Abstract
The granulocyte-macrophage colony-stimulating factor (GM-CSF) gene is part of a cytokine gene cluster and is directly linked to a conserved upstream inducible enhancer. Here we examined the in vitro and in vivo functions of the human GM-CSF enhancer and found that it was required for the correctly regulated expression of the GM-CSF gene. An inducible DNase I-hypersensitive site appeared within the enhancer in cell types such as T cells, myeloid cells, and endothelial cells that express GM-CSF, but not in nonexpressing cells. In a panel of transfected cells the human GM-CSF enhancer was activated in a tissue-specific manner in parallel with the endogenous gene. The in vivo function of the enhancer was examined in a transgenic mouse model that also addressed the issue of whether the GM-CSF locus was correctly regulated in isolation from other segments of the cytokine gene cluster. After correction for copy number the mean level of human GM-CSF expression in splenocytes from 11 lines of transgenic mice containing a 10.5-kb human GM-CSF transgene was indistinguishable from mouse GM-CSF expression (99% ± 56% SD). In contrast, a 9.8-kb transgene lacking just the enhancer had a significantly reduced (P = 0.004) and more variable level of activity (29% ± 89% SD). From these studies we conclude that the GM-CSF enhancer is required for the correct copy number-dependent expression of the human GM-CSF gene and that the GM-CSF gene is regulated independently from DNA elements associated with the closely linked IL-3 gene or other members of the cytokine gene cluster.
Granulocyte-macrophage colony-stimulating factor (GM-CSF) was first identified as a cytokine that stimulates the in vitro growth and differentiation of colonies of GM lineage cells from bone marrow progenitor cells (1). In vivo, GM-CSF functions predominantly in the recruitment and activation of myeloid lineage cells at sites of inflammation. In both humans and mice the GM-CSF gene is located just downstream of the IL-3 gene and these closely related genes appear to have arisen by gene duplication. The human GM-CSF and IL-3 genes reside just 10 kb apart in a compact locus that includes the two genes (2) and numerous associated regulatory elements (3–11), all within a span of about 30 kb. This pair forms part of a larger cytokine gene cluster that also includes the IL-4, IL-5, and IL-13 genes several hundred kb downstream of the GM-CSF gene (12).
IL-3 and GM-CSF expression is tightly regulated in a tissue-specific manner (1, 3, 7, 13). In T cells the two genes are coregulated as in most cases they both are expressed on activation of T cell receptor (TCR) signaling pathways. Although they respond to similar signaling pathways, the IL-3 and GM-CSF genes also can be differentially regulated. Although IL-3 expression is restricted primarily to T cells, GM-CSF is produced in response to proinflammatory agents such as bacterial lipopolysaccharide and tumor necrosis factor α by a wide range of additional cell types that includes monocytes, endothelial cells, and fibroblasts (1).
It is of general interest to determine whether conserved gene clusters use shared regulatory elements, as is clearly the case in the globin loci, or whether the regulatory apparatus is replicated along with the genes in conserved gene clusters. In the α and β globin loci, clusters of conserved genes are expressed stepwise during development and are regulated by upstream locus control regions that exist as DNase I-hypersensitive (DH) sites in erythroid cells (14–16). Our previous studies of the human IL-3/GM-CSF gene cluster revealed the existence of an extensive array of DH sites that encompasses DNA elements likely to be required for the correct inducible, tissue-specific, and differential regulation of the IL-3 and GM-CSF genes (3–6, 17) (summarized in ref. 6). The best-studied of these elements are two inducible DH sites located 3 kb upstream of the GM-CSF gene (3–5, 17) and 14 kb upstream of the IL-3 gene (6), which exist within inducible enhancers that respond to activation of TCR signaling pathways. Each enhancer encompasses an array of binding sites for the Ca2+-inducible transcription factor NFAT (nuclear factor of activated T cells) and is suppressed by CsA that inhibits induction of NFAT (3, 4, 6, 13).
In this study we explored the in vitro and in vivo functions of the GM-CSF enhancer. We determined that the GM-CSF enhancer functions in essentially all cell types that express GM-CSF and that the GM-CSF locus can be correctly regulated in vivo independently of DNA elements in the IL-3 locus.
Materials and Methods
DH Site Analyses.
