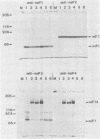
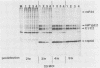
Abstract
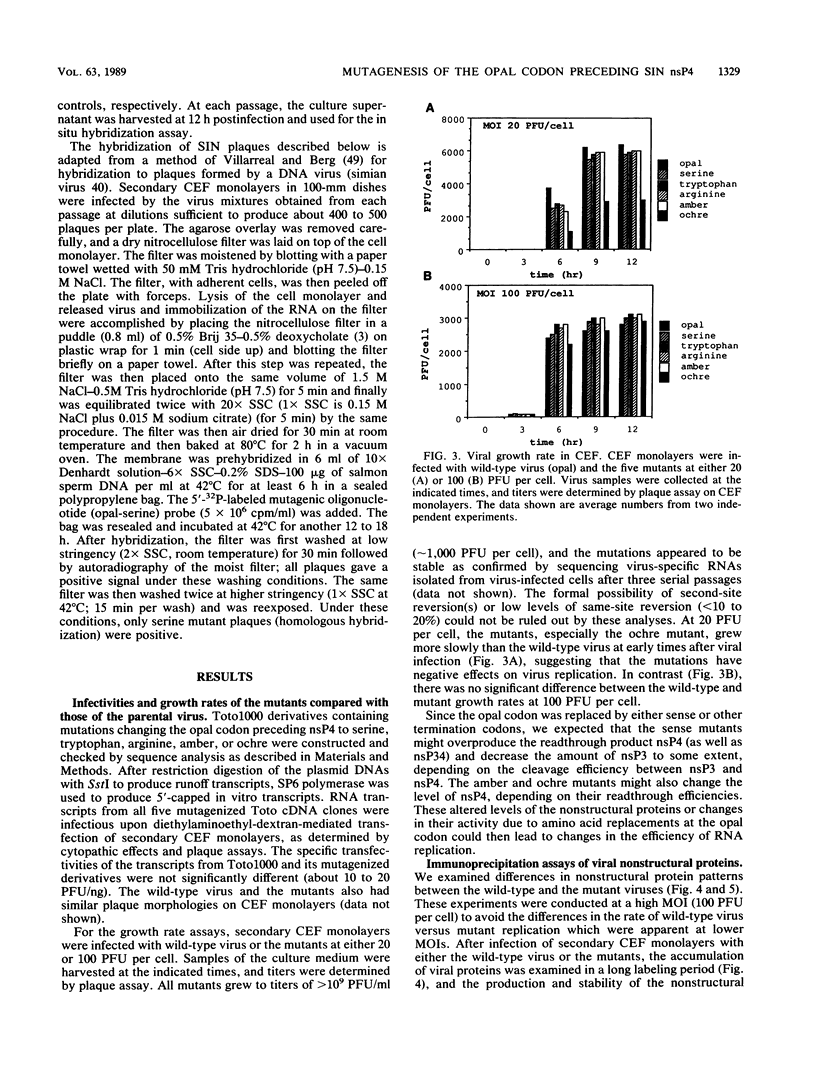
Sindbis virus (SIN) contains an in-frame opal termination codon in the nonstructural protein-coding region separating nsP3 and nsP4 and provides a useful tool to study the readthrough phenomenon of the termination codon in host cells and its role in viral replication. We have changed the opal codon by site-directed mutagenesis of a full-length SIN cDNA clone to either sense amino acids (serine, tryptophan, or arginine) or the other two translation termination codons (amber or ochre). Transcripts from all of the mutant cDNA clones were infectious when used to transfect chicken embryo fibroblasts. The resulting progeny virus stocks were then used to study the effects of these mutations on viral protein and RNA synthesis, growth properties, host range, and fitness compared with the parental strain. None of the mutants showed temperature sensitivity in plaquing efficiency or plaque morphology on chicken embryo fibroblast monolayers. Relative to the wild-type parent, the mutants containing sense replacements overproduced nsP34 but not nsP4 and made slightly decreased levels of nsP3, with a delay in its appearance. This indicates that the cleavage separating nsP3 and nsP4 occurs in these mutants and also that the level of nsP4 is not regulated solely by readthrough of the opal codon. The amber and ochre mutants produced decreased levels of nsP34, and the ochre mutant grew significantly more slowly than the other mutants or wild-type virus. For all five mutants, RNA synthesis early in infection was inhibited compared with that of the parental virus. This effect was apparent at multiplicities of infection of 20 PFU per cell but not at 100 PFU per cell. Using in situ hybridization to distinguish between mutant and wild-type plaques, we have studied the behavior of the serine mutant in a high-multiplicity growth competition experiment with wild-type virus. The wild-type virus eventually outcompeted the mutant after several passages, and these results indicate that this mutation has resulted in effects that are at least partially cis acting. Furthermore, by studying the growth, plaque formation, and protein synthesis of the mutants in various cell types, we have observed host range effects of the mutations, especially in mosquito and human cells. In addition, we have demonstrated, at least indirectly, that opal, amber, and ochre termination codons in the SIN nucleotide context can be suppressed in cultured cells of chicken, human, hamster, and mosquito origin.
