Abstract
The structural dynamics of the DNA binding domains of the human papillomavirus strain 16 and the bovine papillomavirus strain 1, complexed with their DNA targets, has been investigated by modeling, molecular dynamics simulations, and nuclear magnetic resonance analysis. The simulations underline different dynamical features of the protein scaffolds and a different mechanical interaction of the two proteins with DNA. The two protein structures, although very similar, show differences in the relative mobility of secondary structure elements. Protein structural analyses, principal component analysis, and geometrical and energetic DNA analyses indicate that the two transcription factors utilize a different strategy in DNA recognition and deformation. Results show that the protein indirect DNA readout is not only addressable to the DNA molecule flexibility but it is finely tuned by the mechanical and dynamical properties of the protein scaffold involved in the interaction.
INTRODUCTION
Papillomaviruses are small double-stranded DNA viruses that infect both mucosal and cutaneous epithelial tissues. High-risk human papillomavirus (HPV-1) is strongly associated with the development of malignant lesions and promote cervical cancer in >95% of cases (1). HPV-16 and HPV-18 are the most common types in invasive cervical squamous cell carcinomas, accounting for >65% of these cancers. The E2 proteins regulate expression of all viral genes (2) and viral replication through association with the E1 helicase (3).
The structure of the E2 protein consists of three domains: the well-conserved N-terminal trans-activation domain, a variable intermediate hinge region, and a C-terminal DNA-binding/dimerization domain (4). The crystal and nuclear magnetic resonance (NMR) structures of the bovine papillomavirus type 1 (BPV-1) E2 DNA binding domain alone (5,6), and in complex with an oligonucleotide (7), have been solved.
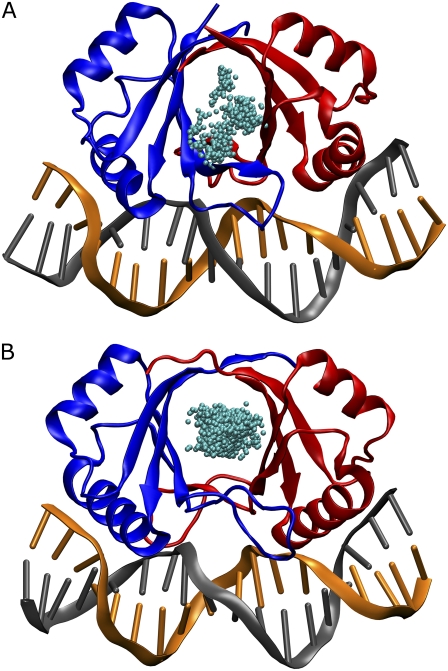
For the type 16 of human papillomavirus, the structure of the E2 DNA binding domain (HPV-16) alone (8,9) and in complex with DNA (10) is available. These structures reveal that the protein forms a dimeric β-barrel with surface recognition α-helices (Fig. 1).
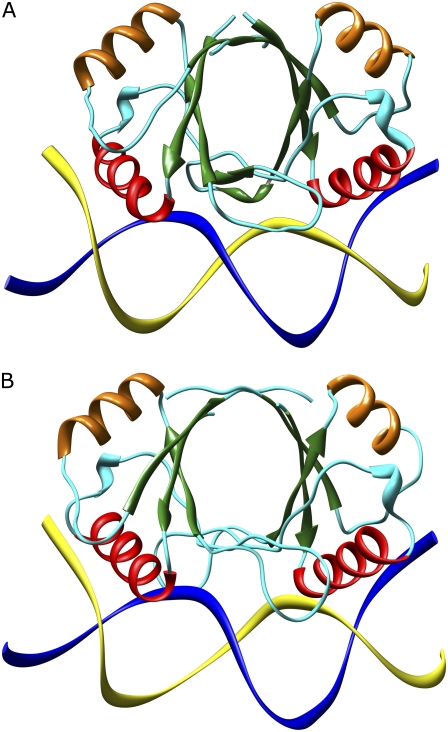
FIGURE 1.
Side view of the HPV-16-DNA complex (A) and BPV-1-DNA complex (B). The α-helices involved in the DNA recognition are shown in red while the other α-helices are represented in orange. The β-strands are shown by green arrows. The cyan wire indicates the random-coil structure and the turns. The DNA strands are shown as blue and yellow ribbons. The picture was produced using the Chimera program (43).
Although the tertiary structure of all characterized E2 DNA binding domain is similar, there is an interesting variation in the relative orientation of the two subunits (2). On this basis, the E2 proteins are divided into two distinct classes, one including HPV-16 and HPV-31 and the other BPV-1 and HPV-18 (2). The differences in quaternary structure are likely to induce a different DNA deformation upon E2 binding.
The transcriptional regulation, growth inhibition, and replication functions of E2 are mediated through its interaction with a palindromic consensus sequence ACCgN4cGGT, where N4 indicates the spacer nucleotides and small letters represent preferred but not totally conserved nucleotides. Multiple E2 binding-sites that differ in the sequences of the central N4 spacer nucleotides are present in the viral genomes (17 in BPV-1 and 4 in HPV-16) (8). The structure of the spacer region, which is not contacted by the protein, is critical for the formation of the high-affinity sequence-specific protein-DNA complex, and the differential binding affinity has been proposed to be regulated by the intrinsic structure and deformability encoded in the base sequence of the DNA target (11). The two proteins display differential affinity also toward binding-sites possessing nicked or gapped spacers, indicating distinct differences in their sensitivity to DNA structure and/or flexibility (11). Despite these differences, the residues involved in direct base interactions are identical (11), indicating that direct interaction is not the mechanism discriminating the DNA binding-site sequence. As a matter of fact, the specificity of papillomavirus E2 protein-DNA binding depends critically upon the sequence of a region of the DNA not in direct contact with the protein, and represents one of the best known examples of indirect readout (12).
