Abstract
In an effort to identify nuclear receptors important in retinal disease, we screened a retina cDNA library for nuclear receptors. Here we describe the identification of a retina-specific nuclear receptor (RNR) from both human and mouse. Human RNR is a splice variant of the recently published photoreceptor cell-specific nuclear receptor [Kobayashi, M., Takezawa, S., Hara, K., Yu, R. T., Umesono, Y., Agata, K., Taniwaki, M., Yasuda, K. & Umesono, K. (1999) Proc. Natl. Acad. Sci. USA 96, 4814–4819] whereas the mouse RNR is a mouse ortholog. Northern blot and reverse transcription–PCR analyses of human mRNA samples demonstrate that RNR is expressed exclusively in the retina, with transcripts of ≈7.5 kb, ≈3.0 kb, and ≈2.3 kb by Northern blot analysis. In situ hybridization with multiple probes on both primate and mouse eye sections demonstrates that RNR is expressed in the retinal pigment epithelium and in Müller glial cells. By using the Gal4 chimeric receptor/reporter cotransfection system, the ligand binding domain of RNR was found to repress transcriptional activity in the absence of exogenous ligand. Gel mobility shift assays revealed that RNR can interact with the promoter of the cellular retinaldehyde binding protein gene in the presence of retinoic acid receptor (RAR) and/or retinoid X receptor (RXR). These data raise the possibility that RNR acts to regulate the visual cycle through its interaction with cellular retinaldehyde binding protein and therefore may be a target for retinal diseases such as retinitis pigmentosa and age-related macular degeneration.
Nuclear receptors are ligand-activated transcription factors involved in a wide variety of physiological and regulatory processes. They share a tripartite domain structure including a variable N-terminal domain (A/B domain), a highly conserved DNA-binding domain (DBD) (C domain), and a less conserved ligand-binding domain (LBD) (E/F domain) (1–3). Many novel nuclear receptors have been found in the past decade, mainly through low stringency hybridization screening of cDNA libraries using the DBD as probes, and through degenerate PCR based on consensus regions (4–8). The rapid increase in the number and diversity of partially sequenced cDNA clones (ESTs) has provided an alternative approach for the discovery of novel members of gene families (9–11) but has been less productive in identification of novel nuclear receptors (F.C., unpublished work).
Our laboratory is interested in finding new targets for treating degenerative retinal diseases, including age-related macular degeneration (AMD) and retinitis pigmentosa (RP). AMD affects 10 million patients in the U.S. alone with progressive loss of central vision; pathologically, it is a disorder of the sensory retina, retinal pigment epithelium (RPE), and choriocapillaris (12). Most AMD is sporadic, but the genes responsible for several inherited forms of the disease have recently been identified (13). RP is a heterogeneous group of retinal degenerative diseases that affects ≈100,000 individuals in the U.S. and is characterized by night blindness, progressive loss of peripheral vision, and decline in the electroretinograph (ERG) (14). There are at least 23 genetic loci associated with dominant, recessive, X-linked, and digenic forms of retinitis pigmentosa, but the majority of RP is the autosomal dominant form (ADRP). Approximately 25% of all cases of ADRP are caused by point mutations in the rhodopsin gene (14), and to date over 100 different ADRP associated mutations in rhodopsin have been identified (15).
In both AMD and ADRP, there is thought to be dysfunction in the retinal pigmented epithelium, which serves critical functions subserving normal vision, including photoreceptor outer segment phagocytosis and cycling of visual pigments (16). These visual pigments are retinoids derived from dietary vitamin A, and the RPE is one of the major sites of retinoid uptake and metabolism in the body (17). It has been suggested from epidemiologic or interventional data that vitamin A consumption may affect both AMD and ADRP incidence (18). The actions of vitamin A are mediated by a series of retinoic acid receptor (RAR) and retinoid X receptor (RXR) retinoid nuclear receptors, expressed at high levels in the retina (19, 20). We reasoned that the many retinoid derivatives present in the retina could function as ligands for additional nuclear receptors, which may mediate the physiological and therapeutic effects of the retinoids.
In an effort to identify new members of the nuclear receptor superfamily expressed in the retina, we screened both mouse and human retinal cDNA library for novel nuclear receptors. We report here the cloning of a nuclear receptor that is expressed exclusively in the pigmented epithelium and Müller glial cells of the mammalian retina. This novel nuclear receptor, which we designate retina-specific nuclear receptor (RNR), was found to be identical to the recently reported photoreceptor-specific nuclear receptor (PNR) (NR2E3) (21), although we did not detect RNR expression in mouse or primate photoreceptors. Preliminary data suggest that this receptor is a transcriptional repressor in the absence of ligand and that it may regulate transcription of cellular retinaldehyde-binding protein (CRALBP), an important component of the visual cycle. We suggest that RNR may be an attractive target for the development of novel therapeutics for retinal disease.
Materials and Methods
Isolation of Human and Mouse RNR.
