Abstract
Background
Despite the importance of the shoot apical meristem (SAM) in plant development and organ formation, our understanding of the molecular mechanisms controlling its function is limited. Genomic tools have the potential to unravel the molecular mysteries of the SAM, and legume systems are increasingly being used in plant-development studies owing to their unique characteristics such as nitrogen fixation, secondary metabolism, and pod development. Garden pea (Pisum sativum) is a well-established classic model species for genetics studies that has been used since the Mendel era. In addition, the availability of a plethora of developmental mutants makes pea an ideal crop legume for genomics studies. This study aims to utilise genomics tools in isolating genes that play potential roles in the regulation of SAM activity.
Results
In order to identify genes that are differentially expressed in the SAM, we generated 2735 ESTs from three cDNA libraries derived from freshly micro-dissected SAMs from 10-day-old garden peas (Pisum sativum cv Torsdag). Custom-designed oligonucleotide arrays were used to compare the transcriptional profiles of pea SAMs and non-meristematic tissues. A total of 184 and 175 transcripts were significantly up- or down-regulated in the pea SAM, respectively. As expected, close to 61% of the transcripts down-regulated in the SAM were found in the public database, whereas sequences from the same source only comprised 12% of the genes that were expressed at higher levels in the SAM. This highlights the under-representation of transcripts from the meristematic tissues in the current public pea protein database, and demonstrates the utility of our SAM EST collection as an essential genetic resource for revealing further information on the regulation of this developmental process. In addition to unknowns, many of the up-regulated transcripts are known to encode products associated with cell division and proliferation, epigenetic regulation, auxin-mediated responses and microRNA regulation.
Conclusion
The presented data provide a picture of the transcriptional profile of the pea SAM, and reveal possible roles of differentially expressed transcripts in meristem function and maintenance.
Background
Organ formation is not limited to embryonic development, but can occur throughout the lifetime of a plant. The potential to develop new organs post-embryonically is attributed to meristems located at the growing tips of the plants, with the root apical meristem generating the underground part of the plant and the shoot apical meristem (SAM) giving rise to the entire shoot system after seed germination.
Like its root counterpart, the SAM contains a pool of pluripotent stem cells that can self-maintain as well as produce the cells that can differentiate into multiple cell and tissue types [reviewed in [1]]. While lateral organs such as the leaves are initiated from the peripheral regions of the SAM, the basal regions of the SAM contribute to the formation of the stem. The stem cells of the SAM must thus replenish areas where cells have been recruited and at the same time maintain the population of stem cells. This is generally attributed to an active process of communication among neighbouring SAM cells in the microenvironment of the stem cells [2-4].
Elegant genetic work carried out in the model plant, Arabidopsis thaliana, has enhanced our understanding of this vital developmental process [reviewed by [5]]. This is exemplified by the identification in Arabidopsis of WUSCHEL (WUS), a homeodomain transcription factor essential for maintaining the pools of stem cells in an undifferentiated state [6], and the CLAVATA group of genes that act together to restrict the proliferation of stem cells [7]. While the Arabidopsis genus provides invaluable model plants for enhancing our understanding of plant biology, it does not represent all the diverse developmental, environmental and physiological processes operating in the plant kingdom. There thus remains a need to extend the knowledge gained to other plant species especially crop plants.
Applying modern genomics research techniques to improving crops requires new knowledge and the development of new genomics resources. Legume species belonging to the family Fabaceae are cultivated for seeds rich in proteins, and represent important components of the diet in many parts of the world, especially pea, lentil and soybean. Legumes have attracted the attention of biologists because of their unique characteristics such as nitrogen fixation, secondary metabolism and pod development, and these are among the various processes that cannot be studied in Arabidopsis species. On the other hand, garden pea (Pisum sativum) has been a classic model species used in genetics [8,9] and plant-development studies. Its extensive use in studies of flowering initiation and development has provided important insights into this transition process [10-13]. In addition, the availability of various developmental and flowering pea mutants [11,14-18] makes this tractable for genomics studies.
In this study, we applied a transcriptomics approach to investigate the gene expression profiles associated with the SAM of the garden pea, an agriculturally and commercially important model legume. We also investigated the use of micro-dissected SAMs in unravelling the transcriptome profile of the SAM. By identifying genes that exhibit differential expression between SAMs and non-meristematic tissues (NM), we aimed to elucidate the transcriptional signature of the SAM and thereby identify genes that might play important roles in regulating SAM activity.
To this end, three directional cDNA libraries were constructed using SAMs that were carefully micro-dissected from garden peas. These libraries comprised the standard cDNA library plus a normalized library and a subtracted library, in order to increase the likelihood of recovering rare cDNAs, allowing the sampling of the wide diversity of genes expressed in the pea SAM. The EST sequences derived from these libraries were used in the subsequent design of a CombiMatrix CustomArray™ 4 × 2 K oligonucleotide array that was representative of the gene content of the SAM.
In this paper, we present the EST and transcriptional profiling data from this genomics project. The transcriptional profiling experiment represents the first analysis of genes that exhibit differential expression between the pea SAM and NM. The data reveal that transcripts putatively annotated as being associated with cell division and proliferation, epigenetic regulation, auxin-mediated responses and microRNA (miRNA) regulation are more abundant in the SAM than in the NM. In contrast, sequences related to photosynthesis, abiotic or biotic stress responses, reactive oxygen species (ROS) homeostasis and general cell-wall maintenance are down-regulated in the SAM.
Results and Discussion
Features of generated ESTs
A total of 3000 clones from three cDNA libraries were single-pass sequenced from their 5' ends. Sequence cleaning processes as outlined in the Materials and Methods yielded 2735 ESTs. These sequences had an average trimmed length of 519 base pairs and were assembled into 348 clusters and 1332 singlets, resulting in the final annotation of 1686 unigenes. Clusters ranged in membership from 67 ESTs (one) to two ESTs (253). The redundancy levels were 15.0%, 20.3% and 62.8% within the normalized, standard and subtracted libraries, respectively. A high redundancy level of cDNA libraries constructed using a similar subtraction method from Thellungiella plants under abiotic stress conditions has also been observed previously [19].
The translated products of the 1686 unigenes were searched against the non-redundant protein database provided by GenBank [20] to putatively assign their functions. At the time of writing, 918 (54.4%) unigenes showed significant similarity (E value cut-off of 10-5) to genes of known or putative function, whereas 549 (32.6%) ESTs were assigned to transcripts with unknown function, which includes hypothetical genes predicted in genomes of model organisms (Table 1). The remaining 219 ESTs (13.0%) had no homologues in the public protein sequence database. A further BLASTN search against GenBank EST collections revealed that 62 of the 219 ESTs were likely to be novel sequences.
Table 1.