Molecular dynamics (MD) simulations have been extensively carried out to characterize this system and test the hypothesis of a structural code for DNA recognition by these regulatory proteins.
Conformational properties of the E2 DNA oligomers in absence of protein, containing different spacers, have been investigated through molecular dynamics simulations (13,14). Other MD studies, carried out on the BPV-1-DNA complexes and compared with free DNA simulations, have indicated that despite a severe slowing-down of motions, the DNA geometrical parameters are preserved (15). A recent simulation of HPV-16 and BPV-1 E2, carried out in the absence of DNA, have identified different dynamical features in the two proteins (16). The HPV-16 E2 has a higher flexibility on the recognition helices but also a higher compactness of the β-barrel when compared with the BPV-1 E2 domain. Consequently, it has been proposed that in HPV-16 protein deformation is prevented by a rigid β-barrel and deformable spacers are the preferred targets in the complex formation, while in BPV-1 a more deformable β-barrel confers a larger adaptability to the protein, allowing the binding of less flexible DNA regions (16).
In this work, we have investigated, through MD simulation, the structural-dynamical properties of the HPV-16 and BPV-1 E2 proteins bound to DNA. The results show that the E2 from different species with a sequence identity of ∼30%, having approximately the same secondary and tertiary structure, show a different distribution of molecular flexibility. The mechanical properties that characterize the two proteins, together with the different structural and conformational characteristics of the spacer regions in the DNA target sequences, confer a diverse strategy for the DNA recognition and deformation. This feature provides to the protein the ability to discriminate between spacer sequences, since the formation of the various E2-DNA complexes is not only based on the rigidity of the base sequences in the DNA spacers, but also on the intrinsic deformability properties of each E2 DNA binding domain.
MATERIALS AND METHODS
Starting structures
The HPV-16 (10) and BPV-1 (7) E2 binding domain coordinates have been obtained by nuclear magnetic resonance (NMR) and stored in the Protein Data Bank (PDB www.rcsb.org/pdb) with the PDB entry codes 1ZZF and 2BOP, respectively.
HPV-16-DNA complex modeling procedure
The HPV-16 PDB file 1ZZF does not include the DNA coordinates. The HPV-16-DNA complex has been generated by using the structure of HPV-16 protein (PDB file 1ZZF), solved in the presence of the target DNA (10), and the DNA molecule included in the NMR HPV-18-DNA complex (PDB file 1JJ4) (17). As a first step, the corresponding Cα atoms of the three-dimensional structures of HPV18-E2 and HPV-16-E2 have been identified and superimposed using the program Swiss PDB Viewer (18) (http://www.expasy.org/spdbv/). The DNA atoms of the HPV18-DNA complex have been then merged with the HPV-16 protein coordinates upon elimination of the HPV-18 protein coordinates. After the DNA merging, the HPV16-DNA complex model was found to display just some contacts that were eliminated by the relaxation procedure.
Both the DNA molecules contained in the PDB files 2BOP and 1JJ4 have been optimized with the InsightII program by Accelrys (www.accelrys.com/products/insight/).
In detail, in the HPV-18 DNA (PDB file 1JJ4), the 5′ phosphates are substituted by hydroxyl groups:
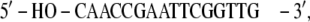 |
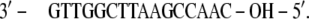 |
The hydroxyl groups have been substituted with phosphates and the DNA strands have been completed by adding a GC basepair (underlined) to each extremity:
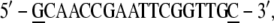 |
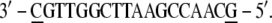 |
In the BPV-1 DNA (PDB file 2BOP), the extremities show unpaired bases,
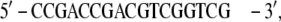 |
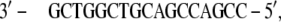 |
and a G base (underlined) has been added to each extremity to complete the strands:
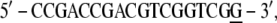 |
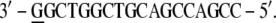 |
The structures of E2-DNA complexes have been relaxed as described in the next paragraph.
Molecular dynamics simulations and analysis
Two simulations of 15 ns have been carried out on the HPV-16 and BPV-1 complexes. The systems topologies have been obtained with the AMBER Leap module (19), and modeled with the all-atoms AMBER95 force field (20,21). The proteins have been immersed in rectangular boxes filled with TIP3P water molecules (22) (Table 1), imposing a minimal distance between the solute and the box walls of 10.0 Å. The two systems have been neutralized through the AMBER Leap module, adding the necessary amount of Na+ ions (Table 1) in electrostatic favorable positions. Optimization and relaxation of solvent and ions were initially performed by means of four energy minimizations and six molecular dynamics simulations (Table 2), keeping the solute atoms constrained to their initial positions with decreasing force constants of 500, 50, 25, and 10 Kcal/(mol Å). Thereafter the systems were simulated without any constraint for 40.0 ps at constant temperature of 300 K using Berendsen's method (23) and at a constant pressure of 1 bar with a 2.0 fs time step. After this procedure, each system has been simulated for 15 ns. Pressure and temperature coupling constants were 0.4 ps. The atomic positions were saved every 250 steps (0.5 ps) for the analysis. The two systems have been simulated in periodic boundary conditions, using a cutoff radius of 9.0 Å for the nonbonded interactions, and updating the neighbor pair list every 10 steps. The electrostatic interactions were calculated with the particle-mesh Ewald method (24,25). The SHAKE algorithm (26) was used to constrain all bond lengths involving hydrogen atoms.
TABLE 1.
Size of the simulated systems
| Protein type | BPV-1 | HPV-16 |
|---|---|---|
| Total atoms | 29,993 | 31,029 |
| Protein atoms | 2674 | 2664 |
| Amino acids | 170 | 162 |
| DNA atoms | 1072 | 1143 |
| Bases | 36 | 36 |
| Water molecules | 8718 | 8661 |
| Na+ ions | 25 | 17 |
| Simulation box side X (Å) | 90 | 97 |
| Simulation box side Y (Å) | 96 | 92 |
| Simulation box side Z (Å) | 103 | 100 |
| Saved configurations | 30,000 | 30,000 |
TABLE 2.
