Abstract
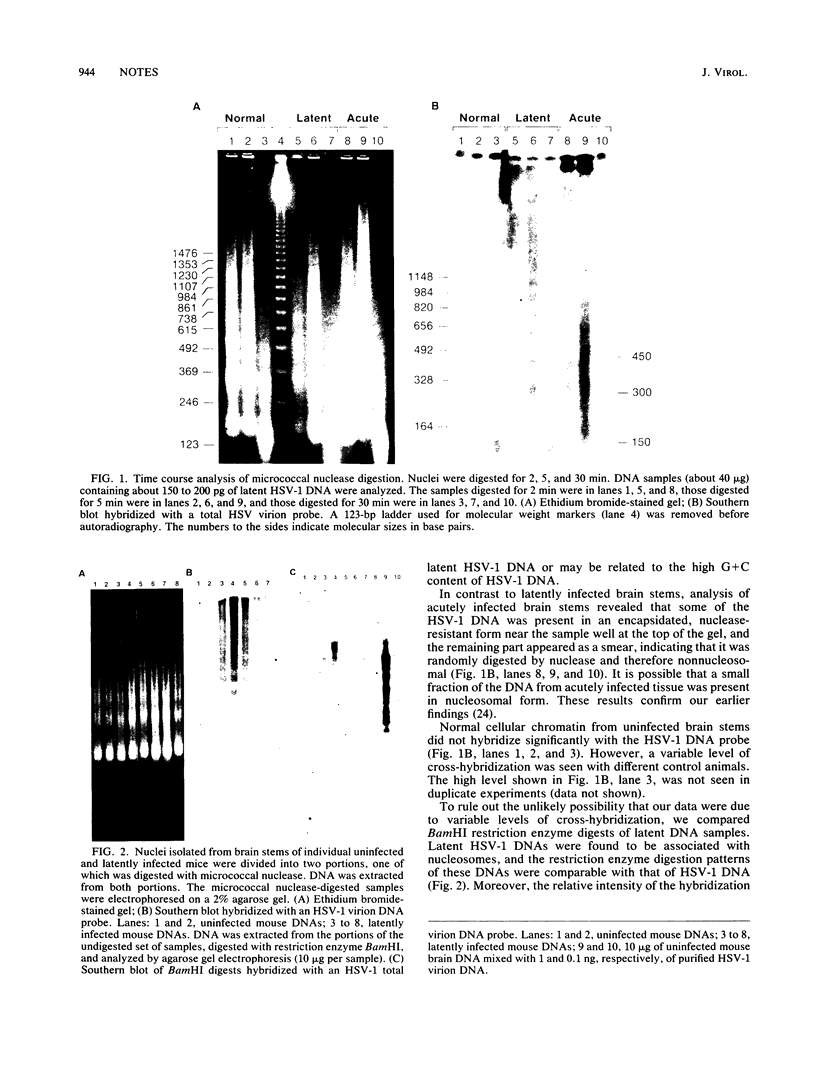
Latent herpes simplex virus type 1 DNA has a nucleosomal structure similar to that of cellular chromatin, as determined by micrococcal nuclease digestion. All of the major regions, including the transcriptionally active region of the genome, were found to be associated with nucleosomes. Such a chromatin structure is likely to be an important element in the control of herpes simplex virus type 1 gene expression during latency.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook M. L., Bastone V. B., Stevens J. G. Evidence that neurons harbor latent herpes simplex virus. Infect Immun. 1974 May;9(5):946–951. doi: 10.1128/iai.9.5.946-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen K. D., Ostrove J. M., Dragovic L. J., Smialek J. E., Straus S. E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene "anti-sense" transcript by in situ hybridization. N Engl J Med. 1987 Dec 3;317(23):1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- Deatly A. M., Spivack J. G., Lavi E., Fraser N. W. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci U S A. 1987 May;84(10):3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R., Rock D. L., Fraser N. W. Latent herpes simplex virus type 1 DNA is not extensively methylated in vivo. J Gen Virol. 1987 Jun;68(Pt 6):1761–1765. doi: 10.1099/0022-1317-68-6-1761. [DOI] [PubMed] [Google Scholar]
- Dyson P. J., Farrell P. J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985 Sep;66(Pt 9):1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- Efstathiou S., Minson A. C., Field H. J., Anderson J. R., Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986 Feb;57(2):446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hall M. R., Aghili N., Hall C., Martinez J., St Jeor S. Chromosomal organization of the herpes simplex virus type 2 genome. Virology. 1982 Dec;123(2):344–356. doi: 10.1016/0042-6822(82)90268-9. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974 Jul;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinbach S. S., Summers W. C. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J Gen Virol. 1980 Nov;51(Pt 1):45–59. doi: 10.1099/0022-1317-51-1-45. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Mellerick D. M., Fraser N. W. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology. 1987 Jun;158(2):265–275. doi: 10.1016/0042-6822(87)90198-x. [DOI] [PubMed] [Google Scholar]
- Mouttet M. E., Guétard D., Béchet J. M. Random cleavage of intranuclear herpes simplex virus DNA by micrococcal nuclease. FEBS Lett. 1979 Apr 1;100(1):107–109. doi: 10.1016/0014-5793(79)81141-2. [DOI] [PubMed] [Google Scholar]
- Muggeridge M. I., Fraser N. W. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J Virol. 1986 Sep;59(3):764–767. doi: 10.1128/jvi.59.3.764-767.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Schröder C. H., Zentgraf H., Franke W. W. Coexistence of nucleosomal and various non-nucleosomal chromatin configurations in cells infected with herpes simplex virus. Eur J Cell Biol. 1980 Dec;23(1):197–203. [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Fraser N. W. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature. 1983 Apr 7;302(5908):523–525. doi: 10.1038/302523a0. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Fraser N. W. Latent herpes simplex virus type 1 DNA contains two copies of the virion DNA joint region. J Virol. 1985 Sep;55(3):849–852. doi: 10.1128/jvi.55.3.849-852.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock D. L., Nesburn A. B., Ghiasi H., Ong J., Lewis T. L., Lokensgard J. R., Wechsler S. L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987 Dec;61(12):3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Shaw J. E., Levinger L. F., Carter C. W., Jr Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979 Feb;29(2):657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. E. The circular intracellular form of Epstein-Barr virus DNA is amplified by the virus-associated DNA polymerase. J Virol. 1985 Mar;53(3):1012–1015. doi: 10.1128/jvi.53.3.1012-1015.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E., Francke B. Organization of herpes simplex virus type 1 deoxyribonucleic acid during replication probed in living cells with 4,5',8-trimethylpsoralen. Biochemistry. 1982 Aug 31;21(18):4484–4490. doi: 10.1021/bi00261a045. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987 Dec;61(12):3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner I., Spivack J. G., O'Boyle D. R., 2nd, Lavi E., Fraser N. W. Latent herpes simplex virus type 1 transcription in human trigeminal ganglia. J Virol. 1988 Sep;62(9):3493–3496. doi: 10.1128/jvi.62.9.3493-3496.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. G. Latent herpes simplex virus and the nervous system,. Curr Top Microbiol Immunol. 1975;70:31–50. doi: 10.1007/978-3-642-66101-3_2. [DOI] [PubMed] [Google Scholar]
- Stevens J. G., Wagner E. K., Devi-Rao G. B., Cook M. L., Feldman L. T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987 Feb 27;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]