A summary of the results obtained from BLASTX search against the NCBI (nr) protein database with expect value cut-off at 1e-5.
| Category | Number of unique ESTs |
| Match to genes with known or putative function | 918 (54.4%) |
| Match to unknown or hypothetical genes | 549 (32.6%) |
| No hits found | 219 (13.0%) |
| Total | 1686 |
Overview of putative genes represented in SAM-derived cDNA libraries
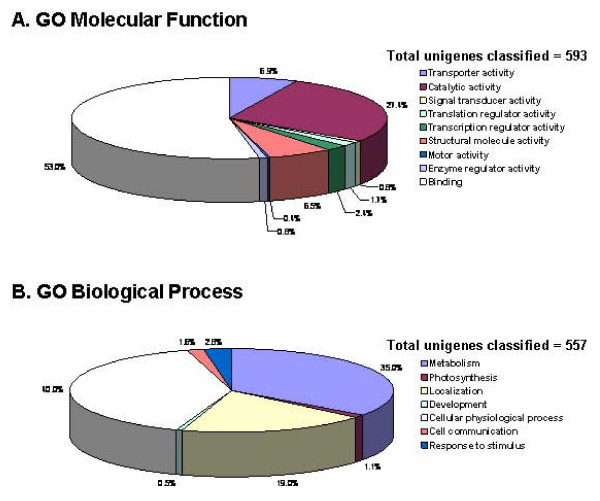
More than 50% of the SAM unigenes could be assigned as genes with a known or putative function based on sequence similarity. However, the lack of information on the encoded products meant that many of these transcripts could not be functionally categorized according to the Gene Ontology Consortium (GO). Using BLAST2GO [21], we successfully classified 593 and 557 unigenes in terms of GO molecular functions (Figure 1A) and biological processes (Figure 1B), respectively. A single gene product might be assigned to more than one GO term, and hence the total number of GO mappings in each of the ontologies exceeded the number of ESTs.
Figure 1.
Categorization of SAM unigenes according to the Gene Ontology (GO). Unigenes with a BLASTX score of < 10-5 were classified using the BLAST2GO automated system [21]. Note that a single gene can be assigned to more than one category in the GO classification system.
The successfully classified unigenes cover a broad range of GO functional categories (Figure 1). Under a molecular-function classification, most of the genes (53%) were assigned to the "binding" class (Figure 1A). This class includes sequences with putative involvement in mainly nucleic acid binding, a substantial number of which are predicted to encode transcription factors that are known to be essential to the regulation of plant development. Unigenes predicted to encode histone subunits and histone-modification proteins, chromatin remodelling factors and DNA methyltransferases represent another group of sequences linked to nucleic acid binding.
To investigate the different types of transcription-factor families represented by our EST libraries, a search using the best-matching Arabidopsis locus for SAM unigenes (based on a BLASTX search against the TAIR Arabidopsis protein database) was performed at the Arabidopsis Gene Regulatory Information Server [22]. There are 50 families of transcription factors currently listed in the database, 19 of these are represented by the SAM unigenes (Table 2). Six members of the family of homeobox transcription factors that play key roles in the regulation of development may represent interesting candidate genes for further studies.
Table 2.
ESTs related to the ontology of nucleic acid binding.
| SAM Clone | Family | Matching Arabidopsis Locusa |
| EX568781, EX569978 | CCAAT-HAP5 | At1g07980, At1g08970 |
| EX569532 | MYB-Related | At1g09770 |
| EX569985, EX569919 | TUB | At1g16070, At2g18280 |
| EX569797, EX569977 | GRAS | At1g21450, At5g66770 |
| EX569951 | AP2-EREBP | At1g28360 |
| EX571097 | NAC | At1g28470 |
| EX570087, EX570238, EX569491 | ARF | At1g30330, At4g23980, At5g20730 |
| EX571252, EX569634, EX570163, EX569945 | WRKY | At1g30650, At2g03340, At2g47260, At2g24570 |
| EX570518, EX569818 | bZIP | At1g42990, At4g34590 |
| EX570905, EX569205, EX570291, EX568744 | C2H2 | At1g55110, At1g75710, At2g23740, At2g27100 |
| EX568999, EX571110, EX570832, EX571147, EX569211, EX570070 | Homeobox | At1g62360, At2g27990, At4g36870, At4g40060, At4g32980, At5g03790 |
| EX570236 | G2-like | At1g69580 |
| EX569752, EX569559 | ARID | At1g76110, At1g76510 |
| EX571125 | BHLH | At2g27230 |
| EX568890 | CCAAT-HAP3 | At2g37060 |
| EX569425 | MADS | At2g45660 |
| EX569354 | C2C2-Gata | At3g06740 |
| EX569807 | SBP | At3g60030 |
Arabidopsis transcription factor family represented by the shoot apical meristem (SAM) unigenes. The matching Arabidopsis locus for each clone is indicated.
a. Annotation is based on the best BLASTX match against TAIR Arabidopsis protein database (Evalue < 1e-5)
In the biological-processes category, 40% and 35% of the unigenes were involved with cellular physiological processes and metabolism, respectively (Figure 1B). The former contains gene products that play an important role in cell organization and biogenesis, while the latter has sequences related mainly to protein metabolism. The sequences relevant to protein metabolism ranged from those associated with protein biosynthesis, such as different ribosomal subunits, to those that modify and degrade proteins, including various sequences involved in the ubiquitin-proteasome pathway.
Detection of differentially expressed transcripts in the SAM using the pea 2 K array
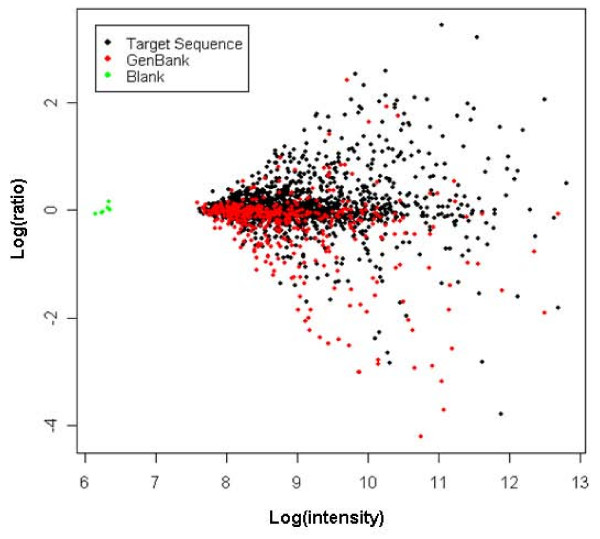
An oligonucleotide microarray has been developed using our EST resource and about 500 pea sequences randomly retrieved from the GenBank pea protein database, which contains approximately 2000 entries (see Materials and Methods). We utilized this array to compare the transcriptional profiles of the SAM and NM. Four independent replications of balanced-block-design dual-label experiments were performed [Materials and methods, [23]] and the resulting data were depicted in Figure 2.
Figure 2.