DH sites in a wide range of cell lines were assayed as described (3, 6). Briefly, for each cell line a DNase I titration was performed and samples that had optimal extents of DNase I digestion were selected for Southern blot hybridization analysis of DH sites. DH sites up to 6.8 kb from the GM-CSF gene were mapped within a 9.4-kb EcoRI GM-CSF gene fragment by using a 1.5-kb BamHI fragment as described (3). The region downstream of the IL-3 gene was mapped by using BamHI-digested DNA and a 1-kb BglII/BamHI fragment of λJ1–16 DNA (2).
Plasmid Construction.
The plasmid pGM contains the 627-bp human GM-CSF promoter upstream of a luciferase reporter gene. pGM-GME and pGM-SV40E consist of a 717-bp BglII fragment containing the GM-CSF enhancer or a 158-bp fragment of the simian virus 40 (SV40) enhancer, respectively cloned upstream of the GM-CSF promoter of pGM (5, 6). Note that the 717-bp GM-CSF enhancer contains an extra G at position 118 from the sequence published in ref. 3.
Cell Culture.
The Jurkat, CEM, and HSB2 T cell lines, KG1a and U937 myeloid cell lines, K562 pro-erythroid cell line, human embryonic lung fibroblasts, HeLa cervical carcinoma cell line, HepG2 hepatic cell line, and Raji B and Ball-1 B cell lines were cultured in RPMI medium containing 10% FBS. The 5637 bladder carcinoma epithelial cell line was cultured in RPMI containing 7% FBS. Human umbilical vein endothelial cells (HUVECs) were cultured as described (17). Human peripheral blood T cells were prepared by isolating mononuclear cells from whole blood and depleting B cells by adhering them to nylon wool.
Transient Transfections and Luciferase Assays.
All cells, with the exception of HUVECs, were transfected with 5 μg CsCl-purified plasmid DNA by electroporation, cultured for 20–24 hr, left either unstimulated or stimulated with 20 ng/ml phorbol 12-myristate 13-acetate (PMA) and 1 μM calcium ionophore A23187 for 9 hr, and assayed for luciferase reporter gene activities as described (6). HUVECs were transfected with 15 μg CsCl-purified plasmid DNA by the DEAE-dextran procedure, cultured for 24 hr, either unstimulated or stimulated with 20 ng/ml PMA and 2 μM calcium ionophore A23187 (PMA/I) for 9 hr, and assayed for luciferase reporter gene activities as described (17).
Generation and Identification of Transgenic Mice.
The human transgene contains a 10.5-kb XhoI/HindIII region encompassing the human GM-CSF gene and extends 5.9 kb upstream of the gene and 2.4 kb downstream of the gene. This transgene was made by fusing the HindIII fragment of pCH5.2 (18) to a HindIII segment of λJ1–16 (2). The 9.8-kb transgene lacking the enhancer was prepared by excising a 717-bp BglII fragment encompassing the enhancer. Transgenic mice were generated by microinjection of DNA into the pronuclei of fertilized mouse eggs, which then were cultured overnight to the two-cell stage and transferred into the oviducts of pseudopregnant females. Transgenic founders were identified and their copy numbers of integrated transgenes were estimated by Southern blot analysis of transgenic mouse DNA. Founders then were bred to establish lines used for GM-CSF expression analyses.
GM-CSF Expression Assay.
Whole spleens were isolated from transgenic mice and squeezed between two glass slides to extract lymphoid cells, and cells were plated out to 1 × 107 lymphocytes per ml in Iscove's modified Dulbecco's medium containing 10% FBS and 5 × 10−5 M β-mercaptoethanol. Cells were either left unstimulated (−) or stimulated with 20 ng/ml PMA and 1 μM calcium ionophore A23187 (PMA/I) for 15 hr (+). Supernatants were harvested and assayed for mouse and human GM-CSF levels by ELISA (R&D Systems).
Results
Chromatin Remodeling Occurs in the GM-CSF Enhancer in All Cells Where It Is Active.
