Abstract
The fast growing Protein Data Bank (PDB) contains a vast amount of 3-dimensional data on proteins, and nucleic-acid structures obtained by X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy. PyDeT is a PyMOL (molecular visualization software system) plug-in that visualize tessellations derived from the protein structure along with the source protein. PyDeT is released under a GNU General Public License (GPL) and is available from the authors.
Keywords: proteins, nucleic acids, structures, visualization, plug-in
Delaunay-Decompositions and QHull
Triangulations on the plane are frequently used for discretizing geometrical data and relations. Delaunay triangulations are of special importance and they are generated easily. The resulting triangles are usually not far from regular triangles. We need the three-dimensional analogue of the triangulations to discretize spatial data for protein-ligand complexes.
Definition
Given a finite set of points A ⊆ R3, and an H ⊆ A such that the points of H are on the surface of a sphere and the sphere does not contain any further points of A, then the convex hull of H is called a Delaunay region.
To find the Delaunay decomposition of a set, we have used the qhull algorithm [1] (the source of which is available elsewhere [2]). The underlying principle of the algorithm is that the Delaunay decomposition is easily traced back to a simple convex hull problem [3] and it is then solved by an incremental algorithm.
PyMol
PyMol is a widely used open-source molecular visualization program maintained by DeLano Scientific LLC as a user sponsored project. PyMol version 1.0 earned a reputation for its ease of use and numerous features. A powerful set of scripts for calculating physical properties of molecules is available among classical molecular models in PyMol.
The core of the program is a full featured Python interpreter, which is then extended by an OpenGL 3D display, a Tcl/Tk based GUI and the PyMol API which gives the possibility of creating plug-ins. Although this later feature was mainly intended for automatic coloring of atoms according to relevant properties, loading and manipulating structures, it is also capable of inserting additional graphical primitives, like spheres, cylinders, triangles and vertices called Compiled Graphic Objects (CGOs).
PyDeT
Usage
PyDeT will adopt any selection of atoms of a molecule already loaded into the PyMol software. The installation of the plug-in is described in a later subsection. Using the plug-in is as easy as typing the pydet [selection], [name] command into PyMol command line, where the two arguments described below is optional.
The first argument is a selection and defaults to ‘all’. The plug-in will take the atoms in this selection as points in 3D space and generate the Delaunay Triangulation of the point set for visualizing it by its edges. The visualization is done by creating a layer called PyDeT, and inserting ‘line like’ cylinders between the endpoints of the edge. The edges are colored by relative length: red corresponds to very short edges and blue corresponds to very long edges. This is relative to other edges in the same tessellation. Hence, there will always be at least one blue and one red edge. Entering a name as the second argument will rename the PyDeT layer to PyDeT [name] to support using multiple tessellations corresponding to different selections.
An example
There are two different point sets that may prove to be of interest for most of the protein structures: the set of all heavy atoms (i.e. non-hydrogen atoms) and the set of carbon alpha atoms. Singh, Tropsha and Vaisman [4] described the later case and concluded that Delaunay tetrahedra situated along helices tend to be smaller than tetrahedra along sheets. This result can be visualized easily by PyDeT. Taking the PDB entry 1CRN as an example, the majority of short edges (colored red by PyDeT) gather around the helices.
Installation
It is necessary to obtain a working copy of QHull, available elsewhere [2] and a copy of PyMol available elsewhere [5] to use PyDet. After the successful installation of QHull, the PyMol modules/pmg_tk/startup directory is located manually. A copy of PyDeT.py is then placed in the directory. The PyDeT.py file is modified using a text editor such that qdelaunay variable contains the path of qdelaunay executable.
Structure of the python code
There are 5 core functions in Pydet.py. The _init_ is the initializations function. The corresponding lines should simply be uncommented to use functions from PyMol command line. PyDeT is a simple wrapper-around-function which defines filenames that serve as the channel for communication with the external qdelaunay program and then fires the remaining 3 functions. Filtering on the tetrahedra before visualizing can be inserted into this function. PyDeT_DExtract extracts spatial data from the selection for writing into the disk.
PyDeT_QDelaunay calls qdelaunay which reads the file created in the previous step for writing the result to disk. PyDeT_TessVisualise reads the result generated by qdelaunay for drawing the edges. A filtering function has to be implemented to dispose certain tetrahedra based on some properties. This is called between PyDeT_QDelaunay and PyDeT_TessVisualise.
An example for co-spherical points of a helix
Most Delaunay regions are tetrahedral. However, if more than 4 points fall on the same sphere, a complex Delaunay region is developed. In case of a random point set, it is assumed that all regions are tetrahedral and 5 points are co-spherical with probability 0.
Aromatic rings will clearly result in hexagonal pyramid shaped Delaunay regions when dealing with point sets derived from protein heavy atom coordinates.
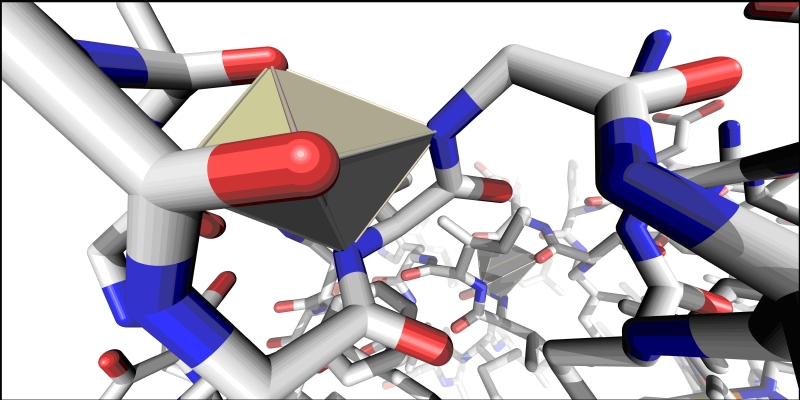
Not so apparent is the fact, that in a large number of cases certain atoms of an alpha helix are arranged on a sphere. On figure 1 this happens twice in one of the α helices of the protein found in the PDB entry 101m. (Only perfectly co-spherical points are shown on the figure: allowing a small computation error would result in most of the Delaunay regions inside the helix, to merge with one of their neighbors).
Figure 1.
Example of co-shperical points in an α helix is shown.
References
- 1.Barber CB, et al. ACM Transactions on Mathematical Software. 1996;22:46. [Google Scholar]
- 2. http://www.qhull.org/
- 3.Brown FD. Informal processing letters. 1978;9:223. [Google Scholar]
- 4.Singh RK, et al. Journal of Computational Biology. 1996;3:213. doi: 10.1089/cmb.1996.3.213. [DOI] [PubMed] [Google Scholar]
- 5. http://pymol.sourceforge.net/