Results of transcript profiling experiments using the custom designed pea SAM Combimatrix 2 K chip. Expression profiles of SAM were compared with those of NM tissues. Average plot were generated from the experimental data of four independent biological replicates after normalization (see Materials and Methods for details). Black dotsrepresents data generated from the sequences derived from this study while red applies to those from the GenBank. Green corresponds to empty spot on the array. log(ratio) = log10(ISAM/INM); log(intensity) = 0.5 log10(ISAM.INM) where ISAM and INM are signal intensities for a transcript in the SAM or the NM tissues, respectively.
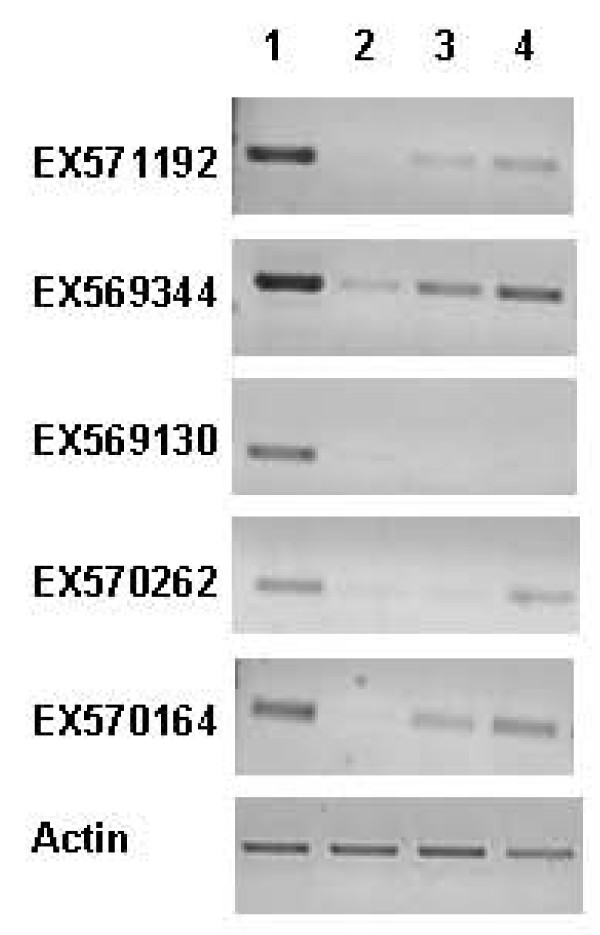
Differentially expressed genes that were detected using LimmaGUI [24] (at an adjusted probability value of < 0.05) were subjected to further selection based on the relative change, with up- and down-regulated transcripts defined using cut-offs of greater than 1.3-fold or less than 0.7-fold, respectively. Based on these criteria, we identified 184 and 175 transcripts that were significantly up- or down-regulated in the pea SAM relative to the NM. These transcripts were annotated based on the best BLASTX match, and exhibited changes relative to the NM of 0.1- to 10.7-fold (see Additional File 1 &2). The change of 0.1-fold was for a gene encoding a type 1 metallothionein (AB176564), while that of 10.7-fold was for a sequence annotated as vegetative lectin (AAA33691). A study of similar genes encoding type 1 metallothionein in cotton revealed their abundant (although not exclusive) expression in the root [25]. Meanwhile, the high expression of a sequence encoding vegetative lectin in the pea apex has been observed [26]. These and other studies listed in Table 3 provide independent verification of our microarray data. We also performed RT-PCR analysis on five selected transcripts and as shown in Figure 3, the outcome is generally in good agreement with the microarray data.
Table 3.
Differentially-regulated transcripts with corresponding orthologues known to be highly expressed in shoot apical meristems (SAMs) or non-meristem (NM) tissues.
| Probe IDa | Annotation | Fold change | References |
| AB176564 | Metallothionein | 0.1 | Hudspeth et al., 1996 |
| AF029242 | Dormancy associated gene 1 (DRM1) | 0.4 | Stafstrom et al., 1998 |
| EX570325 | MERISTEMATIC RECEPTOR-LIKE KINASE | 1.4 | Fujita et al., 2003 |
| EX568912 | F-box protein (STAMINA PISTILLOIDA) | 1.8 | Taylor et al., 2001 |
| EX570203 | Protodermal factor 1 | 1.8 | Abe et al., 2001 |
| EX568701, EX571325, EX569084, EX570634 | Histone subunits | 2.0 | References in Meshi et al., 2000 |
| EX570428 | Mini-chromosome maintenance proteins | 2.2 | Stevens et al., 2002 |
| EX570270, AB008186 | Proliferating cell nuclear antigen | 4.8, 3.8 | Kosugi et al., 1991 |
| AB031227 | PsAD1 | 5.3 | Madoka &Mori, 2000 |
| EX570531 | Vegetative lectin | 10.7 | Dobres & Thompson, 1988 |
The data reported is in good agreement with the transcript profiling data.
a. GenBank accession number that begins with EX is derived from this study.
Figure 3.
Verification of microarray data using RT-PCR analysis. RT-PCR analysis was carried out under linear amplification conditions for five randomly selected transcripts as indicated. The actin gene was used as an internal control. 1, SAM; 2, Leaf; 3, Stem; 4, Root.
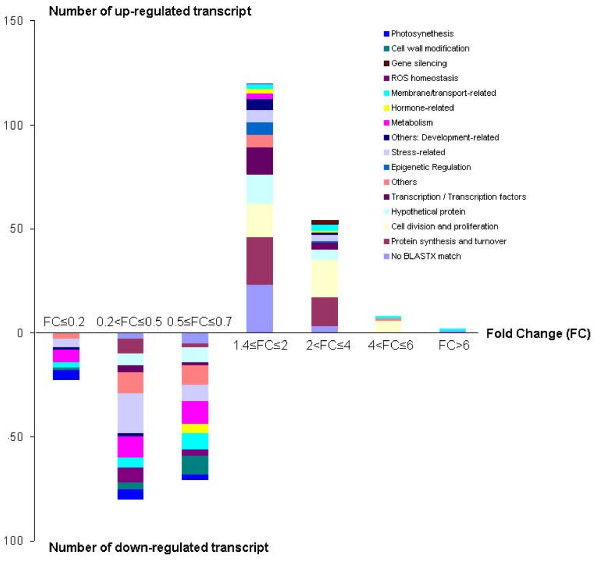
Further comparison of the functional categories identified to be differentially regulated in maize data [25] and this study (Figure 4) revealed that transcripts associated with the categories of transcription, chromatin and cell division are highly represented in the list of up-regulated genes, while sequences categorized as being related to metabolism, stress and photosynthesis are down-regulated in the SAM. However, our data also highlight differences between the transcriptional repertoires of the monocot SAM [maize, [27]] and the dicot SAM (pea, present study). In particular, the high retrotransposon-related transcriptional activity reported in maize [27] does not appear to be a conserved feature of the SAM, since it was not present in the pea SAM ESTs.
Figure 4.