DNA fragments corresponding to the DNA binding domain (DBD) regions of androgen receptor, estrogen receptor β (ERβ), glucocorticoid receptor, and vitamin D receptor (3, 22) were labeled with 32P by nick translation (Promega). Several human primary λZAP cDNA libraries were screened at low stringency with the above probes at a concentration of 106 cpm/ml, essentially as described (8). Purified phage clones were converted to plasmid clones according to the Stratagene protocol. End sequences of all of the candidate clones were obtained through automated sequencing using M13 forward and reverse primers. blast (http://www.ncbi.nlm.nih.gov/blast) search of partial cDNA sequences revealed that one purified clone from a human retina cDNA library was identical to an EST W27871, which both showed 74.2% amino acid sequence identity toTlx DBD region, a chicken homolog of the Drosophila tailless gene (23). Sequencing via primer walking was carried out on the clone to obtain the entire cDNA sequence. The mouse RNR ortholog was isolated through degenerate PCR on phage clones obtained from one round of low stringency screening of a mouse λZAP eye cDNA library. Oligonucleotide 5′-CCC AGG CTT TAC ACT TTA TGC TTC C-3′ corresponding to pBluescript II SK(+) vector (Stratagene) was paired with primer hNR5R (5′-GCT GTC TCC GCA CAC GCG GCA-3′) to PCR amplify the mRNR 5′ cDNA using AmpliTaq Gold in a Perkin–Elmer 9600 (PE Applied Biosystems, Foster City, CA). PCR conditions were 1× PCR buffer, 250 μM dNTP, 0.5 μM each primer, 1 unit/20 μl TaqGold polymerase, and 0.5 μl of the phage plaque elute solution per 20 μl of reaction. PCR profile was: 95°C × 9 min for initial denaturation, 35 cycles at 95°C × 30 sec, 50°C × 20 sec, and 72°C × 30 sec, and a final extension at 72°C × 10 min. Initial PCR products showed 80% DNA sequence identity with human RNR in the 5′ coding region and so were used to design a new primers for vector PCR (mR5F1 5′-TCG GTT GGG CCC AGC AAC TTC-3′, paired with pBluescript II SK(+ )primer 5′-GGG GAT GTG CTG CAA GGC GA-3′), which amplified the remaining mRNR 5′ ORF from phage pools.
Tissue Distribution of RNR.
Northern blot analysis.
A DNA fragment corresponding to the entire LBD of hRNR was labeled with 32P by random priming (Promega) and was hybridized to multitissue Northern blots from CLONTECH and a custom-made blot containing RNA samples of human brain and retina (FRP, San Francisco). A human β-actin probe was used as control.
Reverse transcription–PCR.
By using primers 5′-ATG AGC TCC ACA GTG GCT GC-3′ (NR5F2) and 5′-GCT GTC TCC GCA CAC GCG GCA-3′ (NR5R), reverse transcription–PCR was performed on the following samples: adrenal, bone marrow, brain, fetal brain, heart, fetal kidney, liver, fetal liver, lung, fetal lung, mammary gland, pancreas, placenta, prostate, skeletal muscle, small intestine, spleen, testis, thyroid, thymus, uterus, retina human genomic DNA, and a cDNA library made from passaged, primary fetal human RPE cells. The experimental protocol was essentially as described (24).
In situ hybridization.
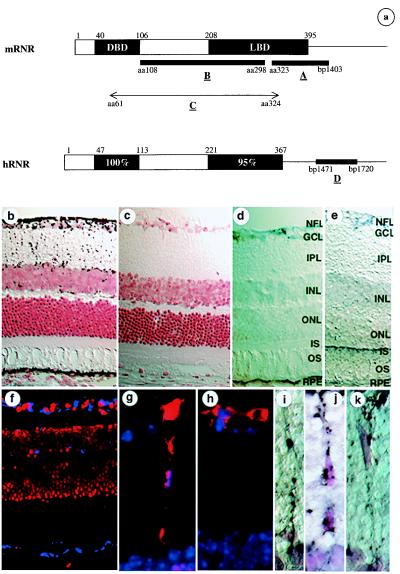
DNA fragments at the 3′-end of human and mouse RNR (Fig. 2a, probes A and D) were generated by using primer pairs 5′-GGC AGT GAC CTC ACT GAA GA-3′ (NR5F9)/5′-ACT GGC AGG AAC CTG TTA TAC-3′ (NR5R10) and 5′-AAG GAT CCT GAG CAC GTG GAG-3′ (mR5F5)/5′-TAG GAT GGC AAT GAA TAT GCC-3′(mR5R3). PCR fragments were subcloned into the vector pCRIITA (Invitrogen) and were used as templates to prepare probes for in situ hybridization. A DNA template corresponding to amino acid residues 108–298 of mouse RNR (Fig. 2a, probe B) was also prepared in the same vector. Biotinylated sense and antisense riboprobes specific for hRNR and mRNR were generated via in vitro transcription using Biotin RNA Labeling Mix (Boehringer Mannheim). Whole eyes from 8-week-old Swiss–Webster albino mice were fixed by intracardiac perfusion with 4% paraformaldehyde, were embedded in paraffin, and were sectioned at 8 μm. In situ hybridization was carried out as described (13) by using the TSA Indirect Kit (NEN). Amplified signal was visualized with diaminobenzidine (Sigma) solubilized in nickel chloride buffer (Digene Diagonstics, Silver Spring, MD). Whole monkey eyes were enucleated and fixed by immersion in 4% paraformaldehyde for 8 hr. After radial cuts and lens removal, the eyes were cryoprotected in graded sucrose solutions and a sucrose/Tissue Tek O.C.T. (optimum cutting temperature) Compound (Miles) mixture and were frozen on dry ice as described (25). Retinal segments were sectioned (6 μm), and in situ hybridization was carried out as above by using TSA Direct Blue (Coumarin) according to manufacturer's instructions. Sections were mounted with Vectashield mounting medium containing Propidium Iodide (Vector Laboratories), and images were digitally acquired by using a Spot CCD camera (Phase 3 Imaging Systems, Milford, MA). An in situ probe corresponding to nucleotides 6,681–7,146 of the mouse ATP-binding cassette transporter gene ABCR (26, 27) was also generated, and in situ hybridization was performed on mouse eye sections following the same procedure.