Systems thermalization phases
| Time (ps) | Thermalization process | Number of steps and ΔT | Position restraint value (Kcal/mol × Å) |
|---|---|---|---|
| 0 | EM1 | 10,000 | 500 |
| 0 | EM2 | 20,000 | 500 |
| 12.5 | MD1 | 25,000 of 0.5 fs | 500 |
| 0 | EM3 | 15,000 | 50 |
| 25.0 | MD2 | 25,000 of 1.0 fs | 50 |
| 0 | EM4 | 10,000 | 25 |
| 20.0 | MD3 | 10,000 of 2.0 fs | 25 |
| 40.0 | MD4 | 20,000 of 2.0 fs | 10 |
| 40.0 | MD5 | 20,000 of 2.0 fs | 10 |
| 40.0 | MD6 | 20,000 of 2.0 fs | — |
EM, energy minimization; MD, molecular dynamics.
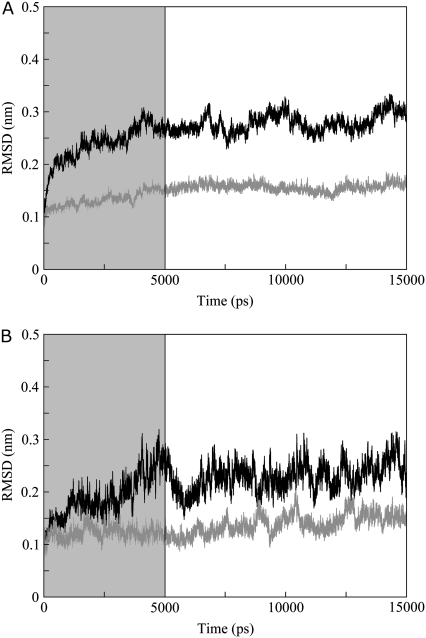
The root mean-square deviation (RMSD) from starting structures of the E2 proteins and their relative DNAs (Fig. 2, parts A and B, respectively) shows a good stability after the first 5 ns of simulation that have been excluded from the analysis.
FIGURE 2.
All atoms RMSD from starting structures of (A) HPV-16 (solid line) and BPV-1 (shaded line) E2 proteins; (B) HPV-16 DNA (solid line) and BPV-1 DNA (shaded line). The shaded box indicates the trajectory fraction not considered in the analysis.
The calculations have been carried out at CASPUR research center of Rome, Italy (Inter Universities Consortium for Supercomputing Applications) on Power 4 IBM parallel computers by using an eight-CPU cluster. The trajectory and principal component analyses for both systems have been carried out using the GROMACS MD package version 3.2.1 program (27) and in housewritten codes. The hydrogen bonds have been analyzed through the g_hbond GROMACS module (27) that has been modified to list the involved atoms. The residue root mean-square fluctuations (RMSFs) have been compared to the residue temperature factor (B) obtained from x-ray diffraction using
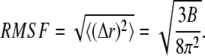 |
(1) |
Volume of the cavities and external crevices was measured using the program SURFNET (28). In this program, full internal cavities, and crevices communicating with the outside, are defined by first filling the internal volumes of the molecule with gap-spheres and then using these to compute a three-dimensional density map which, when contoured, defines the surface of the cavity (28).
DNA curvature and geometrical parameters have been calculated using the program CURVES (29) using the second and penultimate basepair as reference, while the basepairs deformation energies have been computed through the EnergyPDNA program (http://3dna.rutgers.edu/x3dna/user_corner) that uses geometrical parameters calculated by 3DNA (30).
NMR spectroscopy
Uniformly 15N-13C-labeled HPV-16 E2 DNA-binding domain was expressed and purified as previously described (31). The concentration of the DNA-free protein in buffer solution (50 mM sodium phosphate, 5 mM DTT, pH 6.5) was 0.6 mM. The concentration of the DNA-bound protein in buffer solution (50 mM sodium phosphate, 250 mM NaCl, 5 mM DTT, pH 5.6) was 0.9 mM.
The Cα dynamics has been evaluated measuring the crosspeak intensity from the constant time HACACO experiment (32–34). NMR experiments were performed at 30°C for the DNA-free protein and at 45° for the DNA-bound protein on an Avance700 spectrometer (Bruker, Fitchburg, WI) equipped with triple resonance probe incorporating self-shielded gradient coils. NMR relaxation analysis has been used to analyze the mobility of the HPV-16 E2 protein measuring the relaxation properties of the Cα nuclei. The spectral width was 9124 Hz in F3 (1H), 5282 Hz in F2 (Cα) and 2112 Hz in F1 (CO). The carrier frequency was placed at 4.7 ppm in the proton dimension, at 55.5 ppm in the Cα dimension and at 176 ppm in the CO dimension. The experiment was acquired using States-TPPI scheme in F1 and Echo-Antiecho mode in F2, collecting 2048 complex points in F3, 292 points in F2 and 44 complex points in F1. NMR data were processed on Silicon Graphics workstations using NMRPipe and analyzed using NMRView.
RESULTS
Analysis of root mean-square deviations and fluctuations
The all-atoms root mean-square deviations (RMSDs) from the starting structures for the two E2 proteins and for the DNA molecules are reported as a function of time in Fig. 2, parts A and B, respectively.
In both the trajectories the RMSDs reach a stable value for proteins and DNAs after 2–3 ns; however, to guarantee investigation over a well-thermalized system, all the analyses have been carried out discarding the first five nanoseconds, i.e., over the last 10 nanoseconds.