To determine whether chromatin remodeling of the GM-CSF enhancer was a consistent feature of GM-CSF-expressing cells we mapped DH sites in a wide variety of human cell types where we previously had assayed induction of GM-CSF mRNA expression (unpublished data). These cells included T cells that coexpress IL-3 and GM-CSF, myeloid and nonhaemopoietic adherent cell types that express GM-CSF but not IL-3, and cell lines that do not express either gene. Based on these expression patterns the collection of cells was divided into the three groups shown in Fig. 1A. As this panel of cells normally would require a diverse range of agents to activate the GM-CSF locus via cell surface receptors (1), we chose a combination of inducing agents that directly triggers the intracellular signaling pathways that lead to GM-CSF expression. GM-CSF enhancer activation and DH site induction in T cells and endothelial cells are known to involve both Ca2+ and kinase-dependent pathways, which in T cells are linked to the TCR (3–8, 13). Hence, all of the cells used in this study were activated with the combination of the calcium ionophore A23187 and the phorbol ester PMA, which directly activates Ca2+ and kinase pathways.
Figure 1.
Mapping of DH sites between the IL-3 and GM-CSF genes. (A) Mapping of GM-CSF enhancer and promoter DH sites in an EcoRI fragment in DNase I-digested nuclei isolated from either unstimulated cells (−) or cells stimulated for 4–6 hr with 20 ng/ml PMA and 2 μM A23187 (+). On the right, the letters indicate DH sites at the GM-CSF promoter (P), GM-CSF enhancer (E), and a site at −4.4 kb present predominantly in myeloid cells (M). (A and B) The first three lanes from the left hand side are a HindIII digest of λ DNA (molecular weight marker), total non-DNaseI-digested Jurkat DNA, and a partial HindIII digest of Jurkat DNA, respectively. The lanes labeled T cells are human peripheral blood T cells. (B) Mapping of DH sites in the BamHI fragment downstream of the IL-3 gene as in A. * marks an inducible DH site present in some cell lines. (C) Map of identified DH sites together with the probes used to identify them. The transgene constructs used to make transgenic mice also are shown. (D) Mapping of DH sites in Jurkat and K562 cells as in A but at higher resolution.
To induce DH sites cells were stimulated for 4–6 hr with A23187 and PMA. We mapped DH sites from an EcoRI site 6.8 kb upstream of the GM-CSF gene and detected up to three DH sites (Fig. 1 A and C). We observed a striking correlation between the induction of DH sites within the enhancer and the ability of cells to express GM-CSF on activation. Accordingly, the DH site was induced in the enhancer in peripheral blood T cells and in the Jurkat, CEM, and HSB2 T cell lines, all of which express both GM-CSF and IL-3. Within the myeloid lineage the DH site appeared in the cell lines KG1a and K562, which express GM-CSF (Fig. 1A), but not in activated U937 cells that do not express GM-CSF (data not shown). Among nonhaemopoietic cells the enhancer DH site appeared in HUVECs, human embryonic lung fibroblasts, HeLa cells, and HepG2 cells in parallel with induction of GM-CSF expression. We also analyzed the two B cell lines, Raji and Ball-1, that do not express GM-CSF, and no significant DH sites were detected upstream of the GM-CSF gene. The only cells that expressed GM-CSF without a DH site appearing in the enhancer were the 5637 cells. In most cases the DH site spanned a 250-bp region defined by ApaI and PstI sites (4) but in pro-erythroid K562 cells the DH site appeared as a doublet and extended approximately 150 bp further upstream to include additional conserved sequences that include GATA and Ets elements (5). This doublet is seen more clearly in Fig. 1D where the patterns in Jurkat and K562 cells are compared at higher resolution.
We also detected a DH site in the GM-CSF promoter in most cells expressing GM-CSF, but there was no strict correlation between the presence of this site and the state of activity of the GM-CSF gene (Fig. 1A). Furthermore, this site was absent from activated Jurkat cells that express GM-CSF but present in U937 cells that do not express GM-CSF. Interestingly, we detected a strong constitutive DH site in the promoter in 5637 cells, suggesting that the locus is activated via promoter rather than enhancer-dependent mechanisms in these cells. In addition to the promoter and enhancer a third weak pre-existing DH site was detected in some cell lines in the vicinity of the HindIII site 1 kb upstream of the enhancer (M, Fig. 1 A and C). This site was most pronounced in the myeloid cell lines KG1a and U937.