Figure 2.
In situ hybridization on albino mouse and rhesus monkey retina by using RNR-specific probes. (a) Portions of mRNR and hRNR used for generating the in situ probes. (b–e and i–k) In situ hybridizations on albino mouse retina with chromogenic detection and counterstaining with neutral red; hybridization signal is seen as black and retinal cell nuclei are seen as red. (b) Antisense probe A. (c) Sense probe A. (d) Antisense probe B. (e) Antisense ABCR probe. (i–k) Higher-power views of cells having morphology of Müller glia in the inner nuclear layer/inner plexiform layer (IPL). (f) In situ hybridization of antisense probe D on rhesus monkey retina; in situ signal is detected by using coumarin fluorescence and is seen as blue; cell nuclei are counterstained by using propidium iodide and are seen as red. (g–h) Glial fibrillary acidic protein immunohistochemistry on mouse eye sections adjacent to those used in b–e, seen as red fluorescence; cell nuclei are seen as blue. NFL, nerve fiber layer; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; IS, inner segments of photoreceptors; OS, outer segments of photoreceptors; RPE, retinal pigment epithelium.
Transient Transfection Assays.
The Gal4/secreted alkaline phosphatase (SEAP) reporter plasmid was constructed by inserting the promoter region of pG5CAT, which contains five copies of consensus Gal4 binding sites, into pSEAP2-Basic and pSEAP2-Enhancer vectors (CLONTECH) at the KpnI/HindIII site. CMV-Gal4 was constructed by subcloning the Gal4 DNA binding domain derived from pM vector (CLONTECH) into pcDNA3.1(+) (Invitrogen) at the BamHI/EcoRI site. Gal4-LBD fusion vectors for hRNR, mRNR, hPXR (28), and hERβ (22) were made by subcloning the corresponding LBD to the CMV-Gal4 vector. CV1 cells from the American Type Culture Collection (Rockville, MD) were cultured in phenol red free DMEM supplemented with 10% fetal bovine serum and 1× Pen/Strep/l-Glu. RPE-J cells (29) were cultured in phenol red free DMEM supplemented with 4% fetal bovine serum, 1× Pen/Strep/l-Glu, and 1× nonessential amino acids. All tissue culture media were purchased from Life Technologies (Gaithersburg, MD). The day before transfection, 104 CV1 cells/well or 2 × 104 RPE-J cells/well were seeded in 96-well cell culture plates. The next day, 250 ng of DNA including receptor, reporter, and carrier DNA in 10 μl of Opti-MEM I were combined with 10 μl of 0.2 mg/ml LipofectAMINE diluted in Opti-MEM I (Life Technologies). Cells were rinsed once with Opti-MEM I, and 80 μl Opti-MEM I was added to each well followed by 20 μl DNA:LipofectAMINE and incubation for 4 hr at 37°C. The SEAP assay on cell supernatants was performed on the third day after transfection by using the Phospha-Light kit from Tropix (Bedford, MA). The reaction was read in DYNEX microlite plates by using the dynex revelation 3.2 program (DYNEX Technologies, Chantilly, VA). Each experiment was done in triplicate and was repeated at least three times.
Gel Mobility Shift Assays.
pCRIITA vectors containing full-length mRNR, hRARα, and mRXRβ (3) were used for in vitro transcription and translation via the TNT system (Promega). A DNA fragment corresponding to nucleotide position −430 to −200 of human CRALBP promoter region (CR) was obtained through PCR. The fragment was labeled with [α-32P]dCTP through PCR. A DNA fragment corresponding to the Gal4 binding site (GS) was also obtained through PCR using pG5CAT (CLONTECH) as template. GS was used as nonspecific control element. Reaction set-up was essentially as described (8). The labeled DNA fragment CR ≈5 × 104 cpm was incubated with 1.5 μl of each lysate containing the translated receptor in the absence or presence of >10-fold of unlabeled CR or GS.
Results
Isolation of Human and Mouse RNR.