The average RMSD values in the last 10 nanoseconds are 0.26 nm in HPV-16 E2, 0.21 nm in HPV-16 DNA, 0.14 nm in BPV-1 E2, and 0.13 nm in BPV-1 DNA. The full stability of both systems is also guaranteed by the time evolution of: number of residues in α-helix, 3-10 helix, β-strand, and random coil protein secondary structures (see Supplementary Material, Fig. S1, A and B in Data S1); total solvent-accessible surface area (Fig. S1, C and D in Data S1); gyration radius (Fig. S1, E and F in Data S1); and number of hydrogen bonds (Fig. S1, G and H in Data S1).
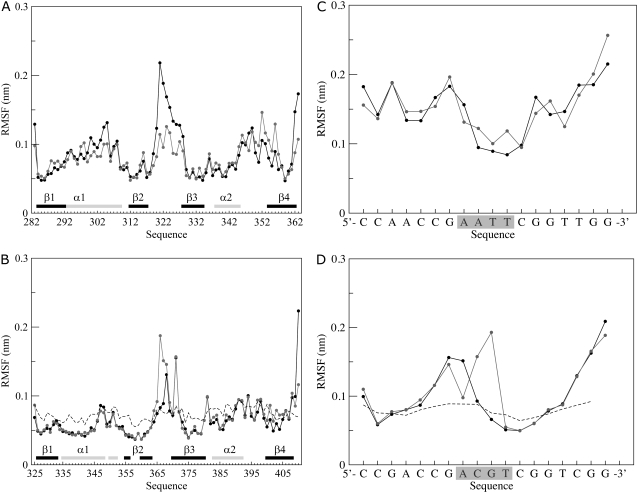
Fig. 3 shows the backbone root mean-square fluctuations (RMSFs), calculated over the trajectories and averaged over each residue or nucleotide, for both the HPV-16 and BPV-1 E2 proteins (Fig. 3, A and B) and their DNA targets (Fig. 3, C and D). The largest fluctuations are observed at the level of the loops connecting strands β2 and β3, being slightly higher in HPV-16 than in BPV-1. In the preceding simulations, in absence of DNA, these fluctuations are larger (16), indicating that the DNA binding induces a partial stabilizing effect on these loops. In line, this region lying in front of the spacer sequence show weak electron density in the crystal structures of unbound E2, suggesting a substantial degree of flexibility (8,17). Previous NMR structural analysis has shown that the β2-β3 loop remains both solvent-exposed and quite flexible in the HPV-16-DNA complex (10), while in the BPV-1-DNA complex the flexibility is reduced (7,17). In line, in the MD simulations the β2-β3 loops of BPV-1 show, on the whole, fluctuations lower than the corresponding HPV-16 loops (Fig. 3). BPV-1 and HPV-16 shows differences at the level of α1-helix: in BPV-1, the α1-helix displays very low RMSF values, comparable to those of the α2-helix (Fig. 3 B), while in HPV-16, helix α1 shows a relative high RMSF value, higher than that of the α2-helix or of the β-strands of both proteins.
FIGURE 3.
Average per-residue backbone RMSF for each subunit of (A) HPV-16 and (B) BPV-1 E2 proteins. The RMSF values are represented by solid and shaded circles for the first and second subunit, respectively. The secondary structure regions in the starting structure are shown by the solid (α-helix) and shaded (β-strand) horizontal bars on the x axis. In panel B, the dotted line shows the corresponding experimental B-factors converted to RMSF values for comparison. Average per nucleotide backbone RMSF for each of the two strands of HPV-16 DNA (C) and BPV-1 DNA (D) complexes. One strand is shown by solid circles and the other one by shaded circles. In panel D, the dotted line shows the corresponding experimental B-factors converted to RMSF values. The shaded box encloses the spacer region nucleotides.
For the BPV-1 protein, the residue RMSF values well reproduce the crystallographic B-factors (7) (Fig. 3 B). This is strictly true for the loops between regular secondary structures segments while the α-helices and the β-strands surprisingly have, in the simulations, fluctuations slightly lower than the corresponding converted B-factors. This is likely due to a relaxation of the protein over the DNA target that permits the occurrence of a high number of protein-DNA hydrogen bonds (Fig. S1, G and H in Data S1).
The RMSF values, averaged over each nucleotide of the DNA strands bound to HPV-16 and BPV-1 DNA targets (Fig. 3, parts C and D, respectively), shows that the DNA interacting with HPV-16 has large fluctuations in the major grooves region and low fluctuations in the spacer region (Fig. 3 C). On the contrary, the BPV-1 DNA shows large fluctuations in the spacer region and low fluctuations in the major grooves region (Fig. 3 D) indicating that the two proteins induce specific deformation in different sections of their DNA binding sites. Also, in this case the converted crystallographic B-factors (7) have a good agreement with the RMSF MD values (Fig. 3 D).
Hydrogen bonds and secondary structure analysis
Tables 3 and 4 list the direct hydrogen bonds, present for >50% of the trajectory time, that show a very good agreement with the hydrogen bonds found in the x-ray (7,17), and NMR (10) structures, used to carry out the simulations.
TABLE 3.
Direct hydrogen bonds between HPV-16 E2 and its DNA target
| Residue identity | Secondary structure location | Identity of most contacted bases | Protein subunit and % of residue occurrence | Hydrogen-bond number |
|---|---|---|---|---|
| Asn294 | α1-helix | A(−6) | A.69 - B.53 | 2 |
| A(−7) | ||||
| Lys297 | α1-helix | G(+4) | A.85 - B.86 | 2 |
| G(+5) | ||||
| Arg300 | α1-helix | C(+3) | A.85 - B.90 | 2 |
| G(+4) | ||||
| Arg302 | α1-helix | C(−8) | A.84 - B.87 | 2 |
| C(−9) | ||||
| A(−7) | ||||
| Lys304 | α1-helix | G(+5) | B.50 | 1 |
| Thr316 | β2-β3 loop | C(+3) | A.100 - B.97 | 2 |
| His318 | β2-β3 loop | T(+2) | A.83 | 1 |
Residues making a direct hydrogen bond in the MD simulation with an occurrence ≥50% of trajectory time.