Functional categories of transcripts differentially expressed in the pea SAM. The number of genes differentially expressed between the pea SAM and NM (p < 0.05) with changes in expression higher than indicated cut offs of the fold change are plotted with different functional categories highlighted by colour coding. Transcripts corresponding to cell division and proliferation, epigenetic regulation, and gene silencing are among the up-regulated categories while stress response and the metabolism class of genes are in down-regulated categories.
Since the SAM we investigated consists of distinct functional zones, the averaging of signals across the whole SAM probably attenuated signals associated with any given stem-cell region. However, our experiment showed that a sequence annotated as encoding PROTODERMAL FACTOR 1 was up-regulated 1.8-fold in the SAM relative to the NM (Table 3). An Arabidopsis counterpart gene has been found to be exclusively expressed in the outermost (L1) layer of the SAM [28]. This indicates the ability to identify transcripts that are specifically expressed in certain domains of the SAM, though the magnitude of the relative change might have been reduced and low abundance transcript could have been missed detection.
A closer inspection of the differentially expressed transcripts revealed that about 60% of the identified down-regulated sequences were derived from GenBank (Table 4). In contrast, only 12% of the genes with higher expression in SAMs were retrieved from GenBank, while the other up-regulated ESTs were from clones derived from our library collection. This is not surprising since the sequences from GenBank were generated primarily from tissues other than the SAM, whereas our EST collection was derived from dissected SAMs. This indicates the significance of constructing a library from specialized tissues and further suggests the utility of our EST collection as a valuable resource in studying the molecular processes underlying the functions of the plant meristem. Meanwhile, 30% of the differentially expressed genes detected were not identified, consisting of genes annotated as hypothetical or expressed protein as well as sequences that have no BLASTX matches in the public database. This list of unknowns could represent intriguing candidate genes for functional analysis.
Table 4.
Representative transcripts that are detected to be significantly up-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.
| Probea | Fold Change | Annotationb |
| Cell division and proliferation | ||
| EX570197 | 2.7 | High mobility group protein (HMGI/Y) |
| EX571067 | 1.6 | SAR-DNA binding protein 2 |
| EX571065 | 1.7 | SAR-DNA binding protein 1 |
| EX570428 | 1.8 | Mini-chromosome maintenance protein 6 |
| EX569064 | 2.0 | Small nuclear ribonucleoprotein associated protein B |
| EX568701 | 2.0 | Histone H1 |
| EX568991 | 2.1 | Delta DNA polymerase |
| EX570486 | 2.1 | Helicase |
| EX570226 | 2.2 | Phosphoesterase |
| EX571345 | 3.3 | Germinal histone H4 |
| EX569084 | 3.7 | Histone H2a.1 |
| EX570634 | 4.2 | Histone H3 |
| EX570270 | 4.8 | Proliferating cell nuclear antigen 2 |
| EX568755 | 5.9 | Histone H4 |
| AB008188 | 1.9 | Cyclin D3.1 protein |
| EX570164 | 1.8 | Mitotic cyclin B1-1 |
| EX570388 | 1.5 | Cell cycle protein kinase |
| Protein synthesis and turnover | ||
| EX569344 | 1.4 | Transducin family of protein (SLOW WALKER 1) |
| EX570594 | 1.9 | 60S ribosomal protein L18a |
| EX569298 | 1.9 | Ribosomal protein L30 |
| EX569956 | 2.0 | 40S ribosomal protein S18 |
| EX568807 | 2.1 | Fibrillarin |
| EX570044 | 2.1 | 60S ribosomal protein L44 |
| EX570565 | 2.1 | Ribosomal protein L23 |
| EX568742 | 2.2 | Ribosomal protein L24 |
| EX570468 | 2.2 | 60S ribosomal protein |
| EX568861 | 2.4 | 40S ribosomal protein S17 |
| EX569048 | 2.6 | Ribosomal protein S2 |
| EX568908 | 2.9 | Ribosomal protein S15-like |
| EX568954 | 3.0 | Ribosomal protein S3 |
| AB021873 | 3.2 | Ribosome sedimenting protein |
| EX570400 | 5.0 | Ribosomal protein S4 |
| EX569708 | 2.3 | Ubiquitin extension protein 2 |
| EX568912 | 1.8 | F-box protein (STAMINA PISTILLOIDA) |
| EX570030 | 1.9 | Chaperonin |
| EX568849 | 1.6 | Elongation factor 1-beta/EF-1-beta |
| EX570437 | 1.4 | Cyclophilin |
| EX568710 | 1.4 | Eukaryotic initiation factor |
| EX570401 | 1.4 | Translation initiation protein |
| Transcription factors or hormonal regulation | ||
| EX569978 | 1.5 | Putative Hap5 transcription factor |
| EX570025 | 1.5 | YABBY family transcription factor |
| EX570238 | 1.5 | Auxin response factor 9 |
| EX570380 | 3.4 | Putative transcriptional co-activator (KIWI) |
| EX571172 | 1.6 | Transcription factor |
| EX570262 | 2.1 | Argonaute protein |
| EX570187 | 2.9 | Indole-3-acetic acid amido synthetase (DWARF IN LIGHT 1) |
| AF325121 | 1.6 | Brassinosteroid biosynthetic protein |
| Epigenetic regulation-related | ||
| EX568764 | 1.4 | Chromomethylase |
| AF034419 | 1.4 | DNA methyltransferase |
| EX569897 | 1.6 | H3 lysine-9 specific SUVH4 |
| EX570366 | 1.9 | SWI/SNF-like ATPase subunits, DDM1 |
| EX570306 | 2.0 | WD-40 repeat protein (MSI3) |
| DQ026703 | 1.5 | WD-40 repeat protein (MSI1) |
| Other developmental regulation-related | ||
| EX569130 | 2.0 | Mandelonitrile lyase family of FAD containing oxidoreductases (HOTHEAD) |
| AY343326 | 1.5 | Late-flowering gene |
| Unclassified/Unknown/No BLASTX match | ||
| EX571192 | 9.0 | No BLASTX match |
| EX569831 | 3.3 | Proline-rich protein |
| EX570071 | 3.7 | Hypothetical protein |
| EX571142 | 3.6 | Pathogenesis-related group 5 protein |
| EX571013 | 2.8 | No BLASTX match |
| EX570256 | 2.4 | Unnamed protein product |
| EX569233 | 2.1 | Serine/threonine dehydratase |
| EX569274 | 4.5 | Lipid transfer protein |
| EX570925 | 3.1 | Kunitz inhibitor ST1-like |
| AB032830 | 3.3 | Endo-1,4-beta-glucanase |
| EX570531 | 10.7 | Vegetative lectin |
| EX569068 | 2.4 | Expressed protein |
| EX569663 | 2.1 | Expressed protein |
| EX570209 | 2.2 | Hypothetical protein |
| EX570712 | 2.0 | Hypothetical protein |
| EX570576 | 1.9 | Cytochrome P450 monoxygenase(CYP78A8) |
| EX569596 | 1.8 | No BLASTX match |
| EX570325 | 1.5 | MERISTEMATIC RECEPTOR-LIKE KINASE 2 |
| EX570536 | 2.4 | Unknown protein |
| EX570361 | 1.5 | No BLASTX match |
| EX570202 | 1.4 | No BLASTX match |
| EX570695 | 1.4 | No BLASTX match |
| EX569023 | 1.6 | No BLASTX match |
| EX570276 | 1.5 | No BLASTX match |
| EX569420 | 1.4 | No BLASTX match |
| EX568900 | 1.6 | No BLASTX match |
a, Probe with GenBank accession number that begins with EX is derived from this study.
b. Annotation is based on the expect value of BLASTX hit that is set at 1e-5
Cell division and proliferation in the SAM
Putative functions could be assigned to 116 of the up-regulated transcripts based on protein sequence similarity. Manual inspection of the corresponding transcripts revealed a high representation of ESTs predicted to encode proteins associated with cell division and proliferation (Table 4; Figure 4).
