Abstract
The nucleotide sequences of the env genes of seven bovine leukemia viruses and the encoded peptide sequence were compared, with the objective of (i) determining the genetic distance separating bovine leukemia virus isolates from different geographical regions, (ii) identifying particular amino acids that contribute to the sequential and conformational epitopes, and (iii) relating such epitopes to their projected position in a three-dimensional model of the structure of the gp51 surface glycoprotein. Two bovine leukemia virus subgroups were clearly identified, a Japanese-American subgroup represented by strains lambda BLV-1, VdM, and FLK-BLV and a European subgroup by strains T15-2, LB285, and LB59. It was possible to identify amino acids that were important in determining three of the epitopes (F, G, and H) recognized by neutralizing monoclonal and polyclonal antibodies. On the model, these epitopes were adjacent and located on the exposed region of the molecule. Amino acid sequences contributing to a fourth cryptic epitope were identified; as predicted by the model, they lay on the opposite side to the neutralizable epitopes in a region involved in glycoprotein subunit association. The fact that this region is not normally exposed on the virion surface provides further evidence for the validity of the model.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bova C. A., Olsen J. C., Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988 Jan;62(1):75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
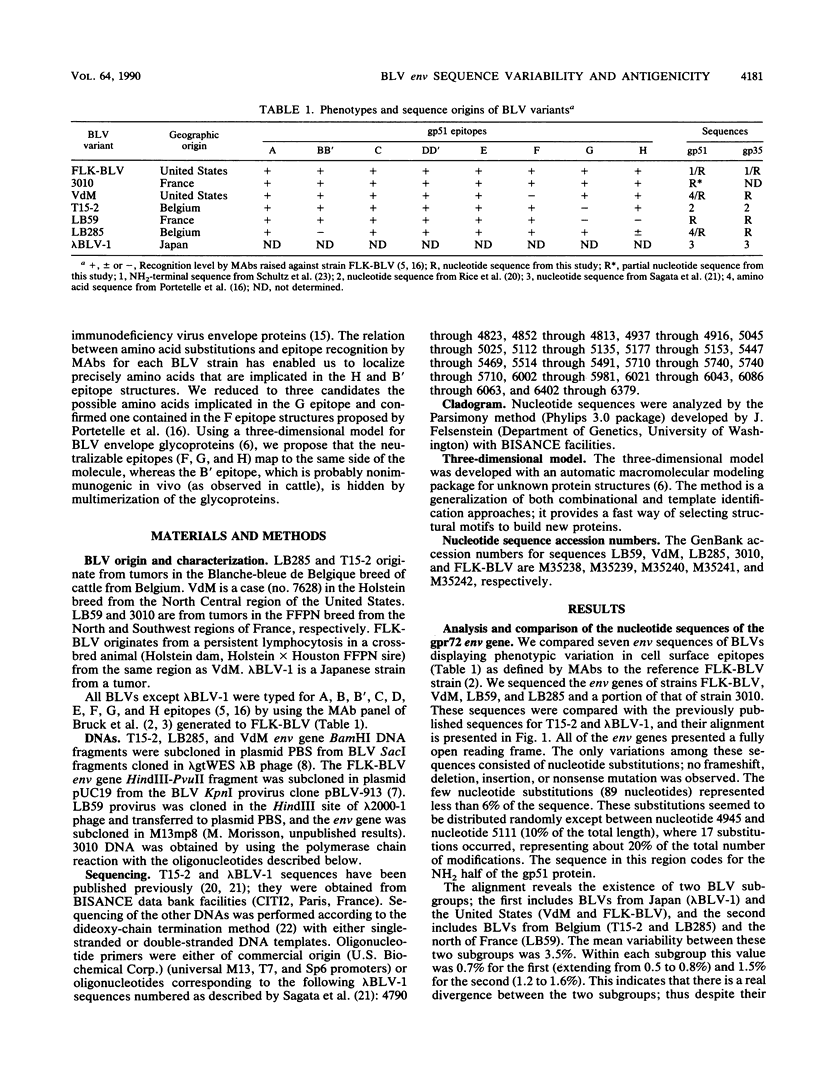
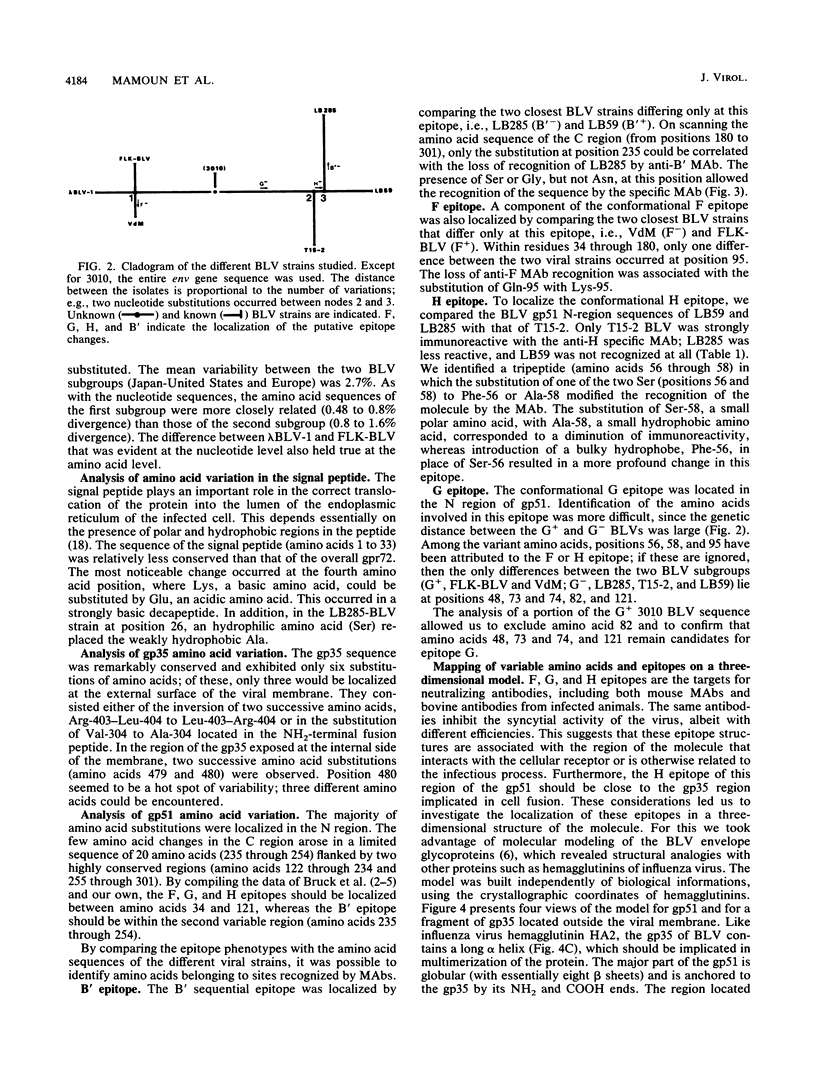
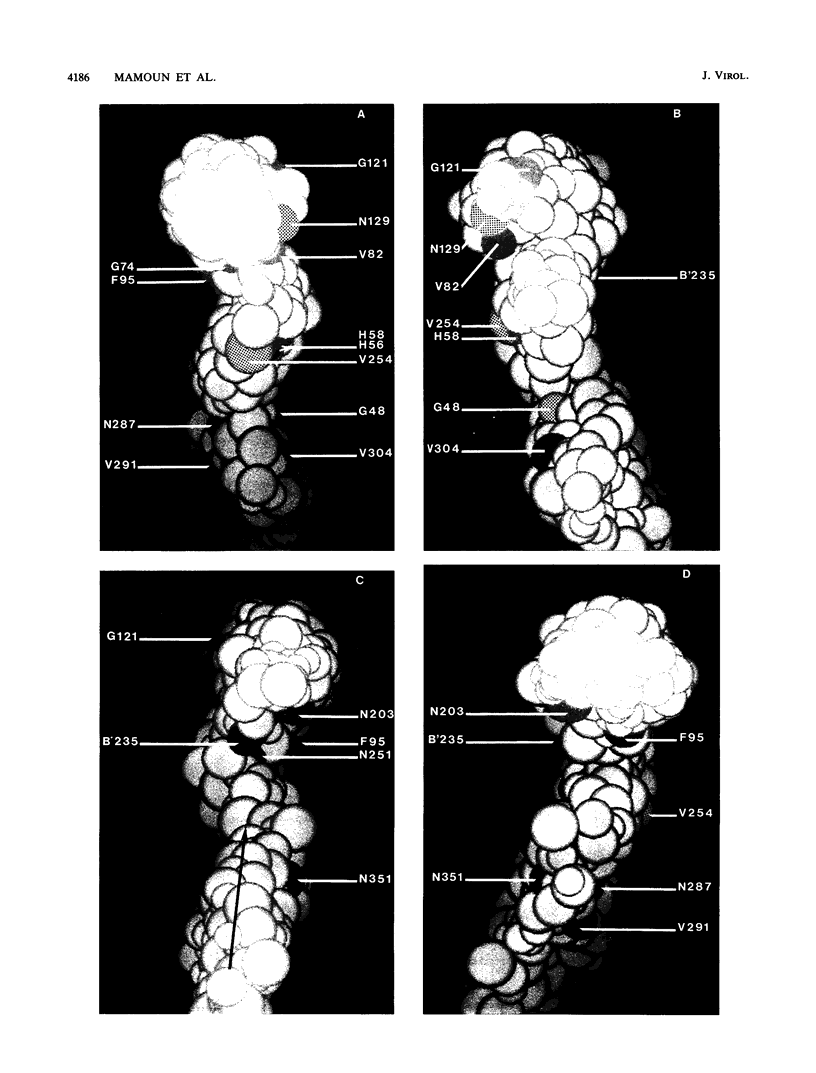
