Abstract
Myofibril assembly and disassembly are complex processes that regulate overall muscle mass. Titin kinase has been implicated as an initiating catalyst in signaling pathways that ultimately result in myofibril growth. In titin, the kinase domain is in an ideal position to sense mechanical strain that occurs during muscle activity. The enzyme is negatively regulated by intramolecular interactions occurring between the kinase catalytic core and autoinhibitory/regulatory region. Molecular dynamics simulations suggest that human titin kinase acts as a force sensor. However, the precise mechanism(s) resulting in the conformational changes that relieve the kinase of this autoinhibition are unknown. Here we measured the mechanical properties of the kinase domain and flanking Ig/Fn domains of the Caenorhabditis elegans titin-like proteins twitchin and TTN-1 using single-molecule atomic force microscopy. Our results show that these kinase domains have significant mechanical resistance, unfolding at forces similar to those for Ig/Fn β-sandwich domains (30–150 pN). Further, our atomic force microscopy data is consistent with molecular dynamic simulations, which show that these kinases unfold in a stepwise fashion, first an unwinding of the autoinhibitory region, followed by a two-step unfolding of the catalytic core. These data support the hypothesis that titin kinase may function as an effective force sensor.
INTRODUCTION
All animals have complex mechanisms to regulate their overall muscle mass. The functional unit of a muscle cell is the sarcomere, a miniature machine comprised of overlapping, interacting filaments. During muscle activity, the sarcomeres act in unison, undergoing rapid contraction/relaxation, transducing work throughout the muscle. The three filaments that give the sarcomere its integrity are thin filaments, thick filaments, and titin filaments. These filaments, together with their multi-protein attachment structures, the M-lines and Z-disks, must undergo assembly and disassembly during myofibril growth and maintenance. Although progress has been made in understanding this intricate system, the precise mechanisms that control these assembly/disassembly processes remain central questions in muscle biology. Data presented by Lange et al. (1) is consistent with a model in which signaling from mammalian titin kinase leads to the building of myofibrils. When in its active conformation, titin kinase transmits a phosphorylation cascade that ultimately leads to the expression of genes involved in myofibril assembly. Specifically, titin kinase phosphorylates nbr1, resulting in less MuRF2 and more SRF in nuclei; SRF directly increases myofibrillar gene expression. However, it has been shown recently that deletion of MuRF2 in mice has no affect on the cellular distribution of SRF (2). Moreover, both knockout and conditional mutants deficient for the titin kinase function have been studied (3). In these murine models, normal myofibrils are assembled, but either appear to have a defect in proper maintenance or have impaired contractile regulation. Thus, the physiological functions for titin kinase are still unknown. What we wish to elucidate is how the normally autoinhibited titin kinase is activated.
In Caenorhabditis elegans there are two titin-like proteins, twitchin and TTN-1. These giant muscle proteins, like those found in other species, are comprised primarily of multiple copies of immunoglobulin (Ig) and fibronectin type-III (Fn) domains (4). Twitchin (∼800 kDa), named for its characteristic twitching mutant phenotype, has 30 Ig domains, 31 Fn domains, and a single kinase domain (5,6). Upon its discovery, twitchin was hypothesized to be an important regulator of muscle contraction, due mainly to its motility phenotype and MLCK-like kinase domain. Later, its role in inhibiting the rate of relaxation was identified in studies of twitchin from Aplysia and Mytilus (7,8). TTN-1 is significantly larger than twitchin, with a molecular mass of 2.2 MDa. In addition to 56 Ig domains and 11 Fn domains, TTN-1 has several regions predicted to be coiled-coil, and two regions consisting of tandem repeats (9). The largest of these tandem repeat regions, called PEVT, is similar in amino acid composition and tandem repeat structure to the main elastic element of vertebrate titin called the PEVK region. Thus, this region of nematode TTN-1 is hypothesized to be elastic. Similar to mammalian titin kinase, TTN-1 and twitchin both have a single kinase and regulatory domain located near the C-terminal end of the giant molecule (9,10). In the primary sequence, the inhibitory/regulatory region is located just downstream of the catalytic core (Fig. 1, A and B) and in 3D space (Fig. 2, A and B) the domain is wedged in between the two subdomains of the catalytic core, making extensive contact with the active site, blocking substrate entry (11,12). The kinases with the highest homology, the vertebrate smooth muscle and nonmuscle myosin light chain kinases (MLCKs), are also autoinhibited by a sequence just C-terminal of the catalytic core. In the case of the MLCKs, binding of the autoinhibitory sequence to Ca+2/calmodulin causes a conformational change sufficient to allow access to its substrate (13). Although the giant kinases can also bind to Ca+2/calmodulin, this alone is not sufficient to activate the enzymes (14). In developing muscle, the combination of Ca+2/calmodulin and phosphorylation of a key tyrosine residue can activate vertebrate titin kinase (15). However, the required tyrosine phosphorylating activity is not found in mature muscle. Furthermore, this tyrosine is not conserved: in the analogous position, in twitchin it is alanine (15), and in TTN-1 it is cysteine (9). Thus, to date, the precise mechanism(s) resulting in the conformational changes that relieve the giant kinases of their autoinhibition remain a mystery.
FIGURE 1.
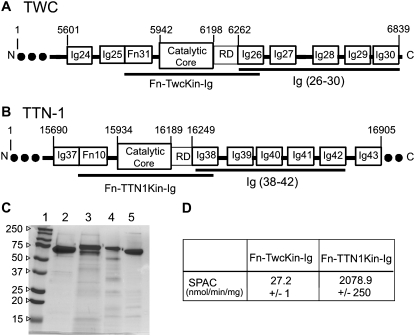
Expression, purification, and enzyme activity of recombinant kinase domains and tandem Ig domain segments from C. elegans giant proteins. (A and B) Twitchin and TTN-1 recombinant protein constructs for AFM experiments. The kinase constructs have an autoregulated kinase domain flanked by Fn and Ig domains. The Ig constructs consist of five tandem Ig domains that immediately follow the kinase domain. The numbers denote the position of the amino acid in the full-length polypeptide. (C) SDS PAGE (12%) stained with Coomassie brilliant blue shows that the proteins were isolated to >95% purity. In each lane, 2 μg of protein was loaded. The lanes are as follows: (1) molecular weight standard; (2) Fn-TwcKin-Ig; (3) Fn-TTN1Kin-Ig; (4) Twc Ig 26-30; (5) TTN-1 Ig 38-42. (D) The purified TTN-1 and twitchin kinases retain phosphotransferase activity in vitro. Using 32P-ATP and a model peptide as substrates the specific activity values (SPAC) obtained are similar to those published previously (9,10). Enzyme activities were determined in triplicate on freshly purified protein.