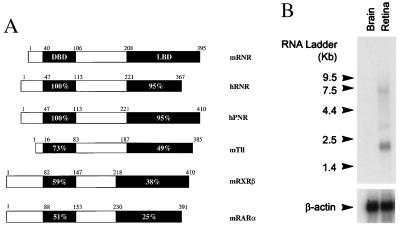
We screened ≈250,000 primary phage clones from an adult human retina cDNA library with DNA probes corresponding to the DBDs of the human androgen receptor, estrogen receptor β, glucocorticoid receptor, and vitamin D receptor, at low stringency (8). Thirty-eight positives were identified and sequenced. The known retinoid receptors RXR and RAR were among these positives. In addition, the 5′-end of one clone was virtually identical to an unannotated EST w27871, which showed >70% DNA sequence homology to Tlx (a chicken homolog of the Drosophila tailless gene) or COUP-TF (chicken ovalbumin upstream promoter transcription factor) in the DBD region (3, 23). Sequencing of the entire clone revealed an ORF encoding a protein of 367 amino acid residues that had an authentic nuclear receptor domain structure (Fig. 1A). The protein sequence of the cDNA shared 73% amino acid sequence identity to mTll [a mouse homolog of Drosophila tailless gene (3) in the DBD (Fig. 1A)]. Further library screening and PCR analysis identified 20 more such clones of the 250,000 primary phage screened. A blast search of the public database using the entire cDNA revealed two additional ESTs (w21793 and w21801) from a human retina cDNA library, which overlapped with our clone in a nonconserved region. The clone was designated retina-specific nuclear receptor (RNR) based on its abundance in retina and the expression pattern described in tissue distribution studies detailed below.
Figure 1.
(A) Comparison of mouse RNR with human RNR, hPNR, and other closely related nuclear receptors. The DBD and LBD regions of each receptor was compared with the corresponding region of mRNR by using gcg bestfit (Genetics Computer Group, Madison, WI). Numbers in the bars show the percentage of amino acid sequence identity to mRNR. The small numbers above the bar indicate the boundaries of different regions based on amino acid residue position. (B) Northern blot analysis on mRNA from human brain and retina. Each lane contains 2 μg of poly(A)+ RNA. β-actin was used to normalize hybridization signal.
A similar low stringency screen was also performed on a mouse eye cDNA library. Eighty-seven positives were obtained, arrayed in 96-well plates, and subjected to degenerate PCR using vector primer and those specific for hRNR N terminus. Sequence analysis on the PCR fragments revealed a cDNA fragment that shared 80% amino acid sequence identity to the hRNR N terminus (Fig. 1A); 16 of the initial 87 positives were this sequence. The new sequence information was used to obtain the remainder of a 1.6 kb cDNA via PCR, sequence analysis of which confirmed that this was a mouse ortholog of hRNR containing the full coding region. mRNR shares 84 and 90% sequence identity with hRNR at the DNA and amino acid levels, respectively. However, mRNR contains an extra 43 amino acids at the C terminus, which is not present in hRNR (Fig. 1A).
RNR was initially found to be most homologous to the chicken and mouse homologs of Drosophila tailless gene (3, 23), with which it shared 73 and 49% sequence identity at amino acid level in the DBD and LBD regions respectively (Fig. 1A). While this manuscript was in preparation, Kobayashi et al. reported the cloning of a human photoreceptor cell-specific nuclear receptor (21). Sequence alignment of hRNR and mRNR with the hPNR cDNA and genomic structure (21) indicated that hRNR is a splice variant of hPNR missing 43 amino acids and that mRNR is a full-length mouse ortholog of hPNR (Fig. 1A).
RNR Is Expressed Specifically in the Retina.
To determine the tissue distribution of hRNR, reverse transcription–PCR was performed on mRNA samples from a large variety of human tissues, using primers directed against the 5′ coding region of hRNR flanking intron A (21). A prominent band of 136 bp was obtained from retina mRNA but from no other tissue, including adrenal, bone marrow, brain, fetal brain, heart, fetal kidney, liver, fetal liver, lung, fetal lung, mammary gland, pancreas, placenta, prostate, skeletal muscle, small intestine, spleen, testis, thyroid, thymus, and uterus. Northern blot analysis further demonstrated that hRNR is specifically expressed in retina, with three transcripts of ≈7.5 kb, ≈3.0 kb, and ≈2.3 kb, the ≈2.3-kb transcript being most abundant (Fig. 1B). Together, these results indicate that hRNR is expressed exclusively in retina.
RNR Is Expressed Specifically in the Retinal Pigment Epithelium (RPE) and Müller Glial Cells.