Table 4 indicates that ESTs encoding all five subtypes of histones, minichromosome-maintenance proteins and cell-proliferating nuclear antigens were among the transcripts whose expression was higher in the SAM than in the NM. The expression of these genes is known to be associated with DNA synthesis and cell proliferation, and they are thus abundant in the meristematic tissue [29,30] since this region consists of actively dividing cells.
In the same category, there is a transcript predicted to encode HIGH MOBILITY GROUP (HMG) protein. HMG are proteins that are known to play an architecture role in modifying DNA conformation to facilitate the assembly of multiprotein-DNA complexes. They may serve only to maintain physical orders but the involvement of these proteins in the network regulating SAM activity seems plausible. This is in view of recent studies that demonstrate the binding of HMG proteins to functionally important regions of plant gene promoter and stimulate transcriptions [31].
Other up-regulated sequences included transcripts predicted to encode cyclin D (AB008188) and cyclin B (EX570164), which are involved in the progression of the cell cycle. D-cyclins are one of the main rate-limiting factors for cell proliferation, and several of them may play key roles in the association between the cell cycle and meristem function, in particular primordia formation [reviewed in [32]].
Epigenetic regulation of the SAM activity
Increases in transcript abundance were also found for genes predicted to encode histone-modification protein (EX569897), chromatin remodelling factors (EX570366, EX570306 and DQ026703) and DNA methyltransferases (EX568764 and AF034419; Table 4). These proteins are known to play a role in the epigenetic regulation of gene expression by participating in mechanisms that alter chromatin structure so as to activate or repress particular sets of genes [reviewed in [33]].
There is an emerging recognition of the significance of the chromatin remodelling process in regulating the activity of plant stem cells [reviewed in [34]]. For instance, mutation of the Arabidopsis FASCIATA1 (FAS1) and FASCIATA2 (FAS2) genes that encode subunits of the chromatin assembly factor leads to dysfunction of the SAM [35]. This was associated with the down-regulation of WUS gene expression in both mutants, suggesting that regulation of meristem function and organogenesis by chromatin remodelling factors is primarily achieved through regulation of the expression of the homeobox transcription factor [35]. However, direct links between chromatin remodelling factors and the regulation of the expression of key meristem genes remain to be established. Nonetheless, the up-regulation of the expression of these sequences in the SAM suggests that the mechanism of epigenetic regulation is important to maintaining the identity of stem cells in plants, as has been reported in animals [36].
Transcription factors and hormonal regulation of SAM activity
We also identified a few putative transcription factors among the genes whose expression was higher in the SAM than in the NM. These included an EST (EX570025) annotated as a putative plant-specific transcription factor from a YABBY-family protein – members of this family are reportedly involved in the abaxial cell-fate specification in lateral organs of Arabidopsis [37]. Also on the list was a sequence predicted to encode AUXIN RESPONSE FACTOR 9 (ARF9; EX570238). Similar members of the auxin response factor group of proteins are known to regulate auxin-mediated transcript activation or repression. For example, the expression of several genes, such as those encoding members of LATERAL ORGAN BOUNDARIES domain proteins and AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, are disrupted in the double mutant of ARF7 and ARF19, implicating their roles in auxin-mediated plant development [38].
Other gene products that might be related to auxin include a putative auxin-efflux carrier (PIN1), auxin-conjugating protein (DWARF IN LIGHT1) and a ribosomal protein L24B (Additional File 1, Table 4). Similar genes in Arabidopsis have been implicated in auxin-mediated developmental regulation [39-41]. For example, DWARF IN LIGHT1 is involved in auxin signal transduction, and inhibits shoot and hypocotyl-cell elongation [39]. The presence of several auxin-related transcripts on our list reflects the well-established roles of auxin in organ initiation and positioning at the meristem [42,43].
Intriguingly, miRNAs that are endogenous 21-nucleotide riboregulators have been shown to target several mRNAs implicated in auxin responses, including DWARF IN LIGHT1 [44]. There is increasing amount of evidence indicating that miRNA-mediated repression plays an important role in the spatial expression of plant cell-fate regulatory genes [e.g. [45]]. A protein called ARGONAUTE is known to function as a catalytic component of the RNA-induced silencing complex, which targets mRNA for degradation using miRNA as a guide [46]. Nevertheless, the precise role for the up-regulation of a transcript annotated as encoding a similar ARGONAUTE in our dataset (Table 4) awaits further study.
Stress responses in the SAM
Many of the down-regulated transcripts were potentially associated with biotic and abiotic stress responses (Table 5), including transcripts predicted to encode dehydrin-related protein (AY065655), pathogenesis-related protein (AJ586324), disease-resistance-response protein (AF139018), antimicrobial defensin (AF525685) and chitinase (AB037832). This might be attributable to the stems, leaves and roots generally being exposed to greater biotic or abiotic stress than the well-shielded SAM, with the former therefore requiring the constitutive presence of these gene products at a higher level than in the SAM in order to maintain successful defence responses. However, we found at least one other sequence (EX571142) potentially related to stress responses whose expression was higher in the SAM (Table 4). Although the molecular basis for this is unknown, it is possible that the corresponding encoded product plays dual roles in both stress responses and development. This is supported by a recent study finding a network of rice genes associated with stress responses and seed development [47].
Table 5.
