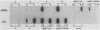Abstract
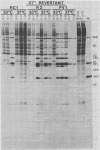
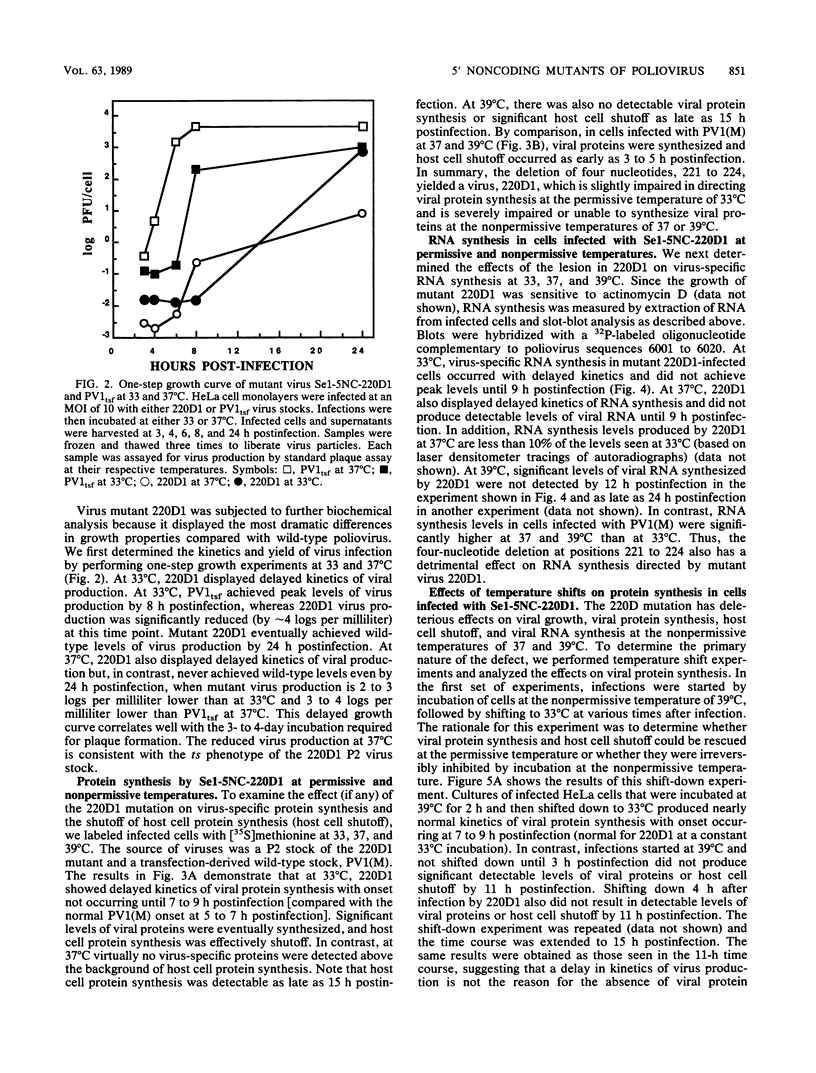
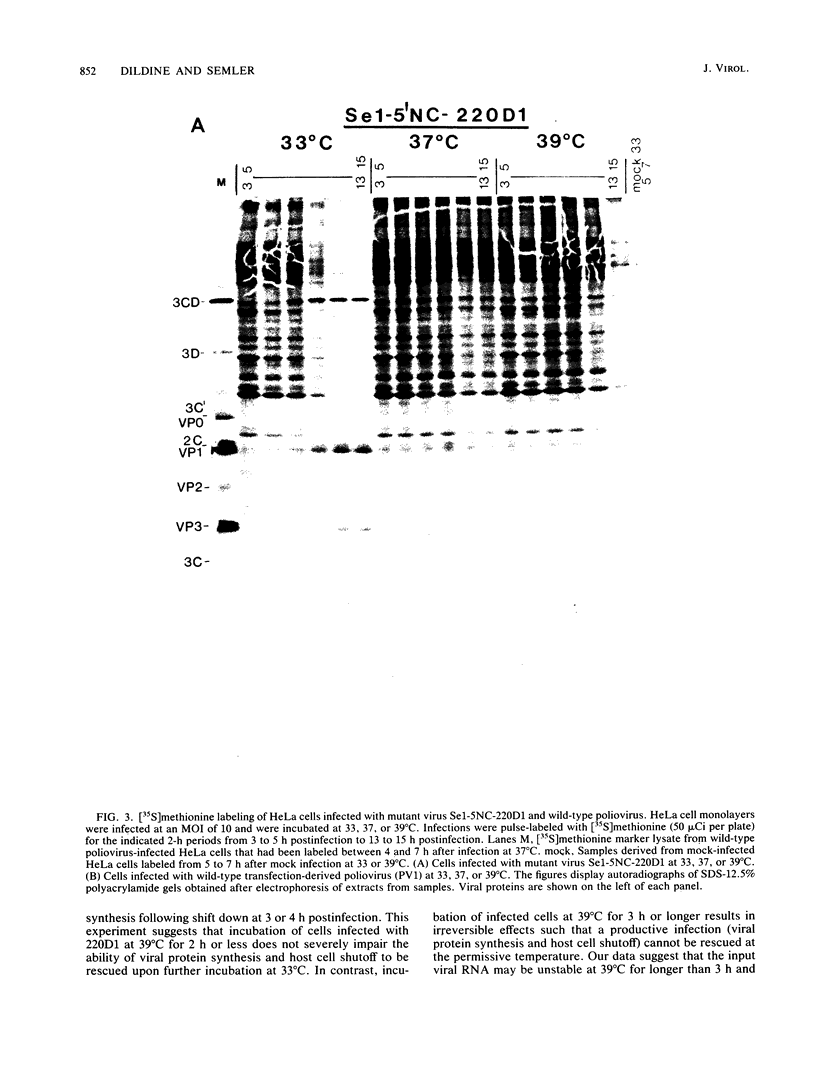
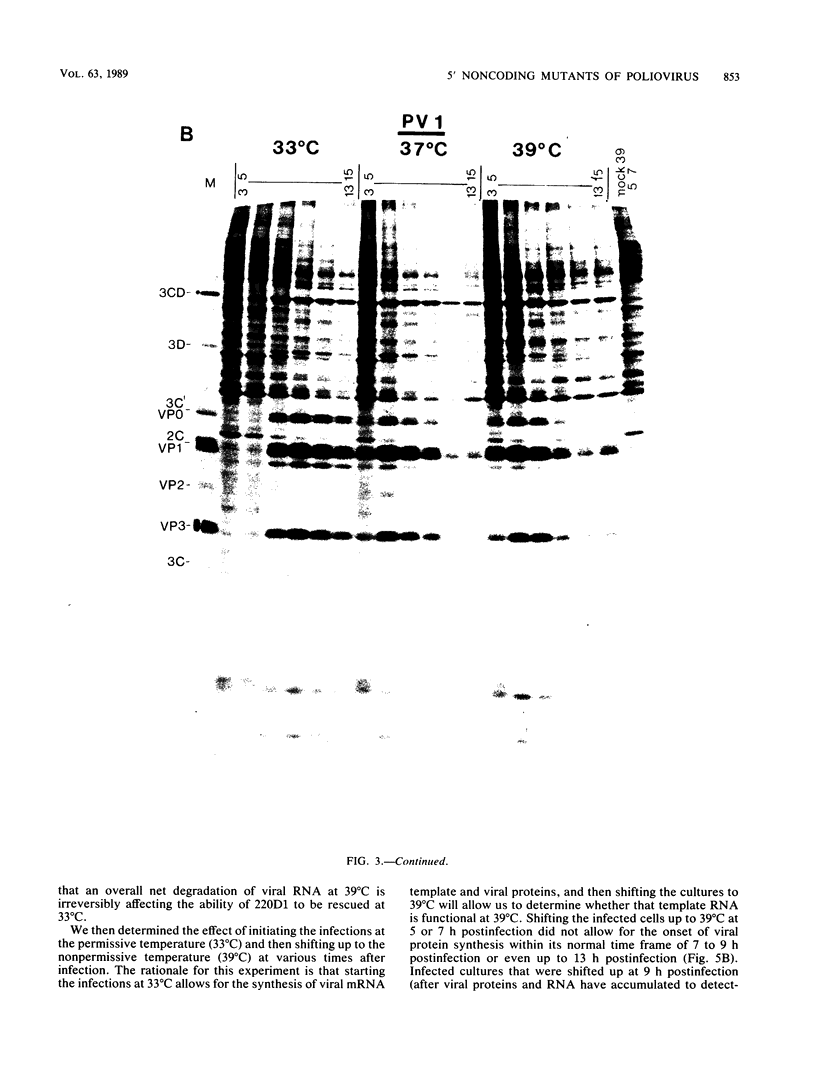
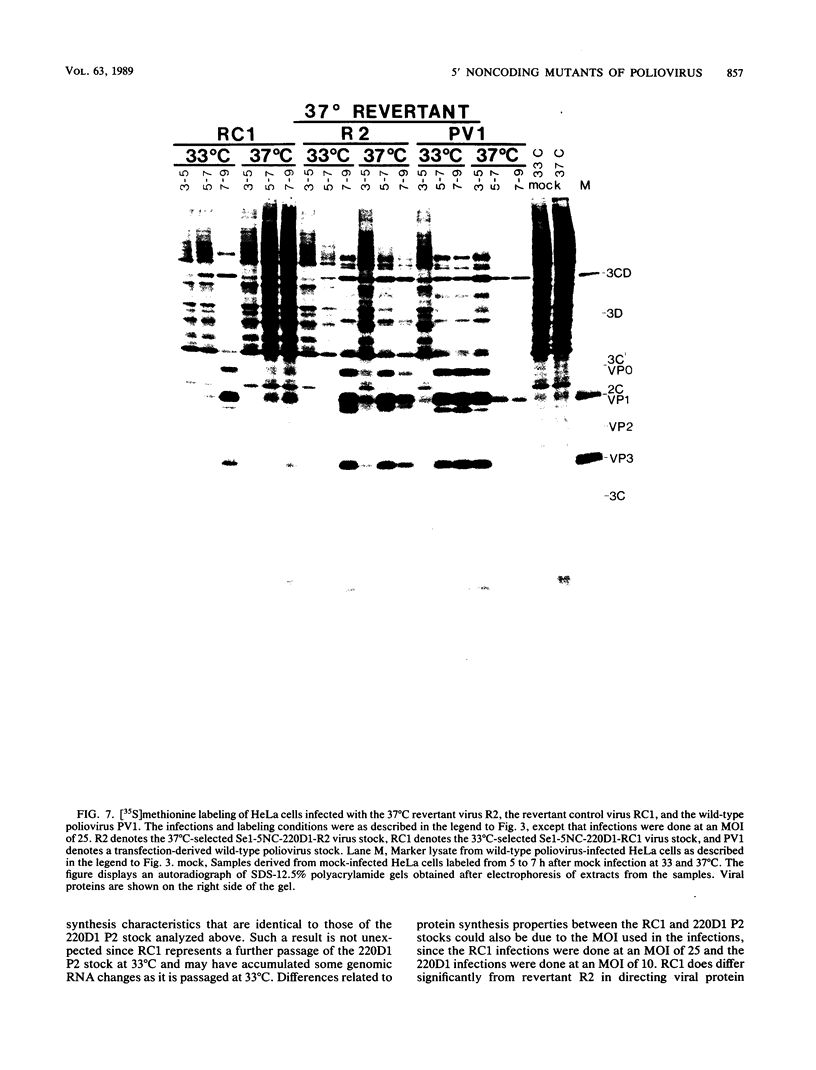
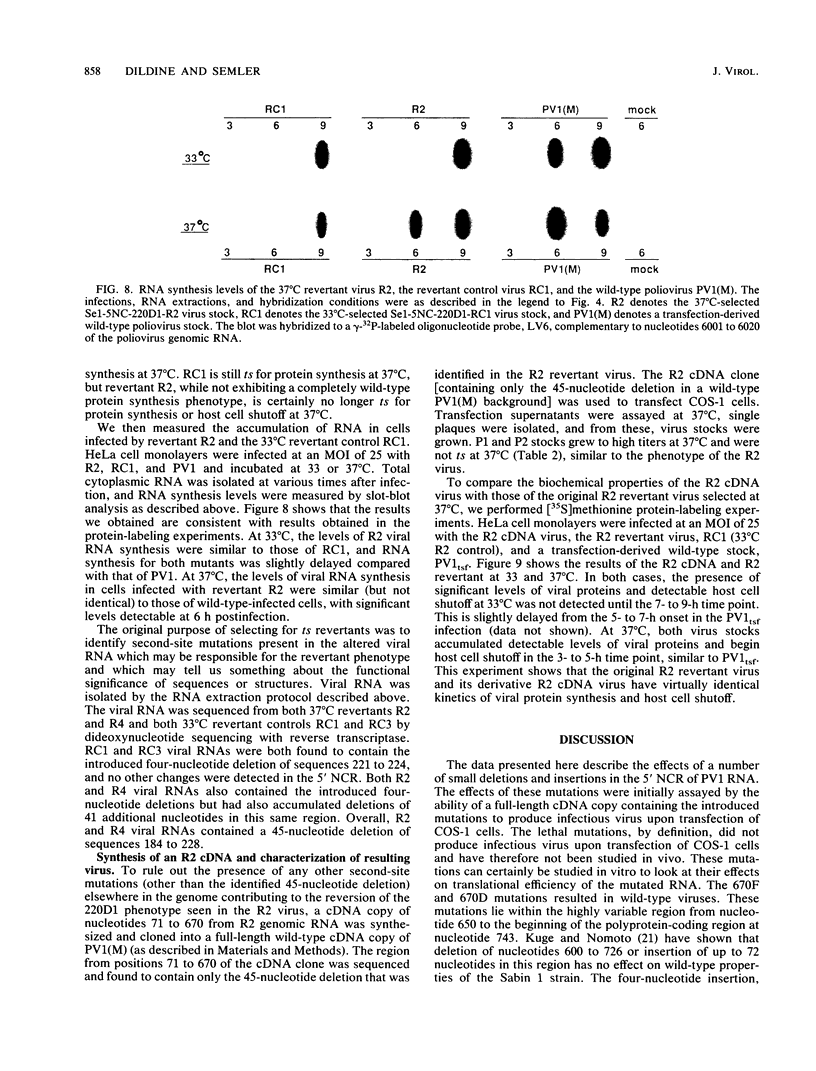
We generated a number of small deletions and insertions in the 5' noncoding region of an infectious cDNA copy of the poliovirus RNA genome. Transfection of these mutated cDNAs into COS-1 cells produced the following phenotypic categories: (i) wild-type mutations, (ii) lethal mutations, (iii) mutations exhibiting slow growth or low-titer properties, and (iv) temperature-sensitive (ts) mutations. The deletion of nucleotides 221 to 224 produced a ts virus, 220D1. Mutant 220D1 was found to have a dramatic reduction in growth, virus-specific protein and RNA synthesis, and the shutoff of host cell protein synthesis at 37 or 39 degrees C compared with 33 degrees C. Temperature shift experiments showed that the mutant viral RNA is not an effective template for protein or RNA synthesis at 39 degrees C and suggested a decreased stability of the 220D1 RNA at 39 degrees C. Selection for a non-ts revertant of 220D1 yielded the virus R2, which was no longer ts for growth or viral protein and RNA synthesis. Sequencing the 5' noncoding region of the genomic RNA from R2 revealed the deletion of 41 proximal nucleotides for an overall deletion of nucleotides 184 to 228. These data suggest that the deleted sequences are nonessential to the poliovirus life cycle during growth in HeLa cells. According to computer-predicted RNA secondary structures of the 5' noncoding region of poliovirus RNA, the R2 revertant virus has deleted an entire predicted stem-loop structure.