Additional DH sites exist just downstream of the IL-3 gene (3) so we probed the same DNaseI-digested samples for DH sites within a 4.6-kb BamHI fragment that overlaps the EcoRI fragment (Fig. 1 B and C). Mapping of these DH sites identified three ubiquitous pre-existing sites plus one previously unidentified inducible site within 2.4 kb of the IL-3 gene. It was apparent that there was no link between the appearance of these DH sites and the activity of the locus. As previous studies of this region had failed to find any regulatory elements in this region (3) these sites were not considered further. These mapping studies effectively conclude the analysis of DH sites in the GM-CSF locus as no sites were found in either the 12-kb region downstream of the GM-CSF gene or the region where the EcoRI and BamHI fragments mapped in Fig. 1 overlap (data not shown). These data now can be used as a blueprint to develop hypotheses on the regulation of this locus.
The GM-CSF Enhancer Functions in Transfected Cells Only When the Endogenous Enhancer Can Form a DH Site.
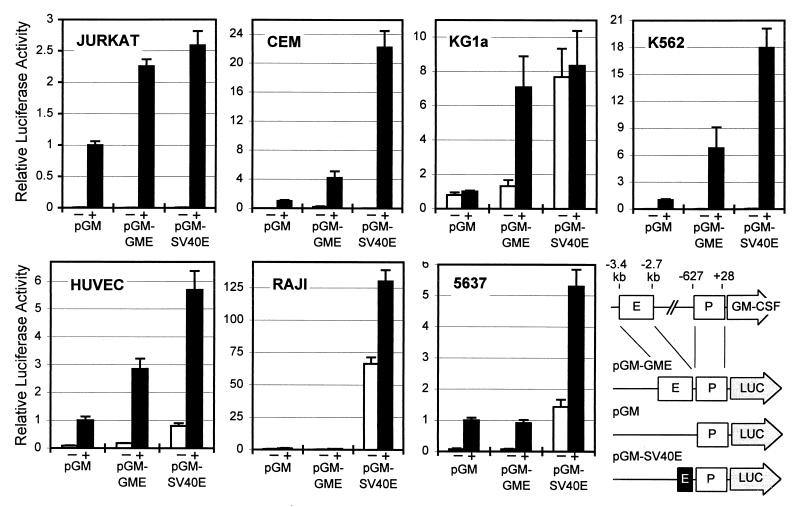
The consistent appearance of the DH site in the enhancer in GM-CSF-expressing cells suggested a strong link between chromatin remodeling, enhancer function, and GM-CSF gene activation. To further explore this link we tested the GM-CSF enhancer for function in transfection assays in seven of the cell lines illustrated above (Fig. 2). A 717-bp BglII fragment of DNA containing all of the defined GM-CSF enhancer elements (3–5, 19) was inserted upstream of the full-length GM-CSF promoter (−627 to +28) in a luciferase reporter gene plasmid (pGM-GME). This plasmid was assayed in parallel with the luciferase plasmid pGM containing the GM-CSF promoter. The plasmid pGM-SV40 containing the GM-CSF promoter plus the SV40 enhancer also was included to provide a comparison with a powerful enhancer that should function in most cell types.
Figure 2.
Tissue-specific regulation of the GM-CSF enhancer. DNA fragments encompassing either the promoter alone (pGM), promoter plus the GM-CSF enhancer (pGM-GME), or promoter plus the SV40 enhancer (pGM-SV40E) were tested for enhancer function in cells transiently transfected with luciferase reporter gene plasmids. Luciferase activity was determined for both unstimulated (−) cells and after stimulation of the cells for 9 hr with 20 ng/ml PMA and 1 μM A23187 (2 μM A23187 for HUVEC) (+). Error bars represent the SEM of 2–11 transfections. All data points are expressed relative to stimulated pGM = 1. A schematic diagram showing the constructs used also is shown.
Cells were transiently transfected with each plasmid and after 20–24 hr were stimulated for 9 hr with PMA and A23187. The luciferase plasmid pGM containing the GM-CSF promoter alone functioned in a highly inducible manner in Jurkat and CEM T cells, HUVECs, K562 cells, and 5637 cells, all of which express GM-CSF, and was essentially inactive in Raji B cells, which do not express GM-CSF. In KG1a cells, however, pGM was constitutively active even though the endogenous GM-CSF gene is inducible.