TABLE 4.
Direct hydrogen bonds between BPV-1 E2 and its DNA target
| Residue identity | Secondary structure location | Identity of most contacted bases | Protein subunit and % of residue occurrence | Hydrogen-bond number |
|---|---|---|---|---|
| Asn336 | α1-helix | A(−6) | A.100 - B.100 | 2 |
| C(−5) | ||||
| Gln337 | α1-helix | G(−7) | A.98 - B.98 | 2 |
| Lys339 | α1-helix | G(+4) | A.96 - B.91 | 2 |
| G(−5) | ||||
| Cys340 | α1-helix | A(−6) | A.91 - B.94 | 2 |
| Arg342 | α1-helix | C(+3) | A.100 - B.100 | 2 |
| G(+4) | ||||
| Arg344 | α1-helix | C(−8) | A.100 - B.100 | 2 |
| G(−7) | ||||
| Tyr359 | β2-β3 loop | C(+3) | A.100 - B.100 | 2 |
| Arg370 | β2-β3 loop | C(+3) | A.100 - B.100 | 2 |
| G(+4) |
Residues making a direct hydrogen bond in the MD simulation with an occurrence ≥50% of trajectory time.
The HPV-16 MD simulation detects 12 (Table 3) over the 26 direct hydrogen bonds observed in the analogous residues of the x-ray structure of the HPV-18 complex (17).
The NMR structural analysis indicates a relevant role in the DNA binding for the residues between Asn294 and Tyr301 (10) confirmed by a rational site-directed mutagenesis study (35), and for Thr316, located in a bulge of the β2 strand, that forms a strong hydrogen bond with DNA (10). In line in the simulation, Asn294, Lys297, Arg300, Arg302, and Thr316 display a hydrogen-bond interaction for a long percentage of time (Table 3). Mutation into alanine of Lys349, a charged residue located outside the recognition helix, shows a chemical shift perturbation when a 18-mer DNA is used for the interaction, indicating the presence of an additional contact between the HPV-16 protein and the DNA located outside the recognition helix and the β2-β3 loop (10). The HPV-16 simulation, involving a DNA 18-mer, shows that Lys349 forms a hydrogen bond with the DNA for ∼40% of the trajectory time, although only in one protein subunit.
In the case of BPV-1 all the direct hydrogen bonds found in the crystal structure of BPV-1 E2-DNA complex (7) are detected for all the simulation trajectory (Table 4), indicating they are very stable being present in all the conformations sampled by the protein DNA complex.
The secondary structure analysis, carried out on both the proteins for all the simulation time indicates that, in the HPV-16 E2, some residues belonging to the α-helices and, in particular, to the C-terminus of the DNA recognition helix α1 (2), lose their secondary structure reaching alternative conformations while the β-segments are stable along the trajectory (Fig. S2 A in Data S1). The opposite is observed in the BPV-1 E2 protein where all the α-helices, including the helices α1, are very stable, while the β-barrel is, on the whole, more flexible (Fig. S2 B in Data S1).
Cα dynamics from HACACO intensity measurement
Experimental evidence for the backbone mobility in the HPV-16 E2 protein was obtained by NMR relaxation analysis, measuring the relaxation properties of the Cα nuclei. In this system, the use of Cα, instead of the 15N nuclei as a magnetic probe, provides a way to study a larger number of residues belonging to the β2-β3 loop, where the amide nitrogens are not present in the 1H-15N HSQC measurements, due to rapid water exchange. The Cα dynamics has been monitored measuring the crosspeak intensity from the constant time HACACO experiment (32,33), as already applied to the ApaG protein (34) and to a lipid-acid complex of the chicken-liver-bile acid binding protein (35). In this kind of experiment, residues, showing mobility in the ns-ps timescale, have crosspeak intensity larger than the average, whereas residues showing internal mobility in the μs-ms timescale show a reduced intensity.
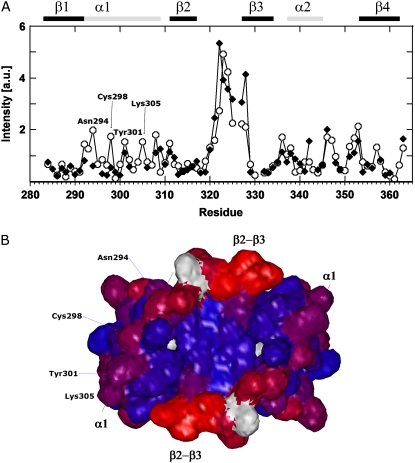
The normalized intensities for the free and complexed forms of HPV-16 E2 are depicted in Fig. 4 A.
FIGURE 4.
(A) Normalized crosspeak intensities observed in the constant time HACACO NMR measurements as a function of the residue number. Open circles and solid diamonds represent the values for HPV-16 E2 in absence and presence of DNA, respectively. The residues of the recognition helix α1 that show a significant enhanced crosspeak intensity are labeled. (B) The DNA facing surface of HPV-16 E2 protein. Red and blue indicate the residues having the highest and the lowest crosspeak intensity, respectively. The residues Asn294, Cys298, Tyr302, and Lys305 belonging to the α1-helix and important for the DNA binding, are labeled.