To determine the cellular localization of RNR within the retina, in situ hybridization studies on mouse and monkey eye sections were performed. Antisense and sense in situ probes corresponding to two different fragments of mRNR (Fig. 2a, probes A and B) were used on albino Swiss–Webster mouse eye sections. Both probes gave the same result, demonstrating that mRNR is expressed in the RPE and in cells (Fig. 2 b and d). Müller glial cell expression was suggested by the expression of mRNR in a regularly spaced subset of cell bodies in the inner nuclear layer, with processes that extended bidirectionally toward the outer and inner limiting membranes, a pattern typical of Müller glia (ref. 30; www.insight.med.utah.edu/webvision/glia.html#müller). Müller glial cell identity was confirmed by colocalization of mRNR by in situ hybridization (Fig. 2 i–k) and glial fibrillary acidic protein and S-100 by immunohistochemistry (Fig. 2 g and h; and data not shown). Given the difference between this expression pattern and that seen by Kobayashi et al. (21), who saw expression of this gene in the outer nuclear (photoreceptor) layer, an extensive series of control experiments was performed to evaluate the accuracy and specificity of the mRNR expression pattern. First, a mRNR sense control probe produced no signal (Fig. 2c). Second, a series of hybridizations at decreasing stringency were performed, and no mRNR signal was seen in the outer nuclear layer under any condition (data not shown). Third, in situ hybridization using a probe virtually identical to that used by Kobayashi et al., but lacking the highly conserved DBD (Fig. 2a, probe B), gave the same pattern of expression in RPE and Müller glial cell (Fig. 2d). Fourth, an irrelevant antisense riboprobe, specific for the mouse rod photoreceptor ATP-binding cassette transporter gene (mABCR), demonstrated the expected expression in the inner segments of the outer nuclear layer photoreceptors (Fig. 2e), consistent with the findings of Allikmets et al. (26) but distinct from the mRNR pattern. Fifth, the ABCR sense probe gave no signal (data not shown). Sixth, in situ hybridization for RNR was performed on rhesus monkey retina by using an hRNR probe (Fig. 2a, probe D) and demonstrated an identical expression pattern (Fig. 2f) to that seen in the mouse retina (Fig. 2 b and d). Importantly, detection of the in situ signal in the monkey was with coumarin to avoid possible confusion with lipofuscin autofluorescence (31). Finally, PCR on a cDNA library made from passaged, primary fetal human RPE cells (K.P., unpublished work) detected the presence of hRNR, further confirming expression of hRNR in the RPE.
RNR-LBD Is a Strong Inhibitor of Transcriptional Activity.
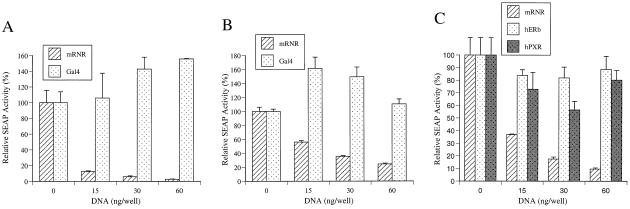
Nuclear receptors have separable DBDs and LBDs. It is therefore possible to create chimeric receptors that contain the yeast Gal4 DBD but a nuclear receptor LBD. These chimeric receptors will bind to and activate transcription of Gal4-responsive reporter genes but are regulable by particular nuclear receptor ligands. This chimeric receptor/reporter system has provided a cell-based approach to orphan nuclear receptor ligand identification without knowledge of DNA binding characteristics (32). To identify the ligand that activates RNR, a Gal4 chimeric receptor/reporter system with low basal activity and high induction was established for this receptor. CV-1 cells were transfected transiently with SEAP reporter and chimeric mouse and human RNR receptor plasmids. Compounds including all-trans-retinoic acid, 9-cis-retinoic acid, and 11-cis-retinaldehyde were added on the second day after the transfection at a final concentration of 10 μM. Rather than observing the expected up-regulation of promoter activity on ligand binding, repression of promoter activity was consistently observed and was independent of ligand. Transcriptional repression was seen in samples transfected with the Gal4-reporter/RNR-LBD, compared with those transfected with the Gal4-reporter alone, using both mouse and human RNR. To confirm the inhibitory effect of the RNR ligand binding domain, a reporter plasmid with high basal activity was made by including an SV40 enhancer in the SEAP reporter. The high basal transcriptional activity of SEAP enhancer reporter was repressed drastically with as little as 15 ng of Gal4-mRNR-LBD plasmid, in the absence of added ligand. The inhibition was plasmid concentration-dependent, with 60 ng of Gal4-RNR-LBD reducing transcriptional activity to <5% of control (Fig. 3A). This inhibition did not result from the CMV mammalian expression vector itself or overexpression of the yeast Gal4 DNA binding domain, as the CMV-Gal4-DBD expression plasmid alone had no inhibition effect on the reporter (Fig. 3A).
Figure 3.
Gal4 chimeric receptor and SEAP reporter cotransfection assays. (A) On CV1 cells. (B) On RPE-J cells. Striped bar indicates relative activity of Gal4-mRNR-LBD, and dotted bar indicates Gal4 vector control. (C) Comparison study on Gal4 chimerics of mRNR-LBD (striped bar), ERβ-LBD (dotted bar), and hPXR-LBD (shaded bar) was carried out on CV1 cells by using a SEAP reporter. The above experiments had been repeated at least three times. The data here represent the mean ± SD of one experiment.