FIGURE 2.
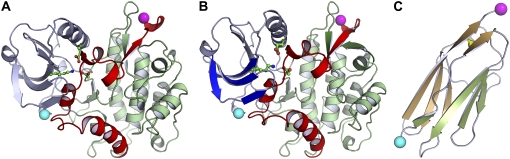
3D structures of C. elegans twitchin Ig and kinase domains and homology model for TTN-1 kinase. (A) Twitchin kinase is composed of three subdomains: an α-helical rich large lobe (green), a small lobe of mainly -sheets (dark blue), and the autoregulatory tail (red) (structure taken from (12)). The autoregulatory tail is situated between the two lobes making extensive contact with the active site. To orient the active site a conserved lysine residue helping to neutralize the ATP binding pocket is shown in a ball and stick interpretation. (B) 3D homology model of TTN-1 kinase. The N-terminal β-sheet (strands βC1-C3) predicted to be parallel to the pulling force is colored dark blue. This sheet is thought to stabilize the kinase on activation by force. The C-terminal β-sheet is predicted to be perpendicular to the pulling force and is composed of βC10 (dark green), βR1 (red), and βC11 (dark green) strands. The autoinhibitory domain is colored red and includes the βR1 strand and αR1-R2 helices. Shown in ball and stick representation is a key lysine residue from the N-terminal β-strands interacting directly with the ATP binding pocket of the active site. (C) Twitchin Ig domain 26, which immediately follows the twitchin kinase domain (structure taken from (12)). The two β-sheets, characteristic of an Ig fold are depicted in green and brown. All three structures have their N-terminal α-carbons marked with cyan spheres and their C-terminal α-carbons marked with magenta spheres.
One hypothesis is that the giant kinases may act as force sensors; that the forces generated from the contraction/relaxation cycles of muscle activity are sufficient to unleash the regulatory domain from the catalytic core (16). Thus, after experiencing a certain threshold of force, the kinase would become active. Grater et al. (16) used molecular dynamics simulations to pull the human titin kinase from its amino and carboxy termini to simulate the strain the molecule would undergo during muscle activity. The authors found that not only can the kinase withstand expected forces, but that the kinase domain is also positioned in perfect orientation within the “molecular spring” of the titin molecule, making the kinase an ideal force sensor. They conclude that the strain on the autoinhibitory domain leads to an ordered sequence of conformational changes that open the catalytic cleft, while maintaining the structural integrity of the enzyme. This remarkable stability is a function of the orientation of the molecule with respect to the pulling force; the pulling geometry of activation is such that the β-sheets most responsible for the force resistance are located parallel to the pulling force, but the β-sheets responsible for exposing the active site are oriented perpendicular to the force. This simple, yet elegant, difference in the orientation of β-sheets sets the stage for the ability of the kinase to simultaneously remove the autoinhibitory region from the catalytic core, while maintaining the structural integrity of the active site. The molecular dynamics simulations support a model in which human titin kinase will sense force and pass along the message by phosphorylation of its substrate.
To begin to test this hypothesis experimentally, we recombinantly expressed twitchin and TTN-1 kinases, the human titin kinase homologs, from C. elegans. Using single-molecule atomic force microscopy (AFM) we analyzed the mechanical strength of the kinase and their flanking Ig/Fn domains, along with a tandem repeat of five Ig domains that immediately follow the kinase domains in the primary structure. Our results show that these kinase domains have remarkably high mechanical stability. The kinase domain unfolds at a force range of ∼30–150 pN, a value that is similar to the unfolding range of nematode Ig and Fn domains (40–180 pN). Further, in contrast to the Ig/Fn domains, which unfold in an all-or-none highly cooperative fashion, the unfolding of the kinase is sequential, first an unwinding of the autoinhibitory region, followed by the biphasic rupture of the catalytic core. These data provide support for the hypothesis that the kinase domains of the giant muscle proteins function as effective force sensors.
MATERIALS AND METHODS
AFM
The mechanical properties of single proteins were studied using a home-built single molecule AFM as described previously (17–22). The spring constant of each individual cantilever (MLCT-AUHW: silicon nitride gold-coated cantilevers; Veeco Metrology Group, Santa Barbara, CA) was calculated using the equipartition theorem (23). The cantilever spring constant varied between 10–50 pN/nm and rms force noise (1-kHz bandwidth) was ∼10 pN. Unless noted, the pulling speed of the different force–extension curves was in the range of 0.4–0.6 nm/ms.
Single protein mechanics
In a typical experiment, a small aliquot of the purified proteins (∼1–50 μl, 10–100 μg/mL) was allowed to adsorb to a clean glass coverslip (for ∼10 min) and then rinsed with PBS pH 7.4. Proteins were picked up randomly by adsorption to the cantilever tip, which was pressed down onto the sample for 1–2 s at forces of several nanonewtons and then stretched for several hundred nm.
Analysis of force extension curves
The elasticity of the stretched proteins were analyzed using the worm-like chain (WLC) model of polymer elasticity (24,25):
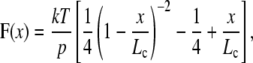 |
where F is force, p is the persistence length, x is end-to-end length, and Lc is contour length of the stretched protein. The adjustable parameters are the persistence length, p, and the contour length, Lc.