High intensity are observed in the two forms for residues belonging to the β2-β3 loop, confirming the high flexibility detected from the RMSF analysis (Fig. 3 A). This graph shows that for the protein alone, some residues of the recognition helix, Asp292, Ala293, Asn294, Cys298, Tyr301, and Lys305, show intensities larger than the average. This behavior is attenuated upon DNA binding. The NMR analysis, as also observed by the simulation (Fig. S2 A in Data S1), indicates that the α1 recognition helix in the HPV-16 complex presents two regions: the N-terminus (residues 296–300), having an α-helix conformation stabilized upon DNA binding; and the C-terminus (residues 301–309), which shows deviations from the α-helical character (10) (Fig. 4 A). Both MD and NMR points to a conformational adaptability of the HPV-16 E2 α1-helices, as already proposed in the simulations carried out in the absence of the target DNA (16), likely to permit an optimized fit into the DNA major groove recognition site. Fig. 4 B displays the relative position of the residues belonging to the helix, directly facing the DNA, showing the surface of the protein exposed to the DNA interaction. Mutation of these surface residues (Asn294, Cys298, Tyr301, and Lys305) into alanine causes a considerable ΔΔG variation of the DNA binding (36).
Principal component analysis
The principal component analysis (PCA), or essential dynamics (37,38), has been applied to both the HPV-16-DNA and BPV-1-DNA complex trajectory to identify the main 3N directions (eigenvectors) along which the majority of the complex motion is defined. The analysis is based on the diagonalization of the covariance matrix built from the atomic fluctuations after the removal of the translational and rotational movement, and permits the identification of the main 3N directions along which the majority of the motion is defined. The analysis, carried out on the Cα atoms of the two proteins plus the phosphorus atoms of the DNA backbone, indicates that, although the motion is dispersed over >600 eigenvectors, ∼80% (HPV-16), and 70% (BPV-1) of the motion depends on the first 30 eigenvectors (Fig. S3 in Data S1) as usually found for many different systems (39,40). The convergence of the simulations has been probed by the cosine content of the first principal component according to the Hess method (41). The obtained values are 0.03 for the HPV-16 and 0.07 for the BPV-1 protein indicating a good simulation convergence.
Dynamical differences between the HPV-16 and BPV-1 E2 proteins and their DNA targets can be appreciated looking at the Cα and phosphorus atoms projections of the MD motions along the first eigenvector, which contain ∼13% (BPV-1) and 27% (HPV-16) of the total motion (Fig. S3 in Data S1). The projections of the motion are shown in Fig. 5, A and B, where the width of the ribbon indicates the amplitude of the motion, the direction going from the blue to the red color. In HPV-16 E2 (Fig. 5 A), the β-barrel undergoes a rotation over an axis perpendicular to the double helix axis and asymmetrically deforms the DNA geometry mainly in the major groove regions through the α1 recognition helices (Movie S1). In BPV-1 (Fig. 5 B), the β-barrel undergoes a distortion, involving the inter-subunits strands β4, and through the β2-β3 loops alters the DNA geometry mainly on one strand of the spacer region (Movie S2).
FIGURE 5.
Tube representation of the motion projections along the first eigenvector for HPV-16 (A) and BPV-1 (B) E2 proteins. The direction of the motion is indicated by the flanked tubes, the versus being defined from the blue to the red color. The picture was produced using the program Chimera (43).
The shift in the hydrogen-bond register at the inter-monomer β4-β4 sheet, which generates a significant change in the relative orientation of the two recognition helices in the HPV-16 and the BPV-1 strains (11,17), may also modulate the different β-barrel motions detected by the PCA analysis.
Cavities analysis
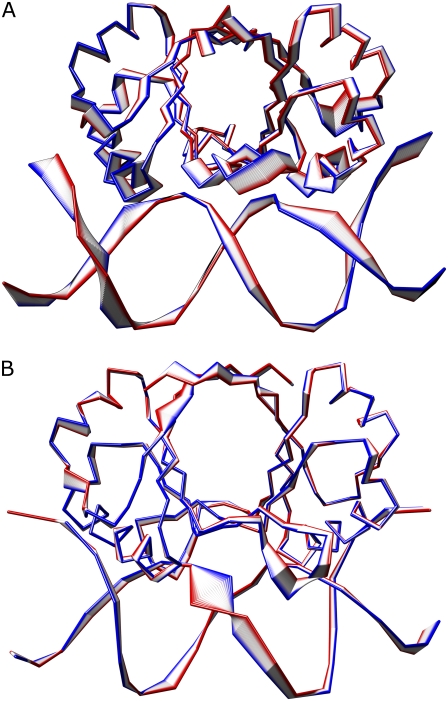
The total number of cavities, that occur along the entire trajectory in the β-barrel of the two E2 proteins, has been evaluated applying the program SURFNET (28) and the results are shown in Fig. 6, A and B.
FIGURE 6.
Distribution of the geometric centers of the cavities (cyan spheres) inside the β-barrel of HPV-16 (A) and BPV-1 (B) E2 proteins. The picture was produced by using the program PyMol (44).
HPV-16 shows the presence of ∼500 peripheral cavities, with an average volume of ∼2890 ± 770 Å3, mainly located at the interface with DNA (Fig. 6 A), while BPV-1 shows the presence of ∼18,000 internal cavities, with an average volume of ∼1350 ± 225 Å3, located at the center of the barrel (Fig. 6 B). The geometric center of these cavities is located inside the β-barrel but their volume extends to the exterior, justifying the large value detected by SURFNET (28). This result indicates that the BPV-1 β-barrel is more flexible and water-exposed than the HPV-16 β-barrel. The flexibility of the protein scaffold allows BPV-1 to adapt itself to a large number of DNA spacers, while the HPV-16 protein must select flexible spacers to compensate its low β-barrel adaptability.
DNA analysis
To better analyze the deformations experienced by the DNA targets, induced by the E2 proteins, the geometric and energetic changes of DNA molecules along the trajectories have been monitored and averaged for each basepair over the entire trajectory. The average DNA curvature measured in the two simulations is equivalent (∼50° ± 10°), indicating a DNA bending decrease in HPV-16 if compared with the corresponding x-ray value of the HPV-18 DNA (29°), and a DNA bending increase in BPV-1 if compared with its starting x-ray value (65°).