Because the in situ hybridization results indicated that RNR is expressed in RPE cells, the inhibitory effect of the mRNR-LBD was also examined in an RPE cell line. The same set of cotransfection assays was carried out on the rat RPE-J cell line (29), and the same inhibition profile was observed (Fig. 3B). We then investigated whether this inhibition was particular to RNR-LBD. Comparison studies of the mRNR, hERβ, and hPXR LBDs were performed on CV1 cells. The same dose-dependent inhibition of SEAP enhancer reporter transcriptional activity was observed on mRNR-LBD (Fig. 3C). However, only minimal and non-dose-dependent inhibition was observed for hERβ-LBD and hPXR-LBD (Fig. 3C). Therefore, we conclude that, in the absence of exogenous ligand, RNR-LBD is a strong inhibitor of transcriptional activity.
Interaction of RNR and the CRALBP Promoter.
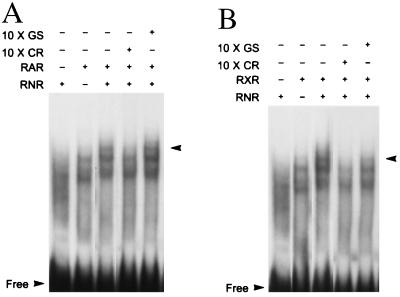
A potential nuclear receptor binding site AGGTCA has been reported in the human CRALBP gene at position −218 to −214 (3, 33). Detailed sequence analysis revealed that several more AGGTCA like sequences are also located near that region (data not shown). To test the binding of RNR to these potential sites, a fragment of the CRALBP promotor corresponding to nucleotide positions −430 to −200 (CR) was generated via PCR and was used in gel shift assays. Both RXR and RAR were identified in our screening of the retina cDNA library, and both are stimulated by retinoids, so we wondered whether either RXR or RAR may dimerize or interact with RNR. To test this, in vitro-translated RAR and RXR were included in the gel shift assays. RNR alone did not bind to CR whereas both RAR and RXR were found to bind to CR (Fig. 4 A and B). When either RAR or RXR were tested together with RNR, an additional up-shifted band could be observed compared with RAR or RXR alone (Fig. 4 A and B). To verify that the additional band depended on binding to CR, the gel-shift was competed with a 10-fold excess of cold CR or the unrelated Gal4 binding site (GS) (Fig. 4 A and B). The additional band was clearly competed by unlabeled CR but not by GS (Fig. 4 A and B). Thus, RNR can interact with the CRALBP promoter in the presence of RAR or RXR.
Figure 4.
Gel mobility shift assay of RNR, RAR, and RXR on CRALBP promoter region. The arrows indicate the extra up-shifted band. “Free” indicates unbound probe. A shows data from RNR in combination with RAR. B shows RNR in combination with RXR.
Discussion
We have cloned and characterized a novel nuclear receptor, RNR, which is expressed exclusively in the pigment epithelium and Müller glial cells of the mammalian retina. Using transcriptional cotransfection assays, we have shown that RNR is a repressor of transcription in the absence of ligand. Furthermore, we have shown that RNR binds to a region of the CRALBP promoter and that this binding depends on the interaction with members of the retinoid responsive nuclear receptor RAR and RXR families. Taken together, these data suggest that RNR may play a role in the regulation of the visual cycle by regulating the expression of genes important to retinoid transport and processing.
Database searching revealed that the human RNR coding region is identical to that of the human photoreceptor specific nuclear receptor, a novel nuclear receptor recently identified by Kobayashi et al. (21). Sequence alignment among hPNR, hRNR, and mRNR shows that mRNR is a mouse ortholog of hPNR whereas hRNR is a shorter form of hPNR (Fig. 1A). It was important to determine whether hRNR is an authentic splice variant or simply the product of an incompletely processed mRNA. Our studies, using the entire human RNR LBD to probe a human Northern blot, demonstrated three transcripts of ≈7.5 kb, ≈3.0 kb, and ≈2.3 kb, but a hRNR 3′-UTR probe (Fig. 2a, probe D) detected only the ≈7.5-kb transcript. In contrast, Kobayashi et al. used a human PNR LBD probe on a rat Northern blot and demonstrated an ≈2.0-kb transcript (21). These data argue that there is more than one RNR mRNA form. Examination of the published genomic structure of hPNR (21) revealed that our hRNR is missing the last exon of hPNR and includes the last intron (intron G) of hPNR (21). However, we were able to obtain cDNA fragments of hRNR via reverse transcription–PCR using primers specific to the hRNR 3′-UTR and coding regions, demonstrating that hRNR is an authentic mRNA generated by this gene. Chromosomal localization via radiation hybridization mapping confirmed that hRNR maps to chromosome 15, in agreement with Kobayashi et al. (21, 34). It is therefore clear that hRNR and hPNR represent two splice variants of the same gene.