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Ticehurst J. R., Miele T. A., Maizel J. V., Jr, Purcell R. H., Feinstone S. M. Sequence analysis of hepatitis A virus cDNA coding for capsid proteins and RNA polymerase. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2143–2147. doi: 10.1073/pnas.82.7.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K., Ehrenfeld E. An internal 5'-noncoding region required for translation of poliovirus RNA in vitro. J Virol. 1988 Aug;62(8):3068–3072. doi: 10.1128/jvi.62.8.3068-3072.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Callahan P. L., Mizutani S., Colonno R. J. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci U S A. 1985 Feb;82(3):732–736. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos R., Villarreal L. P. An SV40 deletion mutant accumulates late transcripts in a paranuclear extract. Virology. 1982 May;119(1):1–11. doi: 10.1016/0042-6822(82)90059-9. [DOI] [PubMed] [Google Scholar]
- Carroll A. R., Rowlands D. J., Clarke B. E. The complete nucleotide sequence of the RNA coding for the primary translation product of foot and mouth disease virus. Nucleic Acids Res. 1984 Mar 12;12(5):2461–2472. doi: 10.1093/nar/12.5.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Dewalt P. G., Semler B. L. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J Virol. 1987 Jul;61(7):2162–2170. doi: 10.1128/jvi.61.7.2162-2170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner A. J., Dorner L. F., Larsen G. R., Wimmer E., Anderson C. W. Identification of the initiation site of poliovirus polyprotein synthesis. J Virol. 1982 Jun;42(3):1017–1028. doi: 10.1128/jvi.42.3.1017-1028.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forss S., Strebel K., Beck E., Schaller H. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 1984 Aug 24;12(16):6587–6601. doi: 10.1093/nar/12.16.