Monte Carlo simulations
The folding and unfolding of a domain was modeled as a two state Markovian process where the probability of unfolding was Pu = Nf*α*Δt where Nf is the number of folded domains and Δt is the polling interval (21,22,26). The folding probability was Pf = Nu*β*Δt where Nu is the number of unfolded domains. The rate constants for unfolding, α, and refolding, β, are given by α = αoexp(FΔxu/kT) and β = βoexp(−FΔxf /kT) where F is the applied force and Δxu and Δxf are the unfolding and folding distances.
Homology modeling of TTN1 kinase and regulatory domains
Molecular modeling of C. elegans TTN-1 was completed using Modeler (version 7v7; University of California San Francisco; http://salilab.org/modeler/) for model construction and SYBYL (version 7.0; Tripos, http://www.tripos.com) for analysis and refinement. The TTN-1 model was built and minimized based on the existing structural information available from C. elegans twitchin (PDB entry 1KOA) (12). Initial sequence alignments were obtained from ClustalW (using TTN-1, human titin, C. elegans, Aplysia, and Mytilus twitchin, chicken smooth muscle MLCK, and Drosophila myosin MLCK, http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html), NCBI BlastP (http://www.ncbi.nlm.nih.gov/), CPHmodel (using TTN-1 and C. elegans twitchin, www.cbs.dtu.dk), SwissModel (using TTN-1 and C. elegans twitchin, http://swissmodel.expasy.org//SWISS-MODEL.html), CDDomain (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, hits = KOA, KOB, 1TKI), and 3Djigsaw (using TTN-1 and C. elegans twitchin, http://www.bmm.icnet.uk/∼3djigsaw/). The alignment was optimized manually after each round of modeling using visual structural comparison of the previously published structural data of C. elegans twitchin (PDB entry 1KOA (12)), Aplysia twitchin (PDB entry 1KOB) (12), and human titin (PDB entry 1TKI) (15). The final alignment is shown in Fig. S1 in Supplementary Material, Data S1. The proteins showed ∼50% (for C. elegans and Aplysia twitchin) and ∼40% (for titin) similarity against TTN-1 residues 15907–16373 (Fig. 1 B), the approximate boundaries of TTN-1 kinase and the flanking Ig in the full-length giant polypeptide.
We specifically studied the sequence alignment at the terminal β-sheets and the regulatory α-helices, as these are the regions predicted to be most important in the force activation mechanism (16), and are also the regions with some of the highest variability (9,10). The N-terminal β-sheets, βC1–C3, are located at residues: 24–33 (βC1), 46–53 (βC2), and 48–56 (βC3) in TKI; 53–62 (βC1), 65–72 (βC2), and 77–85 (βC3) in KOB; 5942–5951
 |
(βC1), 5954–5961 (βC2), and 5966–5974 (βC3) in KOA; 28–37 (βC1), 40–47 (βC2), and 52–60 (βC3) in TTN-1. The C-terminal β-sheets, βC10–11 and βR1 are located at residues: 174–182 (βC10), 193–197 (βC11), and 328–336 (βR1) in TKI; 204–212 (βC10), 224–228 (βC11), and 362–370 (βR1) in KOB; 6093–6100 (βC10), 6113–6117 (βC11), and 6253–6260 (βR1) in KOA; 179–186 (βC10), 199–203 (βC11), and 334–341 (βR1) in TTN-1. αR1 and αR2 are located at residues: 292–306 (αR1) and 312–318 (αR2) in TKI, 321–335 (αR1) and 344–351(αR2) in KOB, 6211–6225 (αR1) and 6234–6241 (αR2) in KOA, and 292–307 (αR1) and 315–323 (αR2) in TTN-1. All protein structure images were generated by PyMOL version 1.1beta1 (http://www.pymol.org).
Steered molecular dynamics simulations
The giant kinase structures were analyzed by steered molecular dynamics (SMD) as implemented in NAMD (27,28). The CHARM22 force field was used throughout. The structural coordinates for each kinase structure (1KOA and the homology model for TTN-1) were solvated in a 65 Å × 65 Å × 65 Å box. Eighteen Na+ ions were added, corresponding to a concentration of 0.11 M. The system was then minimized with 1000 steps of conjugate gradient minimization from an initial temperature of 310 K. This was followed by a 400 ps MD simulation to equilibrate the entire system (protein, water, and ions). The backbone RMSD was evaluated at the completion of the equilibration step. The SMD protein-ion-water system contained ∼30,000 atoms. Forces were applied by restraining a fixed termini point harmonically and moving the SMD atom with constant velocity (0.5 Å/ps) along a predetermined vector. Kinase domains were stretched at a constant speed of 0.5 Å ps−1 until their extension exceeded 99% of the contour length. The trajectories were recorded every 2 fs and analyzed with VMD. Coulombic forces were restricted using the switching function from 10 Å to a cutoff at 12 Å. A spring constant (κ) of 10 kBT/Å2 was used during each simulation. We ran three simulations of the extension of twitchin kinase and the homology model for TTN-1 kinase domains with similar results. To validate the accuracy of our in silico experiments we carried out SMD simulations on other protein domains, such as titin I27, ubiquitin, and synaptotagmin C2A. Our SMD results are very similar to those published previously (Fig. S2 in Data S1).
Cloning and expression of TTN-1 and Twitchin constructs
The TTN-1 and twitchin constructs were amplified from the C. elegans random primed cDNA library RB2 (kindly provided by Robert Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK) using the primer pairs listed below. Each construct was first subcloned into the cloning vector bluescript pKS (+), sequenced and subcloned into the expression vector pET 28, fusing an in frame 6-his tag to the N-terminus.
The following restriction sites were inserted and used for the subcloning procedure: TTN-1 Ig (38–42) 5′ BamHI, 3′ EcoRI; TTN-1 FnKinIg 5′ EcoRI, 3′HindIII; twc Ig (26–30) 5′SacI, 3′XbaI; twc FnKinIg 5′BamHI, 3′XhoI.