Although our results and those of Kobayashi et al. (21) are easily reconciled with respect to hRNR and hPNR sequence, chromosomal localization, and overall tissue distribution (both groups found expression restricted to the retina), results on the cell specificity of expression differ substantially. Although Kobayashi et al. (21) demonstrated photoreceptor specific expression by in situ hybridization, we found RNR expressed only in RPE and Müller glial cells. Several possible methodological differences between the studies may explain this difference. First, the strain of mouse used differed between the two studies; substantial gene expression differences are common among even closely related strains (35). Second, although visualization of RNR/PNR expression in the unpigmented RPE of the albino Swiss–Webster mouse retina used here was straightforward, such visualization in the pigmented RPE of the mouse strain used by Kobayashi et al. would have been difficult; the dark blue chromogenic in situ reaction product is routinely obscured by the brown-black RPE melanin granules in such pigmented retinas (26). Third, our in situ hybridizations used a substantially more sensitive detection technique (36), which allowed more stringent reaction conditions; these are often required for resolution of less abundant transcripts, such as RNR. Fourth, our in situ probes did not contain the highly conserved DBD included in the probe of Kobayashi et al. (Fig. 2a, probe C); elimination of this domain may have avoided cross hybridization to other nuclear receptor genes expressed in the retina. Because of the considerable difference in biological function implied by the expression patterns seen in the two studies, we confirmed the RPE/Müller glial expression pattern by several independent methods. An extensive series of hybridizations in mouse and monkey eyes using multiple RNR probes, multiple sense control probes, and a different gene control probe consistently showed that RNR levels of expression were in the RPE and Müller glial cells. Perhaps most persuasively, the expression pattern of RNR was identical in both mouse and monkey using probes for the orthologous genes in the two species.
Our Gal4 chimeric cotransfection assays indicated that the LBD of RNR functions as a transcriptional repressor in the absence of exogenous ligand, and our gel shift assays indicate that RNR can interact with the promoter region of CRALBP. CRALBP is, like RNR, specifically expressed in RPE cells and Müller cells in the eye (33). Both cell types play an important role in maintaining and nourishing the rod and cone photoreceptor cells, and the RPE is the sole site where all-trans-retinol (AT) isomerization to 11-cis-retinol occurs (16, 37). Mutations in CRALBP, 11-cis-retinaldehyde dehydrogenase, and RPE-65, all proteins involved in the visual cycle, cause early onset autosomal recessive retinitis pigmentosa (38–40). CRALBP and RNR are both expressed specifically in the RPE and Müller glia (33), and our gel-shift studies showed that RNR interacts with the CRALBP promoter in the presence of the retinoid receptors RAR and RXR; RNR may thus regulate CRALBP expression. As CRALBP functions as a carrier of 11-cis-retinal and/or 11-cis-retinol in the RPE, RNR is the first retina specific nuclear receptor identified that could regulate the visual cycle.
In summary, we have identified a retina-specific nuclear receptor, RNR, that is expressed in RPE and Müller glial cells. RNR is a transcriptional inhibitor in the absence of ligand. Our data provide evidence that RNR interacts with the CRALBP promoter as part of a heterologous complex, including RAR and/or RXR, which are well characterized retinoid responsive nuclear receptors. Future studies should address the contribution of RNR to RPE and Müller cell identity and function, the identification of a ligand for RNR, and the potential of RNR as a therapeutic target in AMD, RP, and other retinal degenerative diseases.
Acknowledgments
We thank the staff at the Human Genetics Sequencing Core Facility for sequencing both human and mouse RNR clones, artists in the Merck Visual Communications Department for preparation of figures, Dr. Jeremy Nathans (Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine) for providing the mouse eye cDNA library, Dr. Dean Bok (Department of Anatomy and Cell Biology, University of California at Los Angeles School of Medicine) for providing the primary RPE cells, Dr. Rosalie Crouch (Medical University of South Carolina) for providing 11-cis-retinal, and Dr. Dwight Towler (Merck Department of Bone Biology) for valuable suggestions and comments on the manuscript.