- Bruck C., Mathot S., Portetelle D., Berte C., Franssen J. D., Herion P., Burny A. Monoclonal antibodies define eight independent antigenic regions on the bovine leukemia virus (BLV) envelope glycoprotein gp51. Virology. 1982 Oct 30;122(2):342–352. doi: 10.1016/0042-6822(82)90234-3. [DOI] [PubMed] [Google Scholar]
- Bruck C., Portetelle D., Burny A., Zavada J. Topographical analysis by monoclonal antibodies of BLV-gp51 epitopes involved in viral functions. Virology. 1982 Oct 30;122(2):353–362. doi: 10.1016/0042-6822(82)90235-5. [DOI] [PubMed] [Google Scholar]
- Bruck C., Portetelle D., Mammerickx M., Mathot S., Burny A. Epitopes of bovine leukemia virus glycoprotein gp51 recognized by sera of infected cattle and sheep. Leuk Res. 1984;8(3):315–321. doi: 10.1016/0145-2126(84)90070-5. [DOI] [PubMed] [Google Scholar]
- Bruck C., Rensonnet N., Portetelle D., Cleuter Y., Mammerickx M., Burny A., Mamoun R., Guillemain B., van der Maaten M. J., Ghysdael J. Biologically active epitopes of bovine leukemia virus glycoprotein gp51: their dependence on protein glycosylation and genetic variability. Virology. 1984 Jul 15;136(1):20–31. doi: 10.1016/0042-6822(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Busetta B. The use of folding patterns in a multisolution approach of the all-beta protein three-dimensional structure. A beta-roll structure is predicted in the retroviral glycoprotein. Biochim Biophys Acta. 1989 Oct 19;998(3):301–309. doi: 10.1016/0167-4838(89)90289-6. [DOI] [PubMed] [Google Scholar]
- Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987 Aug;61(8):2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J., Kettmann R., Burny A. Experiments with cloned complete tumor-derived bovine leukemia virus information prove that the virus is totally exogenous to its target animal species. J Virol. 1981 Nov;40(2):605–609. doi: 10.1128/jvi.40.2.605-609.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B., Kaaden O. R., Frenzel B. Subunit and fine structure of the glycoprotein of bovine leukemia virus. Ann Rech Vet. 1978;9(4):613–617. [PubMed] [Google Scholar]