The recombinant plasmids were transformed into Escherichia coli BL21 (DE3) RIL (Stratagene, La Jolla, CA). The cells were grown to mid-log phase in the presence of 25 μg/mL kanamycin and 34 μg/mL chloramphenicol at 37°C. Protein expression was induced with 0.5 mM IPTG and continued overnight at 23°C. The cells were harvested and resuspended in 20 mM TRIS, 500 mM NaCl, and 5 mM imidazole (pH 7.9) with the addition of Roche (Indianapolis, IN) complete EDTA-free protease inhibitor pellets. Lysis was completed by passage through a French press at 1000 psi. Detergents PEI (to 0.1%) and NP40 (to 0.01%) were added to the cell free extract. The soluble fraction was collected and loaded onto a precharged Novagen nickel column (Novagen, Gibbstown, NJ). The column was first washed with 25 column volumes of 20 mM TRIS, 750 mM NaCl, 5 mM imidazole, and 0.01% NP40 (pH 7.9) followed by a second wash with 5 column volumes of 20 mM TRIS, 750 mM NaCl, 20 mM imidazole, and 0.01% NP40 (pH7.9). The proteins were eluted with 20 mM TRIS, 500 mM NaCl, and 1 M imidazole. Protein purity of ∼95% was confirmed using Coomassie brilliant blue staining of 12% SDS PAGE.
Kinase assays
The purified kinases were dialyzed against 20 mM TRIS (pH 8.0), 20 mM NaCl, and 5 mM βME. The enzymes were added to reaction buffer (20 mM TRIS pH 7.4, 10 mM magnesium acetate, 0.05% Triton, 0.2 mg/mL BSA) to a final concentration of 30 pg/μL. The model substrate was a derivative of chicken smooth muscle regulatory myosin light chain (kMLC 11–23) with the sequence KKRARAATSNVFS (14) (synthesized by the Microchemical Facility, Emory University). The peptide substrate was added to the reaction mixture in excess (0.2 mg/mL). The reactions were initiated by the addition of 400 μM γ32P-ATP (0.25 μCi/μL ). Catalysis occurred for 10 min at 30°C. A portion of the reaction (25 μL/40 μL total volume) was removed and spotted onto Whatman P81 filters. The filters were washed in 75 mM phosphoric acid. Once dry, the washed filters were placed into scintillation vials, the counts were measured, and the specific activity was calculated by the following equation:
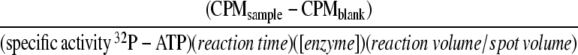 |
Each reaction was done in triplicate and carried out on freshly purified protein.
As expected from earlier results, specific activities were the highest for the reactions with the Fn-TTN-1Kin-Ig and significantly lower for the Fn-Twc kinase-Ig reactions (9,10). The specific activity of the enzymatic labeling was normalized to reactions without the addition of kinase.
RESULTS
3D structures of C. elegans, twitchin, and TTN-1 kinase domains
Crystal structures for both C. elegans and Aplysia twitchin kinases (11,12) and human titin kinase (15) have been solved previously. However, the structure of TTN-1 kinase is unknown. In the interest of developing a better understanding of how the structural similarities among the enzymes might relate to their activation mechanism(s), we constructed a homology model of TTN-1 kinase catalytic core and autoinhibitory region (Fig. 2 B). Overall, the predicted structure is very similar to the C. elegans twitchin kinase. The catalytic region has two lobes, a smaller, β-rich lobe, and a larger α-helical lobe (Fig. 2 B, dark blue and light green, respectively). The regulatory tail (red) wraps between the two lobes, nesting itself into the active site. This inhibited conformation would be maintained as the native structure during catalytic arrest. When the muscle cells require the catalytic activity of the giant protein kinases it is necessary that the regulatory domain be removed for optimal catalysis to be achieved. Closer examination of how the regulatory domain interacts with the catalytic core shows secondary structure elements that are believed to be the fundamental basis for the force activation hypothesis (16).
The molecular dynamic simulations by Grater et al. (16) predicted the N- and C-terminal β-sheets (Fig. 2 B, dark blue and dark green plus red β-strand, respectively) to be the primary mechanical elements responsible for the force resistance. Similar to the known crystal structures, the homology model of TTN-1 kinase maintains the alignment of these important substructures. The C-terminal β-sheet consists of three β-strands: βC10, βC11, and βR1 (“C” for catalytic; “R” for regulatory) (Fig. 2 B; dark green plus red β-strand). Grater et al. (16) showed that during activation, the C-terminal β-sheets are positioned perpendicular to the pulling force and are expected to undergo the initial rupture that leads to the activation of the kinase. When comparing the crystal structures to the model, there is a general conservation along βC11 visualized by comparing the hydrogen-bonding pattern and the side chain interactions. βR1 is sandwiched between βC10 and βC11 and continues to show the consistent bonding pattern between the enzymes.
According to the force activation model, the N-terminal β-sheet (βC1–C3; Fig. 2 B, dark blue) is the region that lies parallel to the pulling force to maintain the active site integrity during force activation (16). This sheet is strikingly similar in all four enzymes. Almost all of the backbone interactions are maintained. The side chain interactions, which further stabilize the β-sheets, are also consistent between the kinases. A conserved lysine residue in the N-terminal βC3 sheet (K82 KOB, K5971 KOA, K57 TTN1, K53 TKI) interacts with the regulatory tail αR2 via van der Waals interactions between the side chain and isoleucine/leucine and valine. This lysine also interacts directly with the active site through electrostatic interactions with an aspartic acid and glutamic acid (11,12). Linked intimately to the N-terminal β-strands, the αR2 helix is a tightly packed 310 helix buried beneath the βC1–C3 sheets, the substrate binding site, and the portion of the protein that lies directly upstream of the helix (the αR1 helix and the linker between αR1 and αR2 helices). The face of the helix that is in contact with the β-strands is notably similar in each of the enzymes; a string of hydrophobic interactions from the nonpolar side chains of the β-strands contacts the nonpolar face of the helix (with the exception of one serine present only in TTN-1 and C. elegans twitchin). On the alternate side of the helix, the residues interact with both the substrate-binding site and the longer of the regulatory helices, αR1. Many of the interactions are conserved between all four kinases, allowing the αR2 helix to retain the overall stability and topology of the enzymes.