Abbreviations
- ADRP
autosomal dominant retinitis pigmentosa
- AMD
age-related macular degeneration
- CRALBP
cellular retinaldehyde-binding protein
- CR
CRALBP promoter region
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- ERβ
estrogen receptor β
- EST
expressed sequence tag
- GS
Gal4 binding site
- PNR
photoreceptor cell-specific nuclear receptor
- RAR
retinoic acid receptor
- RNR
retina specific nuclear receptor
- RP
retinitis pigmentosa
- RPE
retinal pigment epithelium
- RXR
retinoid X receptor
- SEAP
secreted alkaline phosphatase
Footnotes
References
- 1.Evans R M. Science. 1988;240:889–894. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Malley B. Mol Endocrinol. 1990;4:363–369. doi: 10.1210/mend-4-3-363. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giguere V, Yang N, Segui P, Evans R M. Nature (London) 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 5.Issemann I, Green S. Nature (London) 1990;347:645–649. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt A, Endo N, Rutledge S J, Vogel R, Shinar D, Rodan G A. Mol Endocrinol. 1992;6:1634–1641. doi: 10.1210/mend.6.10.1333051. [DOI] [PubMed] [Google Scholar]
- 7.Dreyer C, Krey G, Keller H, Grivel F, Helftenbein G, Wahli W. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen F, Cooney A J, Wang Y, Law S W, O'Malley B W. Mol Endocrinol. 1994;8:1434–1444. doi: 10.1210/mend.8.10.7854358. [DOI] [PubMed] [Google Scholar]
- 9.Wells T N C, Peitsch M C. J Leukocyte Bio. 1997;61:545–550. doi: 10.1002/jlb.61.5.545. [DOI] [PubMed] [Google Scholar]
- 10.Chen H C, Kung H J, Robinson D. J Biomed Sci. 1998;5:86–92. doi: 10.1007/BF02258361. [DOI] [PubMed] [Google Scholar]
- 11.Liu S, Stoesz S P, Pickett C B. Arch Biochem Biophys. 1998;352:306–313. doi: 10.1006/abbi.1998.0608. [DOI] [PubMed] [Google Scholar]
- 12.Zarbin M A. Eur J Ophthalmol. 1998;8:199–206. doi: 10.1177/112067219800800401. [DOI] [PubMed] [Google Scholar]
- 13.Petrukhin K, Koisti M J, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, et al. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 14.Dryja T P, McGee T L, Hahn L B, Cowley G S, Olsson J E, Reichel E, Sandberg M A, Berson E L. N Engl J Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 15.Dryja T P, Li T. Hum Mol Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 16.Bok D. J Cell Sci Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- 17.Blomhoff R, Green M H, Norum K R. Annu Rev Nutr. 1992;12:37–57. doi: 10.1146/annurev.nu.12.070192.000345. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg J, Flowerdew G, Smith E, Brody J A, Tso M O. Am J Epidemiol. 1998;128:700–710. doi: 10.1093/oxfordjournals.aje.a115023. [DOI] [PubMed] [Google Scholar]
- 19.Mangelsdorf D J, Umesono K, Evans R M. In: The Retinoids: Biology, Chemistry, and Medicine. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 319–350. [Google Scholar]
- 20.Hoover F, Seleiro E A, Kielland A, Brickell P M, Glover J C. J Comp Neurol. 1998;391:204–213. [PubMed] [Google Scholar]
- 21.Kobayashi M, Takezawa S, Hara K, Yu R T, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Proc Natl Acad Sci USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosselman S, Polman J, Dijkema R. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 23.Yu R T, McKeown M, Evans R M, Umesono K. Nature (London) 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 24.Chen F, Zhang Q, McDonald T, Davidoff M J, Bailey W, Bai C, Liu Q, Caskey C T. Gene. 1999;228:101–109. doi: 10.1016/s0378-1119(98)00619-2. [DOI] [PubMed] [Google Scholar]
- 25.Barthel L K, Raymond P A. J Histochem Cytochem. 1990;38:1383–1388. doi: 10.1177/38.9.2201738. [DOI] [PubMed] [Google Scholar]
- 26.Allikmets R, Singh N, Sun H, Shroyer N F, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 27.Azarian S M, Travis G H. FEBS Lett. 1997;409:247–252. doi: 10.1016/s0014-5793(97)00517-6. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann J M, McKee D D, Watson M A, Willson T M, Moore J T, Kliewer S A. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marmorstein A D, Gan Y C, Bonilha V L, Finnemann S C, Csaky K G, Rodriguez-Boulan E. J Cell Biol. 1998;142:697–710. doi: 10.1083/jcb.142.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowling J E. The Retina: An Approachable Part of the Brain. Cambridge, MA: Harvard Univ. Press; 1987. [Google Scholar]
- 31.Solbach U, Keilhauer C, Knabben H, Wolf S. Retina. 1997;17:385–389. doi: 10.1097/00006982-199709000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Braselmann S, Graninger P, Busslinger M A. Proc Natl Acad Sci USA. 1993;90:1657–1661. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Intres R, Goldflam S, Cook J R, Crab J W. J Biol Chem. 1994;269:25411–25418. [PubMed] [Google Scholar]
- 34.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 35.Winkler A, Spanagel R. Neuroreport. 1998;9:1459–1464. doi: 10.1097/00001756-199805110-00039. [DOI] [PubMed] [Google Scholar]
- 36.Bobrow M N, Shaughnessy K J, Litt G J. J Immunol Methods. 1991;137:103–112. doi: 10.1016/0022-1759(91)90399-z. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy B N, Goldflam S, Chang M A, Campochiaro P, Davis A A, Zack D J, Crabb J W. J Biol Chem. 1998;273:5591–5598. doi: 10.1074/jbc.273.10.5591. [DOI] [PubMed] [Google Scholar]
- 38.Gu S M, Thompson D A, Srikumari C R, Lorenz B, Finckh U, Nicoletti A, Murthy K R, Rathmann M, Kumaramanickavel G, Denton M J, Gal A. Nat Genet. 1997;17:194–197. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- 39.Wright A F. Nat Genet. 1997;17:132–134. doi: 10.1038/ng1097-132. [DOI] [PubMed] [Google Scholar]
- 40.Burstedt M S, Sandgren O, Holmgren G, Forsman-Semb K. Invest Ophthalmol Vis Sci. 1999;40:995–1000. [PubMed] [Google Scholar]