Results for the X displacement, Y displacement, and roll parameters, diagnostic for double helix regularity in the B-form (42), are shown in Fig. S4, A–F in Data S1, respectively.
In BPV-1 a large X displacement of the spacer region, which is reduced going toward the peripheral regions, is observed (Fig. S4 B in Data S1). On the contrary, a remarkable asymmetrical alteration of the spacer region, as monitored by the Y displacement, is observed in HPV-16 (Fig. S4 C in Data S1). The roll parameter (42), diagnostic of the global helix axis curvature, is altered in the BPV-1 spacer and in its flanking regions (Fig. S4 F in Data S1), but it is deformed in HPV-16 only in the regions flanking the spacer sequence (Fig. S4 E in Data S1).
The DNA deformation energy, i.e., the energy required to alter the basepair geometry starting from the regular B-form, shows values characterized by, on average, low fluctuations in HPV-16 and large fluctuations in the BPV-1 spacer region, indicating a partial destructuration of the DNA double helix (Fig. S5, A and B in Data S1).
DISCUSSION
The results obtained in this work highlight a different mechanical interaction of the two proteins with DNA, indicating that they use a different way of recognition and deformation of their relative target. Both the HPV-16-DNA and BPV-1-DNA complex maintain a good flexibility of the loop connecting strands β2 and β3, as monitored by the RMSFs and, in the case of HPV-16, also by NMR analysis (Fig. 3, A and B, and Fig. 4 A), but HPV-16 displays a more compact β-barrel (Fig. 5, A and B) and more flexible α-helices (Fig. S4 A, Fig. S2 A and B in Data S1). In fact, BPV-1 shows a large β-barrel flexibility, as indicated by the large number of cavities present in its interior (Fig. 6 B), that allow a broad range of movements for the highly structured and stable α1-helices (Fig. S2 B in Data S1), necessary for the nonspecific DNA targets recognition.
The PCA analysis (Fig. 5, A and B), plus the geometrical and energetic DNA analyses (Fig. S4 A–F; and Fig. S5, A and B in Data S1), indicates a different strategy in DNA deformation. HPV-16 alters the DNA geometry deforming asymmetrically the major groove regions contacted by the α1 recognition helices while the loops are only slightly involved in this distortion (Fig. S5 A in Data S1 and Movie S1). BPV-1 deforms the DNA molecule mainly on one strand of the spacer region, the β4 strands and the loops being actively involved in the DNA distortion (Fig. S5 B in Data S1 and Movie S2).
The DNA-protein recognition, in addition to direct interactions (35), relies also on indirect effects such as DNA twisting and bending to better adapt itself to the protein surface (12). In the light of what observed in this work, we suggest that the indirect readout is not only addressable to the DNA molecule flexibility but it is finely tuned by the mechanical and dynamical properties of the protein scaffold involved in the interaction.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Editor: Ruth Nussinov.
References
- 1.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527. [DOI] [PubMed] [Google Scholar]
- 2.Hegde, R. S. 2002. The papillomavirus E2 proteins: structure, function, and biology. Annu. Rev. Biophys. Biomol. Struct. 31:343–360. [DOI] [PubMed] [Google Scholar]
- 3.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giri, I., and M. Yaniv. 1988. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 7:2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegde, R. S., A. F. Wang, S. S. Kim, and M. Schapira. 1998. Subunit rearrangement accompanies sequence-specific DNA-binding by the bovine papillomavirus-1 E2 protein. J. Mol. Biol. 276:797–808. [DOI] [PubMed] [Google Scholar]
- 6.Veeraraghavan, S., C. Mello, E. Androphy, and J. D. Baleja. 1999. Structural correlates for enhanced stability in the E2 DNA-binding domain from bovine papillomavirus. Biochemistry. 38:16115–16124. [DOI] [PubMed] [Google Scholar]
- 7.Hegde, R. S., S. R. Grossman, L. A. Laimins, and P. B. Sigler. 1992. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 359:505–512. [DOI] [PubMed] [Google Scholar]
- 8.Hegde, R. S., and E. J. Androphy. 1998. Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. J. Mol. Biol. 284:1479–1489. [DOI] [PubMed] [Google Scholar]
- 9.Nadra, A. D., T. Eliseo, Y. K. Mok, C. L. Almeida, M. Bycroft, M. Paci, G. de Prat-Gay, and D. O. Cicero. 2004. Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric β-barrel fold. J. Biomol. NMR. 30:211–214. [DOI] [PubMed] [Google Scholar]
- 10.Cicero, D. O., A. D. Nadra, T. Eliseo, M. Dellarole, M. Paci, and G. de Prat-Gay. 2006. Structural and thermodynamic basis for the enhanced transcriptional control by the human papillomavirus strain-16 E2 protein. Biochemistry. 45:6551–6560. [DOI] [PubMed] [Google Scholar]
- 11.Hines, C. S., C. Meghoo, S. Shetty, M. Biburger, M. Brenowitz, and R. S. Hegde. 1998. DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins. J. Mol. Biol. 276:809–818. [DOI] [PubMed] [Google Scholar]
- 12.Gromiha, M. M., J. G. Siebers, S. Selvaraj, H. Kono, and A. Sarai. 2004. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. J. Mol. Biol. 337:285–294. [DOI] [PubMed] [Google Scholar]
- 13.Byun, K. S., and D. L. Beveridge. 2004. Molecular dynamics simulations of papilloma virus E2 DNA sequences: dynamical models for oligonucleotide structures in solution. Biopolymers. 73:369–379. [DOI] [PubMed] [Google Scholar]