Force-extension relationships of C. elegans TTN-1 and twitchin Ig domains
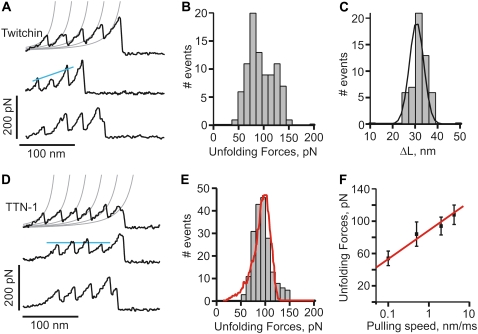
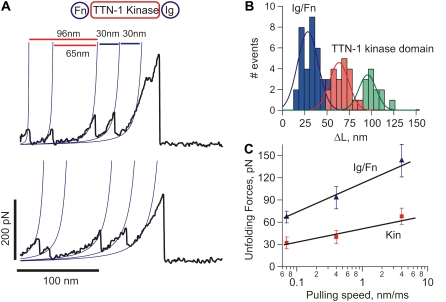
For titin-like proteins, the organization is such that a set of five tandem Ig domains immediately follows the kinase catalytic core and its regulatory sequence (Fig. 1, A and B). If the kinase were to act as a force sensor, we would expect these domains to withstand forces greater than the kinase itself. To test this hypothesis we used single-molecule AFM techniques to analyze the mechanical properties of recombinant proteins containing five tandem Ig domains from twitchin and TTN-1 (Fig. 1, A–C). Random segments of these proteins were picked up by the AFM tip and then stretched with a pulling speed of ∼0.5 nm/ms. The resulting force-extension curves showed a sawtooth-like pattern, characteristic of the unfolding of Ig domains (Fig. 3, A and D) (17,29). To analyze the spacing between peaks in the sawtooth patterns we used the WLC model for polymer elasticity, which predicts the entropic restoring force (F) generated on the extension (x) of a polymer (24,25) (see Materials and Methods). The thin lines in Fig. 3, A and D, correspond to fits of the WLC equation to the curve that precedes each force peak. We found that the separation between force peaks for both proteins is ∼30 nm (30.6 nm ± 3.2 for the twitchin protein; n = 53 force peaks; Fig. 3 C). This value corresponds very well with the expected increase in contour length of a 95 amino acid (aa) Ig domain: 95 aa × 0.35 nm (length of an aa) – 3 nm (size of folded Twc Ig domain 26 (12); and Fig. 2 C) = 30.3 nm.
FIGURE 3.
Force-extension relationships of twitchin and TTN-1 Ig domains. (A and D) Several examples of force-extension curves obtained after stretching twitchin (A) and TTN-1 (D) Ig domains. The gray lines were generated with the WLC equation using a persistence length of 0.4 nm and contour length increments, ΔL, of 30 nm. (B and E) Unfolding force histograms for twitchin and TTN-1 domains. The mean force peak values are 93 ± 25 pN, (n = 88 peaks) and 85 ± 22 pN (n = 193 peaks), respectively. In (E) the red line corresponds to a Monte Carlo simulation of TTN-1 Ig using 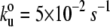 and Δxu = 0.35 nm at a pulling speed of 500 nm/s. (C) Histogram of contour length increments observed on unfolding of twitchin Ig domains shows one main peak centered at ∼30 nm (Gaussian fit: 30.6 ± 3.2 nm). (F) The unfolding forces of TTN-1 Ig domains depend on pulling speed. The experimental data (black symbols) can be well described by Monte Carlo simulations (red line) using
and Δxu = 0.35 nm at a pulling speed of 500 nm/s. (C) Histogram of contour length increments observed on unfolding of twitchin Ig domains shows one main peak centered at ∼30 nm (Gaussian fit: 30.6 ± 3.2 nm). (F) The unfolding forces of TTN-1 Ig domains depend on pulling speed. The experimental data (black symbols) can be well described by Monte Carlo simulations (red line) using 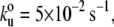 Δxu = 0.35 nm.
Δxu = 0.35 nm.
Unfolding force histograms show that the Ig domains from twitchin and TTN-1 unfold at a similar range of forces and similar average unfolding force values (∼90 pN and ∼85 pN, respectively; Fig. 3, B and E). The unfolding forces of these Ig domains from twitchin and TTN-1 fall within the range or are somewhat weaker than those reported for Ig domains from vertebrate titin (29) and insect projectin (30). It is noteworthy that the recordings from the five Ig domains of twitchin show an ascending pattern of peak heights suggesting a hierarchy in mechanical stabilities for these Ig domains (Fig. 3 A). In addition, we found that mechanical unfolding of TTN-1 Ig domains is fully reversible and that they refold to their native states with a rate at zero force of ∼2 s−1 (not shown).
Our results show that the TTN-1 and twitchin Ig domains unfold at ∼90 pN at a fixed pulling speed of 0.5 nm/ms. However, during normal muscle contraction cycles these domains may experience a wide range of stretching speeds. Because the mechanical stability may not be the same at different pulling speeds, as shown for titin domains (18,26) we studied the rate-dependency of the stability of Ig domains from TTN-1 and twitchin. Fig. 3 F shows the relationship between the unfolding force and the pulling speed (0.1–5 nm/ms) for TTN-1 Ig domains. A 10-fold decrease in pulling speed decreases the unfolding forces by only 20 pN indicating that these domains are mechanically stable over a wide range of pulling speed. The continuous lines correspond to the result of Monte Carlo simulations of two-state unfolding of this sequence at the corresponding range of pulling rates (18,22,26,31). We found that, by using a combination of the rate constant at zero force, 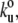 of 5 × 10−2 s−1 and the unfolding distance Δxu between the folded state and transition state of 0.35 nm, we can adequately describe the unfolding force histogram (Fig. 3 E) as well as the speed dependency of unfolding forces (Fig. 3 F). These values are comparable to those reported for several other mechanically stable β-strand rich domains (17,22,29,32). For example human titin Ig domains have Δxu values ranging from 0.25 nm (I27) to 0.35 nm (I1) but it can be as high as 0.4 nm in fibronectin β-sandwich domains. These results are consistent with the hypothesis that these Ig domains are designed to resist stretching forces.
of 5 × 10−2 s−1 and the unfolding distance Δxu between the folded state and transition state of 0.35 nm, we can adequately describe the unfolding force histogram (Fig. 3 E) as well as the speed dependency of unfolding forces (Fig. 3 F). These values are comparable to those reported for several other mechanically stable β-strand rich domains (17,22,29,32). For example human titin Ig domains have Δxu values ranging from 0.25 nm (I27) to 0.35 nm (I1) but it can be as high as 0.4 nm in fibronectin β-sandwich domains. These results are consistent with the hypothesis that these Ig domains are designed to resist stretching forces.