The inclusion of the GM-CSF enhancer significantly increased the inducible activity of the GM-CSF promoter plasmid in each of the cell types where the enhancer formed a DH site (Jurkat, CEM, HUVECs, KG1a, and K562). In the five cell lines where it was active the GM-CSF enhancer increased the activity of the promoter by 2- to 7-fold. The GM-CSF enhancer had an activity similar to that of the SV40 enhancer in Jurkat and KG1a cells and was 20–50% as active as the SV40 enhancer in CEM, HUVECs, and K562 cells. The enhancer was inactive, however, in Raji and 5637 cells where no DH site induction occurs in the endogenous enhancer. The analyses in 5637 cells highlighted distinct differences in the activities of the GM-CSF promoter and enhancer as it was clear that the GM-CSF promoter and the SV40 enhancer were active but the GM-CSF enhancer was nonfunctional. Most importantly, these studies highlighted the fact that the GM-CSF enhancer only functions in those cells where a DH site can form in the endogenous enhancer.
The GM-CSF Enhancer Supports Correctly Regulated Expression of the GM-CSF Gene in Vivo.
The in vitro studies above identified seven DH sites upstream of the GM-CSF gene and highlighted the GM-CSF promoter and enhancer as the elements most likely to be required for correct regulation of the GM-CSF locus in vivo. To determine the minimum requirements for a correctly regulated GM-CSF locus we generated transgenic mice containing a 10.5-kb XhoI/HindIII fragment of the human GM-CSF locus that extends from 5.8 kb upstream of the GM-CSF gene to 2.3 kb downstream of the gene (Fig. 1C). In addition to the promoter and enhancer this gene fragment includes the predominantly myeloid DH site 4.4 kb upstream of the GM-CSF gene (M, Fig. 1 A and C). A similar series of transgenic mice was generated in which the 717-bp BglII fragment containing the enhancer was excised from the 10.5-kb XhoI/HindIII fragment to generate a 9.8-kb transgene (Fig. 1C). An additional aim of this study was to determine whether the GM-CSF gene was regulated independently of elements linked to the IL-3 gene or, alternatively, whether the IL-3/GM-CSF gene cluster was coordinately regulated by shared enhancer elements.
Eleven lines of GM-CSF transgenic mice containing the wild-type human locus and eight lines of GM-CSF transgenic mice lacking the enhancer were created. Transgene function was assessed by stimulating freshly isolated splenocytes for 15 hr with PMA and A23187. In these studies it was assumed that GM-CSF production could be largely attributed to the T cell population as these represent the predominant GM-CSF producers among splenocytes. Transgene activity was assessed by performing ELISAs of both mouse and human GM-CSF produced by activated splenocytes and expressed as a simple ratio of the transgene relative to the endogenous GM-CSF gene.
The wild-type human transgenes appeared to be correctly regulated in a copy number-dependent manner as activity increased progressively with copy number, and the average relative activity of the transgenes was indistinguishable from the endogenous mouse GM-CSF gene in stimulated splenocytes (Fig. 3). When GM-CSF expression levels were expressed per gene copy number, 10 of the 11 lines containing the wild-type gene had an activity within 2-fold of that observed for the mouse GM-CSF gene (Fig. 3B), and the average activity came to 99% of the endogenous GM-CSF gene (Fig. 3C). Linear regression analysis of the copy number dependency showed that it was highly significant with a coefficient of variance (R2) of 0.91. The transgenes were highly inducible and increased in activity by an average of 7,800-fold on stimulation. GM-CSF transgenic mice lacking the GM-CSF enhancer supported a different pattern of GM-CSF gene activity in splenocytes. The average activity after correction for copy number was only 29% of that of the wild-type human transgene, and copy number dependency of expression was reduced (R2 = 0.57). The transgenes lacking the enhancer remained highly inducible but had a lower average fold induction of 900-fold.
Figure 3.
GM-CSF expression in transgenic mouse lines. Human and mouse GM-CSF protein expressed by activated splenocytes. Error bars represent the SEM of 3–5 mice from each line. (A) Relative transgene activity expressed as a ratio of human GM-CSF/mouse GM-CSF from PMA/A23187-stimulated splenocytes (not adjusted for copy number). The copy number for each line is indicated below the graph. Levels of human GM-CSF in unstimulated and PMA/A23187-stimulated splenocytes also are shown. The average levels of mouse GM-CSF produced from all lines was 1 pg/ml from unstimulated cells and 828 pg/ml from PMA/I-stimulated cells. (B) Ratio of human GM-CSF/mouse GM-CSF adjusted for copy number. (C) Average level of transgene relative activity in either the wild-type (WT) mice (SD = 56%, SEM = 18%) or Δ enhancer transgenic (ΔE) mice (SD = 89%, SEM = 34%).