- 14.Djuranovic, D., C. Oguey, and B. Hartmann. 2004. The role of DNA structure and dynamics in the recognition of bovine papillomavirus E2 protein target sequences. J. Mol. Biol. 339:785–796. [DOI] [PubMed] [Google Scholar]
- 15.Djuranovic, D., and B. Hartmann. 2005. Molecular dynamics studies on free and bound targets of the bovine papillomavirus type I e2 protein: the protein binding effect on DNA and the recognition mechanism. Biophys. J. 89:2542–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falconi, M., A. Santolamazza, T. Eliseo, G. de Prat-Gay, D. O. Cicero, and A. Desideri. 2007. Molecular dynamics of the DNA-binding domain of the papillomavirus E2 transcriptional regulator uncover differential properties for DNA target accommodation. FEBS J. 274:2385–2395. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S. S., J. K. Tam, A. F. Wang, and R. S. Hegde. 2000. The structural basis of DNA target discrimination by papillomavirus E2 proteins. J. Biol. Chem. 275:31245–31254. [DOI] [PubMed] [Google Scholar]
- 18.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PDBViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 19.Case, D. A., T. E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K. M. Merz, A. Onufriev, Jr., C. Simmerling, B. Wang, and R. Woods. 2005. The AMBER biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, M. Kenneth, J. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kolman. 1995. A second generation force field for the simulations of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 21.Ponder, J. W., and D. A. Case. 2003. Force fields for protein simulations. Adv. Protein Chem. 66:27–85. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 23.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gusteren, A. Di Nola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Comput. Phys. 81:3684–3690. [Google Scholar]
- 24.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald an N·log(n) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 25.Cheatham, T. E., J. L. Miller, T. Fox, T. A. Darden, and P. A. Kollman. 1995. Molecular dynamics simulation on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA and proteins. J. Am. Chem. Soc. 117:4193–4194. [Google Scholar]
- 26.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 27.Berendsen, H. J. C., D. van der Spool, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 95:43–56. [Google Scholar]
- 28.Laskowski, R. A. 1995. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 13:323–330. [DOI] [PubMed] [Google Scholar]
- 29.Lavery, R., and H. Sklenar. 1989. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 6:655–667. [DOI] [PubMed] [Google Scholar]
- 30.Lu, X. J., and W. K. Olson. 2003. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mok, Y. K., M. Bycroft, and G. de Prat-Gay. 1996. The dimeric DNA binding domain of the human papillomavirus E2 protein folds through a monomeric intermediate which cannot be native-like. Nat. Struct. Biol. 3:711–717. [DOI] [PubMed] [Google Scholar]
- 32.Bazzo, R., D. O. Cicero, and G. Barbato. 1995. A new HCACO 3D pulse sequence with optimized resolution and sensitivity. Application to the 21 kDa Protein Human Interleukin-6. J. Magn. Reson. Series B. 107:189–191. [DOI] [PubMed] [Google Scholar]
- 33.Cicero, D. O., G. M. Contessa, M. Paci, and R. Bazzo. 2006. HACACO revisited: Residual dipolar coupling measurements and resonance assignments in proteins. J. Magn. Reson. 180:222–228. [DOI] [PubMed] [Google Scholar]
- 34.Cicero, D. O., G. M. Contessa, T. A. Pertinhez, M. Gallo, A. M. Katsuyama, M. Paci, C. S. Farah, and A. Spisni. 2007. The solution structure of ApaG from Xanthomonas axonopodis pv. citri reveals a fibronectin-3 fold. Proteins. 67:490–500. [DOI] [PubMed] [Google Scholar]
- 35.Eliseo, T., L. Ragona, M. Catalano, M. Assfalg, M. Paci, L. Zetta, H. Molinari, and D. O. Cicero. 2007. Structural and dynamic determinants of ligand binding in the ternary complex of chicken liver bile acid binding protein with two identical bile salts revealed by NMR. Biochemistry. 46:12557–12567. [DOI] [PubMed] [Google Scholar]
- 36.Ferreiro, D., M. Dellarole, A. D. Nadra, and G. De Prat-Gay. 2005. Free energy contributions to direct readout of a DNA sequence. J. Biol. Chem. 280:32480–32484. [DOI] [PubMed] [Google Scholar]
- 37.Garcia, A. E. 1992. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 68:2696–2699. [DOI] [PubMed] [Google Scholar]
- 38.Amadei, A., A. B. Linssen, and H. J. Berendsen. 1993. Essential dynamics of proteins. Proteins. 17:412–425. [DOI] [PubMed] [Google Scholar]
- 39.Chillemi, G., M. Falconi, A. Amadei, G. Zimatore, A. Desideri, and A. Di Nola. 1997. The essential dynamics of Cu,Zn superoxide dismutase: suggestion of intersubunit communication. Biophys. J. 73:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arcangeli, C., A. R. Bizzarri, and S. Cannistraro. 2001. Concerted motions in copper plastocyanin and azurin: an essential dynamics study. Biophys. Chem. 90:45–56. [DOI] [PubMed] [Google Scholar]
- 41.Hess, B. 2002. Convergence of sampling in proteins simulations. Phys. Rev. E. 65:31910–31920. [DOI] [PubMed] [Google Scholar]
- 42.Dickerson, R. E., M. Bansal, C. R. Calladine, S. Diekman, W. N. Hunter, R. Lavery, H. C. M. Nelson, W. K. Olson, W. Saenger, Z. Shakked, H. Sklenar, D. M. Soumpasis, C. S. Tung, E. V. Kitzing, A. H. J. Wang, and V. B. Zhurkin. 1989. Definitions and nomenclature of nucleic acid structure parameters. J. Biomol. Struct. Dyn. 6:627–634. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 44.De Lano, W. L. 2002. The PyMOL Molecular Graphics System. http://www.pymol.org.