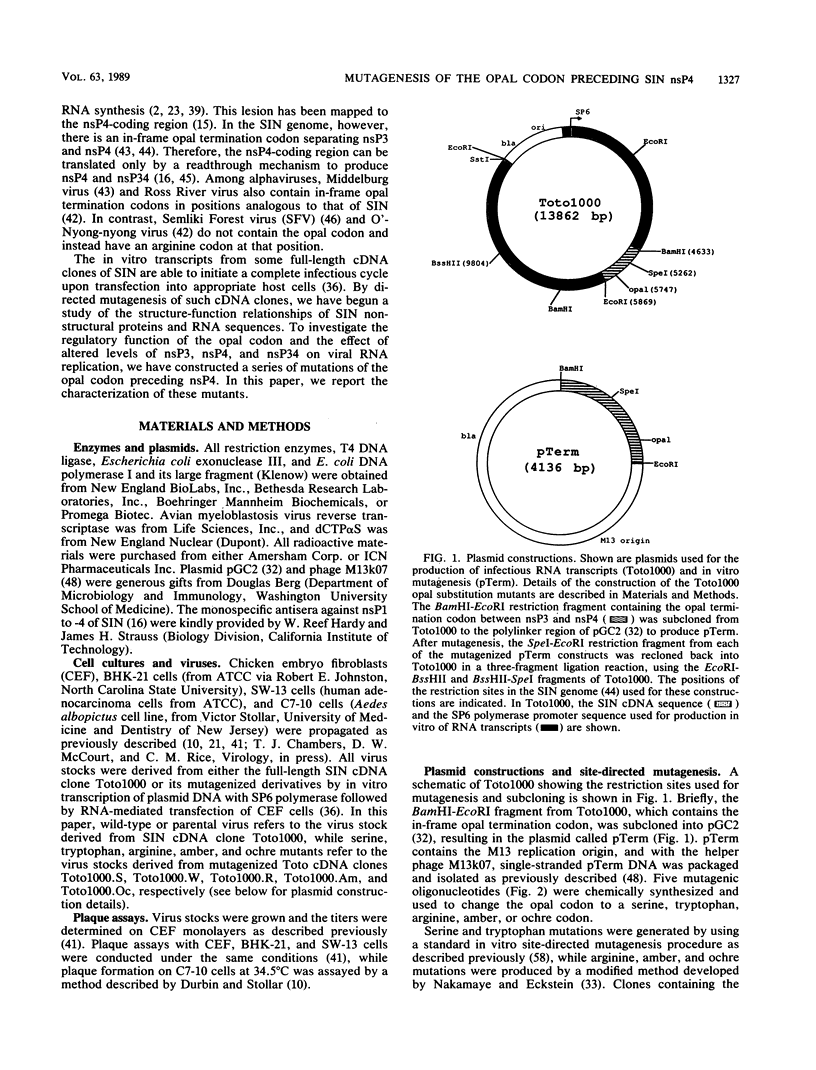
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Blobel G. Immunoprecipitation of proteins from cell-free translations. Methods Enzymol. 1983;96:111–120. doi: 10.1016/s0076-6879(83)96012-3. [DOI] [PubMed] [Google Scholar]
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresser J., Doering J., Gillespie D. Quick-blot: selective mRNA or DNA immobilization from whole cells. DNA. 1983;2(3):243–254. doi: 10.1089/dna.1983.2.243. [DOI] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Complementation between temperature-sensitive mutants of Sindbis virus. Virology. 1966 Oct;30(2):214–223. doi: 10.1016/0042-6822(66)90097-3. [DOI] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology. 1966 Oct;30(2):204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P. R. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the 'termination' codon, TGA. EMBO J. 1986 Jun;5(6):1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark H. F., Cohen M. M., Lunger P. D. Comparative characterization of a C-type virus-producing cell line (VSW) and a virus-free cell line (VH2) from Vipera russelli. J Natl Cancer Inst. 1973 Aug;51(2):645–657. [PubMed] [Google Scholar]
- Diamond A., Dudock B., Hatfield D. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981 Aug;25(2):497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- Durbin R. K., Stollar V. A mutant of sindbis virus with a host-dependent defect in maturation associated with hyperglycosylation of E2. Virology. 1984 Jun;135(2):331–344. doi: 10.1016/0042-6822(84)90190-9. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H. UGA suppression by normal tRNA Trp in Escherichia coli: codon context effects. Nucleic Acids Res. 1981 Feb 25;9(4):983–991. doi: 10.1093/nar/9.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff G. P., Butler P. J., Akam M. E., Gait M. J., Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Carrington J. C., Balàzs E., Jonard G., Richards K., Morris T. J. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 1985 Sep 25;13(18):6663–6677. doi: 10.1093/nar/13.18.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J Virol. 1988 Mar;62(3):998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Goelet P., Zimmern D., Ahlquist P., Dasgupta R., Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D. L., Dudock B. S., Eden F. C. Characterization and nucleotide sequence of a chicken gene encoding an opal suppressor tRNA and its flanking DNA segments. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4940–4944. doi: 10.1073/pnas.80.16.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Meshi T., Motoyoshi F., Takamatsu N., Okada Y. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 1986 Nov 11;14(21):8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. E., Smith J. F. Selection for accelerated penetration in cell culture coselects for attenuated mutants of Venezuelan equine encephalitis virus. Virology. 1988 Feb;162(2):437–443. doi: 10.1016/0042-6822(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S., Käriäinen L. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J Virol. 