the herpes simplex virus type 2 glycoprotein gB gene. J Virol. 1987 Feb;61(2):326–335. doi: 10.1128/jvi.61.2.326-335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]