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost E., Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978 Nov;91(1):39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- HOLLAND J. J., McLAREN L. C. Improved method for staining cell monolayers for virus plaque counts. J Bacteriol. 1959 Oct;78:596–597. doi: 10.1128/jb.78.4.596-597.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlyn P. H., Browniee G. G., Cheng C. C., Gait M. J., Milstein C. Complete sequence of constant and 3' noncoding regions of an immunoglobulin mRNA using the dideoxynucleotide method of RNA sequencing. Cell. 1978 Nov;15(3):1067–1075. doi: 10.1016/0092-8674(78)90290-8. [DOI] [PubMed] [Google Scholar]
- Harris T. J., Dunn J. J., Wimmer E. Identification of specific fragments containing the 5' end of poliovirus RNA after ribonuclease III digestion. Nucleic Acids Res. 1978 Nov;5(11):4039–4054. doi: 10.1093/nar/5.11.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V. H., Semler B. L. Defined recombinants of poliovirus and coxsackievirus: sequence-specific deletions and functional substitutions in the 5'-noncoding regions of viral RNAs. Virology. 1988 Jan;162(1):47–57. doi: 10.1016/0042-6822(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5' noncoding sequence in viral replication. J Virol. 1987 May;61(5):1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen G. R., Semler B. L., Wimmer E. Stable hairpin structure within the 5'-terminal 85 nucleotides of poliovirus RNA. J Virol. 1981 Jan;37(1):328–335. doi: 10.1128/jvi.37.1.328-335.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linemeyer D. L., Menke J. G., Martin-Gallardo A., Hughes J. V., Young A., Mitra S. W. Molecular cloning and partial sequencing of hepatitis A viral cDNA. J Virol. 