Force-extension relationships of C. elegans giant kinases and flanking domains
The protein kinase domains of twitchin and TTN-1 are autoinhibited, containing an endogenous regulatory sequence situated between the two subdomains, making extensive contact with residues essential for ATP binding, substrate recognition, and catalysis. Unlike their closest homologs, the MLCKs, binding to Ca+2/calmodulin does not relieve autoinhibition. It has been hypothesized that the giant kinases may act as force sensors (16); that the forces generated from the contraction/relaxation cycles of muscle activity are sufficient to unleash the regulatory domain from the catalytic core and activate the kinase. The release from a stretching force would restore the inhibited conformation of the kinase.
We used single-molecule AFM to analyze the mechanical strength of the kinase and their flanking Ig/Fn domains. To be sure that the proteins were in their native conformations, the kinase activity of the proteins was measured. Although the proteins are autoinhibited, they have modest activity in vitro (9,10). We found that our recombinant kinase constructs have specific activities comparable to those published previously (Fig. 1 D) (9,10).
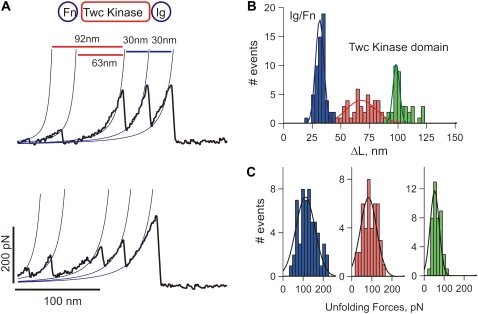
For the force measurements, dilute solutions of the recombinant kinase proteins were nonspecifically attached to a glass coverslip. Random segments of the proteins were picked up by the AFM tip and then stretched with a pulling speed of 0.5 nm/ms. Fig. 4 A shows examples of force-extension curves obtained after stretching single Fn-Twc kinase-Ig molecules. We typically observed recordings with multiple force peaks before the last detachment peak. In the Fn-Twc kinase-Ig construct the Ig and Fn domains have ∼95 aa and should therefore each contribute to an increase in contour length, ΔL, of ∼30 nm (Fig. 3). Hence, we attribute the two force peaks before the detachment peak as the unfolding of Fn and Ig domains and the initial two force peaks to the sequential unfolding of the kinase domain, which has a contour length of ∼95 nm. Fig. 4 B shows a histogram of increases in contour length increments observed on unfolding of Fn-Twc kinase-Ig. There are peaks at ∼30 nm, 65 nm, and 95 nm, which correspond to the unfolding of Ig/Fn domains (blue bars), and kinase domain (red and green bars), respectively. The corresponding unfolding forces are 111 ± 67 pN (n = 57), 83 ± 57 pN (n = 41), and 52 ± 11 pN (n = 43) (Fig. 4 C). The small peak has a contour length of ∼30 nm, a length that would be expected to fit the unwinding of the ∼100 aa small lobe. The second peak gives a contour length of ∼65 nm, fitting nicely to the unwinding of the ∼190 aa large lobe. The regulatory domain (that includes the βR1 C-terminal strand) probably unfolds at very low forces (<10 pN), forces indistinguishable from noise on the AFM. Hence, we interpret the data shown in Fig. 4 as the stepwise unfolding of the two lobes of the kinase. The smaller lobe, composed mainly of β-sheets will unfold first, leaving the larger, α-helical lobe intact. This hypothesized order of rupture, and lack of any unfolding peak from the regulatory domain, is supported by our SMD simulations (see Fig. 6).
FIGURE 4.
Mechanical properties of C. elegans twitchin kinase. (A) Two examples of force-extension curves obtained for the Fn-Twc kinase-Ig construct. The two small force peaks correspond to the stepwise unfolding of the Twc kinase domain and the last two peaks to the unfolding of the flanking Ig/Fn domains. (B) Histogram of increases in contour length increments observed on unfolding, ΔL, of Fn-Twc kinase-Ig. There are peaks at ∼30 nm, 65 nm, and 95 nm (Gaussian fits: 31 ± 5 nm, 67 ± 18 nm, and 97 ± 10 nm, n = 142), which correspond to the unfolding of Ig/Fn domains (blue bars), and kinase domain (red and green bars). (C) Unfolding force distributions for Ig/Fn domains (blue bars) and kinase domain (red and green bars); the respective force peaks are at 111 ± 67 pN (n = 57), 83 ± 57 pN (n = 41), and 52 ± 11 pN (n = 43).
FIGURE 6.
Constant velocity steered molecular dynamics simulation of the mechanical unfolding of twitchin kinase. (Left) Force-extension curve obtained from SMD simulations by stretching the twitchin kinase domain (1KOA) between its C terminus and its N terminus at a pulling speed of 0.5 Å/ps. The total simulation time was 2.4 ns using 33,428 total atoms including 18 Na+ and 9305 water molecules. The fixed atom was Tyr-5915 and the SMD atom Arg-6261. (Right) Four snapshots of twitchin kinase stretched from its termini taken at no extension (rest), after 65 Å (1), 340 Å (2), and 639 Å (3) of extension. At rest, the kinase domain is in a closed conformation. The active site is occupied by the autoinhibitory region (red), which makes extensive contact with the catalytic site, blocking substrate binding. (1) At low forces the regulatory tail will unravel reversibly and expose the active site to its substrates. (2) At high forces the kinase begins to unfold and the integrity of the active site is disrupted. The small lobe (blue), made mainly of β-sheets, unravels first followed by the unfolding of the α-helical rich large lobe (green).