Representative transcripts that are detected to be significantly down-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.
| Probea | Fold Change | Annotationb |
| Stress responses | ||
| AY065655 | 0.1 | Ultraviolet B-repressible dehydrin-related protein |
| AY065659 | 0.1 | Ultraviolet B- inducible protein |
| AJ586324 | 0.1 | Putative basic PR1 protein |
| EX568748 | 0.3 | Dehydrin |
| AF139018 | 0.3 | Disease resistance protein |
| AF525685 | 0.4 | Antimicrobial defensin |
| AJ278699 | 0.5 | Protease |
| AB087832 | 0.4 | Class 1 chitinase |
| AF175278 | 0.7 | Wound-inducible P450 hydrolase |
| AF137351 | 0.2 | Pathogenesis-related protein 4 |
| Reactive oxygen species homeostasis | ||
| EX568770 | 0.3 | Catalase |
| AB026253 | 0.5 | Copper amine oxidase |
| AJ50832 | 0.4 | Germine-like protein |
| AB189165 | 0.4 | Copper zinc superoxide dismutase |
| AB087837 | 0.7 | Glutathione-S-transferase |
| AJ319808 | 0.3 | Thioredoxin H |
| AB087838 | 0.5 | Peroxidase |
| Photosynthesis | ||
| EX569880 | 0.1 | Light harvesting chlorophyll a/b binding protein |
| AY845255 | 0.2 | Light harvesting chlorophyll a/b binding protein 3 |
| EX569551 | 0.2 | Light harvesting chlorophyll a/b binding protein type 1 (CAB) |
| AY292531 | 0.1 | Oxygen-evolving enhancer protein |
| EX569675 | 0.3 | Type II chlorophyll a/b binding protein |
| EX569989 | 0.4 | PSI-K subunit of Photosystem I |
| AY007467 | 0.4 | Photosystem II CP47 protein |
| EX570832 | 0.1 | Ribulose 1,5-biphosphate carboxylase |
| AY065656 | 0.2 | RUBISCO activase |
| Metabolism | ||
| AY112702 | 0.6 | Vacuolar acid invertase |
| AJ012080 | 0.3 | Sucrose synthase |
| Y08728 | 0.6 | ADP-glucose phosphorylase |
| EX569667 | 0.4 | Ribulose-5-phosphate-3-epimerase |
| EX570956 | 0.7 | Phosphofructokinase |
| Cell wall modification | ||
| AF056493 | 0.7 | Pectin methylesterase |
| AB042531 | 0.2 | Xyloglucan endotransglycosylase |
| EX569643 | 0.6 | Cellulose synthase |
| AJ621355 | 0.4 | KORRIGAN |
| AB015428 | 0.6 | Endoxyloglucan transferase 1 |
| Membrane- or transport-related | ||
| AJ243307 | 0.1 | Putative plasma membrane intrinsic protein |
| AJ243309 | 0.3 | Putative tonoplast intrinsic protein |
| AB027616 | 0.5 | Apyrase |
| AF109922 | 0.7 | Sucrose transport protein |
| EX570713 | 0.6 | Aquaporin-like transmembrane protein |
| EX570839 | 0.7 | Outward-rectifying potassium channel |
| EX569706 | 0.7 | Sulfate transporter |
| Unclassified/Unknown/No BLASTX match | ||
| AY371200 | 0.2 | Ripening-related protein |
| AF369889 | 0.5 | Embryo-abundant protein |
| EX568773 | 0.5 | Hypothetical protein |
| EX569332 | 0.6 | Hypothetical protein |
| EX569635 | 0.5 | No BLASTX match |
| EX570920 | 0.7 | No BLASTX match |
| AF515795 | 0.1 | Dormancy-associated protein 3 |
a, Probe with GenBank accession number that begins with EX is derived from this study.
b. Annotation is based on the expect value of BLASTX hit that is set at 1e-5
ROS homeostasis in the SAM
Surprisingly, the down-regulated transcripts included various sequences encoding proteins that scavenge or generate ROS, such as thioredoxin (AJ319808), catalase (EX568770), Cu-Zn superoxide dismutase (AB189165), copper amine oxidase (AB026253) and peroxidase (AB087838). This implies that the level of ROS is lower in the SAM than in the NM, and hence the expression of genes encoding ROS scavengers is lower in the former. The absence of photosynthesis (a source of ROS) in the SAM might explain the lower ROS level therein. However, there is increasing evidence that plants use ROS as signalling molecules for regulating development and various physiological responses, and for mediating abscisic-acid-induced stomatal closure, as well as in auxin signalling and gravitropism in roots [48]. Whether this indicates that the regulatory role of ROS is less prominent in the SAM than in the NM awaits further investigation.
Photosynthesis- and cell-wall-related transcripts
Several of the genes more abundant in the NM than in the SAM were predicted to be related to photosynthesis, including subunits of photosystem I and II and chlorophyll-a/b-binding protein. It is well known that meristematic cells do not contain differentiated plastids, which may explain the lower expression level of genes associated with photosynthesis in the SAM.
Some of the sequences predicted to encode products that play roles in cellulose synthesis, such as cellulose synthase (EX569643), pectin methylesterase (AF056493), xyloglucan endotransglycosylase (AB042531), were found to be down-regulated in the SAM. This probably reflects the cell-wall structure in the stem and leaf being more complex that the thin primary cell wall in meristematic cells [4].
Conclusion
The development of our EST collection from the pea SAM represents an important advance towards understanding SAM function and maintenance, especially due to the under-representation of SAM-related transcripts in the public database as demonstrated in this study. Subsequent transcriptional profiling experiments using the microarray constructed from ESTs yielded the transcriptional signatures of the pea SAM, and we have reported a repertoire of transcripts with putative or unknown functions that are differentially regulated in the SAM. In silico analysis of the predicted gene products has implicated several processes in the complex molecular network that regulates this developmental process. Future studies of these genes should attempt to reveal how they interact in the complex molecular network that maintains and regulates the dynamics of the SAM.
Methods
Plant materials and cDNA libraries synthesis
Garden pea (Pisum sativum) cultivar Torsdag was grown in a greenhouse located at the University of Melbourne, Australia. SAMs were micro-dissected from 10-day-old peas under the dissecting microscope at 40× magnification. Any leaf primordia were excluded in order to create a meristem-enriched tissue collection and the location of tissue sample is indicated in Figure 5. Dissected samples were quickly frozen in liquid nitrogen and stored at -80°C until used for RNA extraction and cDNA library synthesis [49]. For the subtracted library, the driver sequences were derived from an equal mix of RNA extracted from non-meristematic (NM) tissue consisted of primary stem (without axillary meristems), mature leaf lamina, primary roots (without root apical meristem and root hairs), whereas the tester sequences consisted of RNA harvested from dissected SAM. All cDNAs were cloned into pBlueScriptIISK+ plasmid vector.
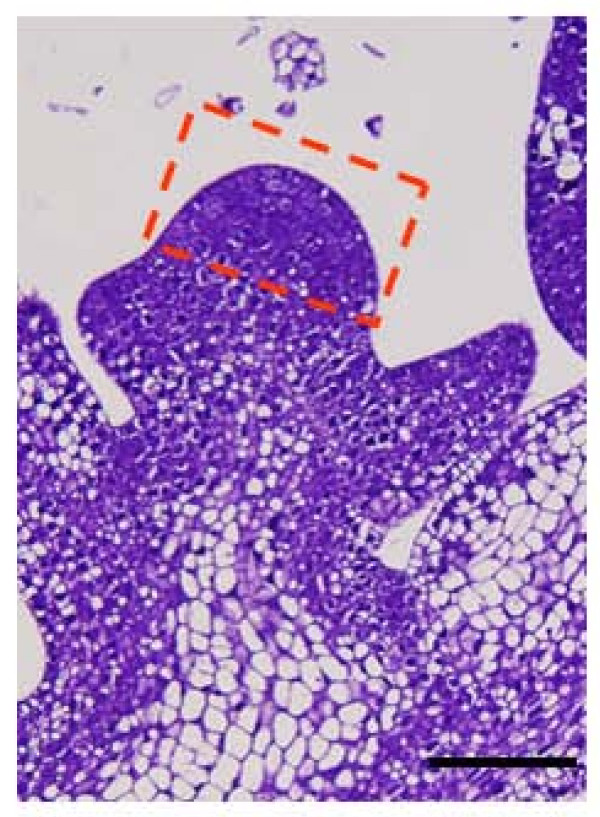
Figure 5.