Discussion
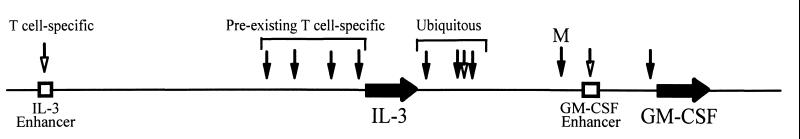
The structural studies presented in this and our previous reports have identified key regulatory elements in the IL-3/GM-CSF locus. The summary of the locations and tissue-specific distributions of the DH sites in this locus (Fig. 4) is most likely a guide to all of the DNA elements required for the correct inducible and tissue-specific regulation of the human IL-3 and GM-CSF genes. This study focused on the GM-CSF enhancer and found that activation of the endogenous gene was almost always accompanied by the appearance of a DH site in the enhancer. In every instance where a DH site was detected within the enhancer, the enhancer had the capacity to function in these cells. Based on these observations we hypothesize that the GM-CSF enhancer is involved in most situations where GM-CSF expression is induced.
Figure 4.
Summary of DH sites and enhancers in the IL-3/GM-CSF locus. Constitutive DH sites are marked as a solid arrow, and inducible DH sites are indicated by hollow arrows. M indicates the DH site present predominantly in myeloid cells.
In this study we investigated the role of the enhancer in Ca2+-dependent activation of gene expression because the enhancer encompasses an array of binding sites for the Ca2+-inducible transcription factor NFAT (3, 5, 13). The most obvious function for the GM-CSF enhancer is as a TCR-response element driven by the array of composite NFAT/AP-1 elements. However, in K562 cells, which express little or no NFAT (unpublished data), the enhancer still functioned efficiently and we found evidence for chromatin remodeling upstream of the array of NFAT sites. It will be interesting to determine in future studies whether the enhancer also is involved in Ca2+-independent activation of GM-CSF by agents such as lipopolysaccharide and tumor necrosis factor α (TNF-α). In a previous study in endothelial cells we observed that Ca2+-dependent activation of GM-CSF gene expression was CsA-sensitive but induction by TNF-α was CsA-resistant (17). This observation raises the possibility that TNF-α works selectively on the promoter by pathways that may be similar to those operating in cells such as 5637 cells where the enhancer does not function.
We also examined the in vivo role of the GM-CSF enhancer in transgenic mice. Because the enhancer was required for efficient activation of the locus in transgenic mice we suggest that the GM-CSF enhancer plus the promoter constitute the minimum amount of DNA required for the correctly regulated activation of the GM-CSF gene. These elements were contained within a 10.5-kb fragment that extended 5.7 kb upstream of the GM-CSF gene and also contained a third DH site 4.4 kb upstream that may have a role in GM-CSF expression in myeloid cells. Remarkably, this short segment of DNA had the capacity to support a level of GM-CSF expression in transgenic mice essentially identical to the endogenous mouse GM-CSF gene. We previously found in transfection studies that the mouse GM-CSF promoter plus enhancer supported the same level of inducible activity as the human GM-CSF promoter plus enhancer (5). This body of work now suggests that the mouse and human GM-CSF genes are indeed expressed at similar levels. This work further implies that the GM-CSF gene does not require any additional regulatory information from either the IL-3 locus or other regions of the cytokine gene cluster. We also identified ubiquitous DH sites downstream of the IL-3 gene but they did not appear to be required for GM-CSF gene activation. They could, however, represent a structural element within chromatin that insulates the IL-3 and GM-CSF genes from each other.
The IL-3 gene likewise may be regulated by a distinct set of elements (summarized in Fig. 4). The IL-3 gene has its own upstream enhancer that resembles the GM-CSF enhancer in its organization (6) and an additional array of at least four DH sites exists immediately upstream of the IL-3 gene (3). As all of the DH sites upstream of the IL-3 gene are for the most part restricted to T cells (unpublished data) we suggest that the 14-kb region extending upstream of the IL-3 gene constitutes a distinct regulatory cassette required for the correct regulation of the IL-3 locus in T cells. Hence, in contrast to most other conserved gene clusters the IL-3 and GM-CSF genes may be independently regulated at the gene level even though they are coregulated at the level of TCR signaling.