1985 May;54(2):247–255. doi: 10.1128/jvi.54.2.247-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarian R., Caput D., Gee W., Potter S. J., Renard A., Merryweather J., Van Nest G., Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985 May;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin M. J., Kräusslich H. G., Toyoda H., Dunn J. J., Wimmer E. Poliovirus polypeptide precursors: expression in vitro and processing by exogenous 3C and 2A proteinases. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4002–4006. doi: 10.1073/pnas.84.12.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T., Kohara M., Sakai Y., Kameda A., Imura N., Nomoto A. Cloned infectious complementary DNA of the poliovirus Sabin 1 genome: biochemical and biological properties of the recovered virus. Gene. 1984 Dec;32(1-2):1–10. doi: 10.1016/0378-1119(84)90026-x. [DOI] [PubMed] [Google Scholar]
- Palmenberg A. C., Kirby E. M., Janda M. R., Drake N. L., Duke G. M., Potratz K. F., Collett M. S. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 1984 Mar 26;12(6):2969–2985. doi: 10.1093/nar/12.6.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B. A., Stark G. R. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus or viral DNA. J Virol. 1979 Aug;31(2):360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Kaplan G., Racaniello V. R., Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5' noncoding region. Mol Cell Biol. 1988 Mar;8(3):1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981 Nov 20;214(4523):916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Meriam C. Poliovirus temperature-sensitive mutant containing a single nucleotide deletion in the 5'-noncoding region of the viral RNA. Virology. 1986 Dec;155(2):498–507. doi: 10.1016/0042-6822(86)90211-4. [DOI] [PubMed] [Google Scholar]