Sampling strategy for the pea SAM. Micrograph of a representative pea SAM, with the box showing the location of the tissue sample. Scale bar = 100 μm.
EST sequencing
A total of about 500, 1000 and 1500 randomly picked clones from each of the standard (C), subtracted (S) and normalized (N) libraries were sequenced at Australian Genome Research Facility (AGRF), Australia and subsequently at Macrogen Korea using T7 primer. These sequences have been deposited in GenBank under the accession numbers EX568682 to EX571416.
Sequence analysis and annotation
Sequence data were trimmed off vector, adaptor and low quality sequences using SEQTools [50]. Trimmed sequences that were shorter than 100 basepairs were excluded from further analysis. Blast score-based clustering method with a score cut-off of 0.6 from SEQTools was then used to assemble the sequences. All clusters and singletons resulting from this automated clustering were considered to be the best estimation of a minimal gene set for our EST library and we have called this set as "unigenes".
All sequences were then imported into Blast2GO, a web-based Gene Ontology (GO) annotation and analysis tool [21] for subsequent analysis. This involved automated retrieval of GO terms associated to the hits obtained after a BLASTX search of the corresponding unigene sequence against NCBI (nr) protein database. The e-value cut off was set at 1e-5.
RNA Extraction for microarray experiments
Total RNA was extracted from dissected SAM (approximately 80 SAMs per extraction) or other plant parts (primary stem, primary roots and mature leaves) using Qiagen RNeasy Mini Kit. Four independent tissue collections and RNA extractions (designated A, B, C and D) were performed for each of the microarray hybridization experiment.
Design of pea SAM Combimatrix CustomArray™ 4X2K
The Combimatrix arrays are semiconductor-based oligonucleotide microarrays and are generated based on CombiMatrix technology (hyperlink) of in situ synthesis [51]. The CustomArray™ 4x2k is a microarray that is divided into 4 sectors, each of which can contain up to 2,240 different oligonucleotide probes (spots) and can be hybridized individually with different targets using a provided sectored hybridization cap. A total of 1686 pea sequences (290 sequences from C library, 300 sequences from S library and 1086 ESTs from the N library) together with 500 pea sequences randomly selected from GenBank pea protein database were submitted for probe design using the open source CombiMatrix probe design system. The length of probes ranged from 35–40 bases in length. A variety of control elements were also arrayed on the slide and these include blank spot, housekeeping genes (actin) as well as non-plant transgenes.
Target preparation and hybridization to microarray
Target preparation and hybridization were performed in Australian Genome Research Facility Ltd (AGRF) according to the standard CombiMatrix protocol described in detail at http://www.combimatrix.com/docs/PTL005_00_4x2K_%20Hyb_%20Imaging.pdf. One microgram of total RNA from SAM or NM was labelled using the Kreatech's ULS™ RNA ampULSe kit to generate Cy5 or Cy3 labelled targets. The Cy5- or Cy3-labelled cDNA was then hybridized to different sector of the chip according to a balanced block design dual label experiment scheme [23]:
Sector 1: Cy3-SAM A vs Cy5-NM A
Sector 2: Cy5-SAM B vs Cy3-NM B
Sector 3: Cy3-SAM C vs Cy5-NM C
Sector 4: Cy5-SAM D vs Cy3-NM D
Image acquisition and data analysis
The Cy5- and Cy3-hybridized chip was then scanned using Genepix 4000B microarray scanner (Axon Instruments, CA, USA) according to manufacturer's instructions. The generated tiff image files were then imported into Combimatrix Microarray Imager to produce intensity data. The LimmaGUI software, which is an implementation of the Empirical Bayes linear modelling approach, was used for subsequent statistical analysis of the resulting data [24]. A robust spline method was chosen for within array normalization and a least-square linear model fit was computed with the p-value adjusted using the Benjamini-Hochberg procedure. The entire differentially-expressed transcripts (p value < 0.05) in the SAM are listed in Additional File 1 and Additional File 2. Microarray data have been deposited in the Gene Expression Omnibus database [20] under accession number GSE9278.
RT-PCR analysis
The one-tube, two enzyme Access RT-PCR system (Promega, Annandale, New South Wales, Australia) was used according to manufacturer's instructions in all RT-PCR analysis. Ten ng of RNA isolated from the SAM, mature stem, mature leaf and primary root of 10-day-old pea seedlings were used as a template in a 10 μl reaction volume. The pea actin gene was used as an internal control. The number of cycles used for the transcripts investigated was routinely between 25–28 and 80 % of the PCR reaction was separated on 1% agarose gel containing 0.1 μg/μl ethidium bromide and visualized under UV light.
Abbreviations
SAM: shoot apical meristem; NM: non-meristem; EST: expressed sequence tag.
Authors' contributions
CEW carried out the EST analysis, participated in the microarray experiment and RT-PCR analysis, and drafted the manuscript. PLB and MBS were responsible for design of the project, standardisation & organization of meristem micro-dissection & making EST libraries as well as overall coordination of the experiments and manuscript editing. HO contributed to EST library characterization and EST sequence analysis. All authors read and approved the final manuscript.
Supplementary Material
Transcripts identified to be up-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.
Transcripts identified to be down-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.
Acknowledgments
Acknowledgements
We thank Andrea Merrell and Cathy Jensen for their assistance in the pea meristem dissections. This work was supported by the Australian Research Council Centre of Excellence (grant no. CEO348212) to the University of Melbourne Node of the Centre of Excellence for Integrative Legume Research.
Contributor Information
Chui E Wong, Email: acewong@unimelb.edu.au.
Prem L Bhalla, Email: premlb@unimelb.edu.au.
Harald Ottenhof, Email: haraldo@aus.biolabgroup.com.
Mohan B Singh, Email: mohan@unimelb.edu.au.