This study provided a unique example of a gene that supports normal levels of gene expression when isolated from a conserved gene cluster. In other gene clusters, such as the α and β globin loci, normal expression of individual genes is governed by a shared locus control region (14–16). The regulation of the IL-3/GM-CSF locus appears to be markedly different, perhaps because unlike the globin genes they are not designed to be expressed in a step-wise manner. Hence, the duplication of the regulatory apparatus may have arisen from the requirements to be able to express the GM-CSF and IL-3 genes simultaneously in T cells but regulate them independently in other cell types.
Acknowledgments
We thank C. Bonifer, G. Goodall, and T. Gonda for helpful discussions and comments on the manuscript. We thank S. Clark and J. Gasson for the gifts of DNA clones and J. Gamble for providing endothelial cells. We thank T. Kuchel and his staff for assistance in our transgenic mouse studies. This work was supported by the National Health and Medical Research council of Australia.
Abbreviations
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- TCR
T cell receptor
- DH
DNase I-hypersensitive
- NFAT
nuclear factor of activated T cells
- SV40
simian virus 40
- HUVEC
human umbilical vein endothelial cell
- PMA
phorbol 12-myristate 13-acetate
References
- 1.Nicola N A. Annu Rev Biochem. 1989;58:45–77. doi: 10.1146/annurev.bi.58.070189.000401. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y-C, Kovacic S, Kriz R, Wolf S, Clark S C, Wellems T E, Nienhuis A, Epstein N. Blood. 1988;71:958–961. [PubMed] [Google Scholar]
- 3.Cockerill P N, Shannon M F, Bert A G, Ryan G R, Vadas M A. Proc Natl Acad Sci USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockerill P N, Bert A G, Jenkins F, Ryan G R, Shannon M F, Vadas M A. Mol Cell Biol. 1995;15:2071–2079. doi: 10.1128/mcb.15.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne C S, Vadas M A, Cockerill P N. J Immunol. 1995;155:226–235. [PubMed] [Google Scholar]
- 6.Duncliffe K N, Bert A G, Vadas M A, Cockerill P N. Immunity. 1997;6:175–185. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- 7.Masuda E S, Naito Y, Arai K, Arai N. Immunologist. 1993;1:198–203. [Google Scholar]
- 8.Masuda E S, Tokumitsu H, Tsuboi A, Shlomai J, Hung P, Arai K, Arai N. Mol Cell Biol. 1993;13:7399–7407. doi: 10.1128/mcb.13.12.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himes S R, Coles L S, Reeves R, Shannon M F. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]
- 10.Mathey-Prevot B, Andrews N C, Murphey H S, Kreissman S G, Nathan D G. Proc Natl Acad Sci USA. 1990;87:5046–5050. doi: 10.1073/pnas.87.13.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor D S, Laubach J P, Nathan D G, Mathey-Prevot B. J Biol Chem. 1996;271:14020–14027. doi: 10.1074/jbc.271.24.14020. [DOI] [PubMed] [Google Scholar]
- 12.Frazer K A, Veda Y, Zhu Y, Gifford V R, Garofalo M R, Mohandas N, Martin C H, Palazzolo M J, Cheng J-F, Rubin E M. Genome Res. 1997;7:495–512. doi: 10.1101/gr.7.5.495. [DOI] [PubMed] [Google Scholar]
- 13.Rao A. Immunol Today. 1994;15:274–281. doi: 10.1016/0167-5699(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 14.Grosveld G, Blom van Assendelft G, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 15.Higgs D R, Wood W G, Jarman A P, Sharpe J, Lida J, Pretorius I M, Ayyub H. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 16.Higgs D R. Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 17.Cockerill G W, Bert A G, Ryan G R, Gamble J R, Vadas M A, Cockerill P N. Blood. 1995;86:2689–2698. [PubMed] [Google Scholar]
- 18.Chan J Y, Slamon D J, Nimer S D, Golde D W, Gasson J C. Proc Natl Acad Sci USA. 1986;83:8669–8673. doi: 10.1073/pnas.83.22.8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cockerill P N, Osborne C S, Bert A G, Grotto R J M. Cell Growth Differ. 1996;7:917–922. [PubMed] [Google Scholar]