The unfolding pattern of TTN-1 Fn-Kinase-Ig parallels that of twitchin Fn-Kinase-Ig. The complete unfolding of TTN-1 kinase has four force peaks (Fig. 5 A) corresponding first to the biphasic rupture of the catalytic core, followed by the unfolding of Ig and Fn domains. There are peaks at ∼30 nm, 65 nm, and 95 nm (Fig. 5 B). To further characterize the mechanical unfolding of the kinase domains, we analyzed the unfolding kinetics by doing experiments at different pulling speeds. Fig. 5 C shows a plot of the average unfolding force versus the pulling rate for TTN-1 Ig/Fn domains and the kinase domain. For the kinase, we analyzed the first force peak that gives a ΔL of ∼30 nm, which we interpret as the unfolding of the small, β-sheet rich lobe. The continuous lines correspond to the result of Monte Carlo simulations of two-state unfolding at the corresponding pulling rates. The parameters used for the Monte Carlo simulation are: 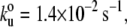 Δxu = 0.35 nm for the Ig/Fn domains and
Δxu = 0.35 nm for the Ig/Fn domains and 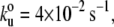 Δxu = 0.6 nm for the TTN-1 kinase. The unfolding rate constant at zero force,
Δxu = 0.6 nm for the TTN-1 kinase. The unfolding rate constant at zero force, 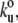 and unfolding distance between the folded and the transition state, Δxu, for the Ig and Fn β-sandwich domains are similar to that for TTN-1 Ig domains 38–42 (Fig. 3 F). However, the transition state distance Δxu for the kinase α-helical lobe is considerable larger than in the β-sandwich domains (0.6 nm vs. 0.35 nm), suggesting different transition state structures.
and unfolding distance between the folded and the transition state, Δxu, for the Ig and Fn β-sandwich domains are similar to that for TTN-1 Ig domains 38–42 (Fig. 3 F). However, the transition state distance Δxu for the kinase α-helical lobe is considerable larger than in the β-sandwich domains (0.6 nm vs. 0.35 nm), suggesting different transition state structures.
FIGURE 5.
Mechanical properties of C. elegans TTN-1 kinase. (A) Two examples of force-extension curves obtained for the Fn-TTN1 kinase-Ig construct. (B) Histogram of increases in contour length increments observed on unfolding, ΔL, of the Fn-TTN-1 kinase-Ig construct (16 molecules). There are peaks at ∼30 nm, 65 nm, and 95 nm (n = 35). (Gaussian fits: 29 ± 16 nm, 63 ± 15 nm, and 95 ± 13 nm). (C) Plot of the average unfolding force versus the pulling rate for TTN-1 Ig/Fn domains and the kinase domain. For the kinase, we analyzed the first force peak with a ΔL of ∼95 nm. The continuous lines correspond to the result of Monte Carlo simulations of two-state unfolding at the corresponding pulling rates. The parameters used for the Monte Carlo simulation are: 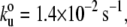 Δxu = 0.35 nm for the Ig/Fn domains and
Δxu = 0.35 nm for the Ig/Fn domains and 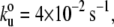 Δxu = 0.6 nm for the TTN-1 kinase.
Δxu = 0.6 nm for the TTN-1 kinase.
From these data, we can conclude that the TTN-1 and twitchin kinase domains show a significant mechanical resistance and they unfold at forces similar to those for Ig/Fn β-sandwich domains (30–150 pN). These results further support the force activation hypothesis for the giant protein kinases.
DISCUSSION
The kinase domain of human titin has been postulated to act as a force sensor (1,16). The kinase domain is thought to be autoinhibited at rest, but during muscle activity, catalysis by the kinase results in a phosphorylation cascade that ultimately causes the expression of genes important in myofibril maintenance and growth. Human titin spans half of the sarcomere, from the M-line to the Z-disk (33). The segment of titin in the A-band ending at the M-line is fixed, whereas the portion of titin in the I-band varies its length in response to the state of muscle contraction (34). The kinase domain, which is located at the periphery of the M-line (35), is in an ideal position and orientation to sense the mechanical strain that occurs during the contraction/relaxation cycle of muscle activity. In C. elegans striated muscle, the proteins of greatest similarity to human titin are TTN-1 located in the I-bands (9), and twitchin located at the non-M-line portions of A-bands (36). Given the high degree of similarity of the kinase catalytic cores and the conservation of organization of surrounding Ig and Fn domains, it is reasonable to hypothesize that force activation might also occur in TTN-1 and twitchin.
If the kinase domains were to act as force sensors, we would expect them to withstand stretching forces. Here we used single-molecule force spectroscopy to test the mechanical properties of C. elegans TTN-1 and twitchin kinases. The proteins were recombinantly expressed and shown to retain activity in their inhibited forms. We found that the mechanical stabilities of the kinase domains are compatible with their postulated function. The kinases unfold in two clearly resolvable steps at forces of ∼50 pN and ∼80 pN. These unfolding forces are lower than most β-strand rich domains, range ∼80–250 pN (20,26,29,30,32,37) but similar to α-helix rich proteins, range ∼30–100 pN (38–40).
Our results show that the TTN-1 and twitchin kinase domains unfold in a biphasic, stepwise fashion, indicated by the presence of two peaks in the force-extension recordings. The first force peak probably represents the unwinding of the smaller lobe of the catalytic core, corresponding to the breakage of β-sheets. After the initial rupture is a second peak, probably representing the rupture of the larger, α-helical lobe of the catalytic core. The small peak has a contour length ΔL of ∼30 nm, a length that is consistent with the unwinding of the ∼100 aa small lobe. The second peak gives a contour length of ∼65 nm, which may correspond to the unwinding of the ∼190 aa large lobe. Unexpectedly, these results contrast with the now large amount of data showing that α-helices are less mechanically stable than β-sheets (41). Finally, the regulatory domain most likely unfolds at very low forces (<10 pN), which is below the resolution of our AFM.