References
- Carraro N, Peaucelle A, Laufs P, Traas J. Cell Differentiation and Organ Initiation at the Shoot Apical Meristem. Plant Molecular Biology. 2006;60:811–826. doi: 10.1007/s11103-005-2761-6. [DOI] [PubMed] [Google Scholar]
- Bhalla PL, Singh MB. Molecular control of stem cell maintenance in shoot apical meristem. Plant Cell Reports. 2006;25:249–256. doi: 10.1007/s00299-005-0071-8. [DOI] [PubMed] [Google Scholar]
- Singh MB, Bhalla PL. Plant stem cells carve their own niche. Trends in Plant Science. 2006;11:241–246. doi: 10.1016/j.tplants.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ. Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends in Plant Science. 2007;12:245–252. doi: 10.1016/j.tplants.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK. Patterning and Polarity in Seed Plant Shoots. Annual Review of Plant Biology. 2008;59:67–88. doi: 10.1146/annurev.arplant.57.032905.105356. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. Shoot apical meristem maintenance: the art of a dynamic balance. Trends in Plant Science. 2003;8:394–401. doi: 10.1016/S1360-1385(03)00164-X. [DOI] [PubMed] [Google Scholar]
- Blixt S. The pea. In: RC K, editor. The Handbook of Genetics. Vol 2. New York: Plenum Press; 1974. pp. 181–221. [Google Scholar]
- Weeden NF, Muffet M. Identification of genes affecting root mass and root/shoot ratio in a JI 1794 x ‘Slow’ RIL population. Pisum Genetics. 2002;34:28–31. [Google Scholar]
- Taylor SA, Hofer JMI, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis THN. PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiology. 2002;129:1150–1159. doi: 10.1104/pp.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell. 2003;15:2742–2754. doi: 10.1105/tpc.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Delgado-Benarroch L, Egea-Cortines M. Genetic control of floral size and proportions. International Journal of Developmental Biology. 2005;49:513–525. doi: 10.1387/ijdb.051998jw. [DOI] [PubMed] [Google Scholar]
- Murfet IC, Reid JB. The control of flowering and internode length in Pisum. In: Hebblethwaite PD, Heath MC, Dawkins TCK, editor. The pea crop. London: Butterworth; 1985. pp. 67–80. [Google Scholar]
- Singer S, Maki S, Sollinger J, Plotz J, Fitzgerald K, Fishbach J, Mullen H. Flower development in pea: Role of Proliferating Inflorescence Meristem (PIM), an AP1 homolog. Developmental Biology. 2002;247:518–518. [Google Scholar]
- Sollinger JD, Singer SR. Unraveling the flower with pea developmental mutants-homologies and hidden potentials. Developmental Biology. 2002;247:444–445. [Google Scholar]
- Gourlay CW, Hofer JMI, Ellis THN. Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA, and TENDRIL-LESS. Plant Cell. 2000;12:1279–1294. doi: 10.1105/tpc.12.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall AD, Turner L, Knox MR, Ambrose MJ, Ellis THN, Hofer JMI. The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell. 2005;17:1046–1060. doi: 10.1105/tpc.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JL, Jablonski W, Reid JB. Leaf and flower development in pea (Pisum sativum L.): Mutants cochleata and unifoliata. Annals of Botany. 2001;88:225–234. doi: 10.1006/anbo.2001.1448. [DOI] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Diaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M. Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Molecular Biology. 2005;58:561–574. doi: 10.1007/s11103-005-6163-6. [DOI] [PubMed] [Google Scholar]
- National Centre for Biotechnology Information http://www.ncbi.nlm.nih.gov/
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Gene Regulatory Information Server http://arabidopsis.med.ohio-state.edu/AtTFDB/
- Dobbin K, Simon R. Comparison of microarray designs for class comparison and class discovery. Bioinformatics. 2002;18:1438–1445. doi: 10.1093/bioinformatics/18.11.1438. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Hudspeth RL, Hobbs SL, Anderson DM, Rajasekaran K, Grula JW. Characterization and expression of Metallothionein-like genes in cotton. Plant Molecular Biology. 1996;V31:701–705. doi: 10.1007/BF00042243. [DOI] [PubMed] [Google Scholar]
- Dobres MS TWF. A Developmentally Regulated Bud Specific Transcript in Pea Has Sequence Similarity to Seed Lectins. Plant Physiology. 1989;89:833–838. doi: 10.1104/pp.89.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, Chen T, Zhang X, Timmermans MCP, Beck J, Buckner B, Janick-Buckner D, Nettleton D, Scanlon MJ, Schnable PS. Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.) The Plant Journal. 2007;52:391–404. doi: 10.1111/j.1365-313X.2007.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M, Takahashi T, Komeda Y. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:571–580. doi: 10.1093/oxfordjournals.pcp.a029579. [DOI] [PubMed] [Google Scholar]
- Meshi T, Taoka K, Iwabuchi M. Regulation of histone gene expression during the cell cycle. Plant Molecular Biology. 2000;V43:643–657. doi: 10.1023/A:1006421821964. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y. Upstream sequences of rice proliferating cell nuclear antigen (pcna) gene mediate expression of pcna-gus chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Research. 1991;19:1571–1576. doi: 10.1093/nar/19.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster CI, Packman LC, Gray JC. HMG-1 enhances HMG-I/Y binding to an A/T-rich enhancer element from the pea plastocyanin gene. European Journal of Biochemistry. 2001;268:3154–3162. doi: 10.1046/j.1432-1327.2001.02191.x. [DOI] [PubMed] [Google Scholar]
- Gegas VC, Doonan JH. Expression of cell cycle genes in shoot apical meristems. Plant Molecular Biology. 2006;60:947–961. doi: 10.1007/s11103-006-0011-1. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Fischer RL. Biology of chromatin dynamics. Annual Review of Plant Biology. 2005;56:327–351. doi: 10.1146/annurev.arplant.56.032604.144118. [DOI] [PubMed] [Google Scholar]
- Guyomarc'h S, Bertrand C, Delarue M, Zhou DX. Regulation of meristem activity by chromatin remodelling. Trends in Plant Science. 2005;10:332–338. doi: 10.1016/j.tplants.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Kaya H, Shibahara K, Taoka K, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Cerny J, Quesenberry PJ. Chromatin remodeling and stem cell theory of relativity. Journal of Cellular Physiology. 2004;201:1–16. doi: 10.1002/jcp.20071. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu GX, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant Journal. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 Protein, Responsible for Translation Reinitiation, Is Required for Auxin-Mediated Gynoecium Patterning. Plant Cell. 2005;17:2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. Auxin transport - shaping the plant. Current Opinion in Plant Biology. 2003;6:7–12. doi: 10.1016/S1369526602000031. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, Nagato Y, Sato Y. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences. 2007;104:14867–14871. doi: 10.1073/pnas.0704339104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, Goff SA, Glazebrook J. A network of rice genes associated with stress response and seed development. PNAS. 2003;100:4945–4950. doi: 10.1073/pnas.0737574100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- VERTIS http://www.vertis-biotech.com/index.php?ip=104
- Rasmussen SW. SEQtools, a software package for analysis of nucleotide and protein sequences. 2006. http://www.seqtools.dk
- Combimatrix Corporation http://www.combimatrix.com
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcripts identified to be up-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.
Transcripts identified to be down-regulated in the pea shoot apical meristem (SAM) in comparison to the non-meristem (NM) tissues.