1979 Oct;32(1):19–29. doi: 10.1128/jvi.32.1.19-29.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S., Ruohonen L. Nonstructural proteins of Semliki Forest virus: synthesis, processing, and stability in infected cells. J Virol. 1983 Sep;47(3):505–515. doi: 10.1128/jvi.47.3.505-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Grosjean H. Usage of the three termination codons: compilation and analysis of the known eukaryotic and prokaryotic translation termination sequences. Mol Gen Genet. 1981;182(3):430–439. doi: 10.1007/BF00293932. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Leake C. J., Varma M. G., Pudney M. Cytopathic effect and plaque formation by arboviruses in a continuous cell line (XTC-2) from the toad Xenopus laevis. J Gen Virol. 1977 May;35(2):335–339. doi: 10.1099/0022-1317-35-2-335. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Nakamaye K. L., Eckstein F. Inhibition of restriction endonuclease Nci I cleavage by phosphorothioate groups and its application to oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1986 Dec 22;14(24):9679–9698. doi: 10.1093/nar/14.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978 Mar 30;272(5652):469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978 Jan;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G., Keränen S., Käriäinen L. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J Virol. 1981 Aug;39(2):348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Lenches E. M., Strauss J. H. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology. 1976 Oct 1;74(1):154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Levinson R., Rice C. M., Dalrymple J., Strauss J. H. Nonstructural proteins nsP3 and nsP4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology. 1988 May;164(1):265–274. doi: 10.1016/0042-6822(88)90644-7. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR R. M., HURLBUT H. S., WORK T. H., KINGSTON J. R., FROTHINGHAM T. E. Sindbis virus: a newly recognized arthropodtransmitted virus. Am J Trop Med Hyg. 1955 Sep;4(5):844–862. doi: 10.4269/ajtmh.1955.4.844. [DOI] [PubMed] [Google Scholar]
- Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986 Jul 25;14(14):5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Villarreal L. P., Berg P. Hybridization in situ of SV40 plaques: detection of recombinant SV40 virus carrying specific sequences of nonviral DNA. Science. 1977 Apr 8;196(4286):183–185. doi: 10.1126/science.191907. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol. 1971 Sep 15;234(50):206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- Weiss B., Rosenthal R., Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980 Jan;33(1):463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Oroszlan S. Translational readthrough of an amber termination codon during synthesis of feline leukemia virus protease. J Virol. 1985 Sep;55(3):870–873. doi: 10.1128/jvi.55.3.870-873.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Leinfelder W., Böck A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci U S A. 1987 May;84(10):3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]