- Ghysdael J., Kettmann R., Burny A. Translation of bovine leukemia virus virion RNAs in heterologous protein-synthesizing systems. J Virol. 1979 Mar;29(3):1087–1098. doi: 10.1128/jvi.29.3.1087-1098.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. V., Berry B. T., Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989 May;63(5):2379–2384. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik K. T., Even J., Karpas A. Molecular cloning and complete nucleotide sequence of an adult T cell leukaemia virus/human T cell leukaemia virus type I (ATLV/HTLV-I) isolate of Caribbean origin: relationship to other members of the ATLV/HTLV-I subgroup. J Gen Virol. 1988 Jul;69(Pt 7):1695–1710. doi: 10.1099/0022-1317-69-7-1695. [DOI] [PubMed] [Google Scholar]
- Mamoun R. Z., Astier T., Guillemain B., Duplan J. F. Bovine lymphosarcoma: processing of bovine leukaemia virus-coded proteins. J Gen Virol. 1983 Dec;64(Pt 12):2791–2795. doi: 10.1099/0022-1317-64-12-2791. [DOI] [PubMed] [Google Scholar]
- Mamoun R. Z., Astier T., Guillemain B. Establishment and propagation of a bovine leukaemia virus-producing cell line derived from the leukocyte of a leukaemic cow. J Gen Virol. 1981 Jun;54(Pt 2):357–365. doi: 10.1099/0022-1317-54-2-357. [DOI] [PubMed] [Google Scholar]
- Portetelle D., Couez D., Bruck C., Kettmann R., Mammerickx M., Van der Maaten M., Brasseur R., Burny A. Antigenic variants of bovine leukemia virus (BLV) are defined by amino acid substitutions in the NH2 part of the envelope glycoprotein gp51. Virology. 1989 Mar;169(1):27–33. doi: 10.1016/0042-6822(89)90037-8. [DOI] [PubMed] [Google Scholar]