- Rivera V. M., Welsh J. D., Maizel J. V., Jr Comparative sequence analysis of the 5' noncoding region of the enteroviruses and rhinoviruses. Virology. 1988 Jul;165(1):42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler B. L., Dorner A. J., Wimmer E. Production of infectious poliovirus from cloned cDNA is dramatically increased by SV40 transcription and replication signals. Nucleic Acids Res. 1984 Jun 25;12(12):5123–5141. doi: 10.1093/nar/12.12.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler B. L., Johnson V. H., Tracy S. A chimeric plasmid from cDNA clones of poliovirus and coxsackievirus produces a recombinant virus that is temperature-sensitive. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1777–1781. doi: 10.1073/pnas.83.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skern T., Sommergruber W., Blaas D., Gruendler P., Fraundorfer F., Pieler C., Fogy I., Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985 Mar 25;13(6):2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Minor P. D., Almond J. W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984 Oct 25;12(20):7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Kohara M., Kataoka Y., Suganuma T., Omata T., Imura N., Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984 Apr 25;174(4):561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- Trono D., Andino R., Baltimore D. An RNA sequence of hundreds of nucleotides at the 5' end of poliovirus RNA is involved in allowing viral protein synthesis. J Virol. 1988 Jul;62(7):2291–2299. doi: 10.1128/jvi.62.7.2291-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D., Pelletier J., Sonenberg N., Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5' noncoding region. Science. 1988 Jul 22;241(4864):445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]