- Portetelle D., Dandoy C., Burny A., Zavada J., Siakkou H., Gras-Masse H., Drobecq H., Tartar A. Synthetic peptides approach to identification of epitopes on bovine leukemia virus envelope glycoprotein gp51. Virology. 1989 Mar;169(1):34–41. doi: 10.1016/0042-6822(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Unity in function in the absence of consensus in sequence: role of leader peptides in export. Science. 1989 Mar 3;243(4895):1156–1159. doi: 10.1126/science.2646712. [DOI] [PubMed] [Google Scholar]
- Rassart E., Nelbach L., Jolicoeur P. Cas-Br-E murine leukemia virus: sequencing of the paralytogenic region of its genome and derivation of specific probes to study its origin and the structure of its recombinant genomes in leukemic tissues. J Virol. 1986 Dec;60(3):910–919. doi: 10.1128/jvi.60.3.910-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice N. R., Stephens R. M., Couez D., Deschamps J., Kettmann R., Burny A., Gilden R. V. The nucleotide sequence of the env gene and post-env region of bovine leukemia virus. Virology. 1984 Oct 15;138(1):82–93. doi: 10.1016/0042-6822(84)90149-1. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schultz A. M., Copeland T. D., Oroszlan S. The envelope proteins of bovine leukemia virus: purification and sequence analysis. Virology. 1984 Jun;135(2):417–427. doi: 10.1016/0042-6822(84)90197-1. [DOI] [PubMed] [Google Scholar]
- Szurek P. F., Yuen P. H., Jerzy R., Wong P. K. Identification of point mutations in the envelope gene of Moloney murine leukemia virus TB temperature-sensitive paralytogenic mutant ts1: molecular determinants for neurovirulence. J Virol. 1988 Jan;62(1):357–360. doi: 10.1128/jvi.62.1.357-360.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]