To further understand the molecular origin for the mechanical unfolding of the C. elegans twitchin kinase we carried out SMD simulations. Molecular dynamics simulations have been extensively used to examine the mechanical unfolding of a wide variety of proteins and in general, there is a good correlation with single-molecule data (27,39,42,43). To validate the accuracy of our in silico experiments we carried out SMD simulations on other protein domains, such as titin I27, ubiquitin and synaptotagmin C2A. The I27, ubiquitin, and C2A results are very similar to those published previously (26–28) (Fig. S2 in Data S1). The magnitude of the forces observed in the SMD simulations does not directly correspond to those measured with AFM. This is partially because the pulling speeds are several orders of magnitude different. However, the simulations are qualitatively consistent with the AFM results. For example, similar to our AFM results twitchin Ig26 unfolds at lower forces than I27. In addition, as shown by AFM data (44) the C2A domain unfolds at much lower forces (∼50 pN) than I27 or ubiquitin (∼200 pN) and does not show an initial force-extension burst.
Fig. 6 shows constant velocity SMD simulation of the mechanical unfolding of the twitchin kinase domain. The force-extension curve was obtained from SMD simulations by stretching the C. elegans twitchin kinase domain (1KOA) between its C terminus and its N terminus at a pulling speed of 0.5 Å/ps. On the right four snapshots of twitchin kinase are shown taken at no extension (rest), after 6.5 nm (1), 34 nm (2), and 64nm (3) of extension. At rest, the kinase domain is in a closed conformation. The active site is occupied by the autoinhibitory region (red), which makes extensive contact with the catalytic site, blocking substrate binding. At low forces, the regulatory tail (that includes the C-terminal β-strand βR1 and αR1-R2 helices) will unravel reversibly and expose the active site to its substrates (snapshot 1; red region in the force-extension plot). According to this model the C-terminal β-sheet (strands βC10, C11 + βR1; dark green and red in Fig. 2 B) should unfold together with the αR1 and R2 helices. Because their different mechanical topologies (zipper versus shear), the C-terminal β-sheet should unfold at much lower forces than the N-terminal β-sheet. As shown by Grater et al. (16), we also observe that after breakage of the C-terminal β-sheet and unraveling of the regulatory domain, the tertiary structure of the kinase catalytic site remains intact. At this point, the kinase is in an open, active conformation and downstream signaling can occur. During muscle activity, low forces may result from the repeated contraction/relaxation of the sarcomeres. At high forces, the kinase begins to unfold and the integrity of the active site is disrupted (snapshot 2; blue region in the force-extension plot). The small lobe (blue), made mainly of β-sheets, unravels first followed by the unfolding of the α-helical rich lobe (green; snapshot 3; green region in the force-extension plot). Eventually, with sustained high forces, the enzyme will completely unravel (white region in the force-extension plot). We also carried out constant velocity SMD simulations of the mechanical unfolding of the model for TTN-1 kinase (Fig. S3 in Data S1). The unfolding trajectory for TTN-1 kinase is nearly identical to that of the twitchin kinase indicating a similar stepwise mechanical unfolding pathway, as seen in the AFM data (Fig. 5).
Grater et al. (16) suggested a similar model from results of molecular dynamics simulations. However, in their studies they predicted that only human titin, but not Aplysia twitchin, would be likely to function as a force sensor. The simulations show that when force is applied to human titin the termini of the protein shift away from the kinase, the autoinhibitory region detaches from the active site, and that this release is accompanied by a shift in the two lobes of the catalytic core that results in an active site that is accessible to substrates. For Aplysia twitchin it was predicted that although applied force would remove the inhibitory region, the lobes of the catalytic core remain static and the enzyme would still be inactive. Our data argues that, in C. elegans, both TTN-1 and twitchin kinases meet the requirements of an enzyme likely to be activated by mechanical forces.
The most common feature of all the giant titin-like muscle proteins is the presence of multiple copies of Ig and Fn domains, either as tandem Ig domains or as super-repeats of Ig and Fn domains (4). Moreover, the arrangement of Fn and Ig domains and their actual sequences are highly conserved between twitchins and titins (5,45). One major challenge in muscle biology is to understand how these modular domains function both at the individual and group scale, and how their mechanical properties vary to suit the type of muscle, or even the location of a domain within a single sarcomere. Single molecule experiments have shown the strength of Ig domains of titin and titin-like proteins to vary between 50–300 pN, depending on the position of the domain within the sarcomere (29,30). We studied the mechanical properties of five tandem Ig domains from C. elegans TTN-1 and twitchin. These five Ig domains are located in the C-terminal regions of the endogenous proteins and immediately follow the conserved kinase domain. We hypothesize that this region is functionally important because the architectural arrangement of the kinase domain plus five Igs is conserved in all of the giant titin-like proteins. Our data shows that these Ig domains, at least in the nematode, are slightly weaker (∼93 pN for twitchin and ∼85 pN for TTN-1) than the average Ig domain and those found in other muscle types such as cardiac titin (∼200 pN) (18), insect kettin (∼125–250 pN), and insect projectin (∼109 pN) (30).
TTN-1 can be regarded as a twitchin/titin hybrid. At the sequence level, the TTN-1 kinase catalytic core is more similar to the kinase catalytic core of twitchin (54.4% identical) than it is to the kinase of human titin (39.2% identical) (9). What makes TTN-1 titin-like is its enormous 2.2 MDa size and the presence of several regions consisting of tandem repeats that are likely to act as molecular springs (these are not found in twitchin, only in human titin). Using known crystal structures of the kinase domains of C. elegans twitchin (Fig. 2 A) (12), Aplysia twitchin (12), and human titin (15), we built a molecular model of TTN-1 kinase (Fig. 2 B) to further compare the proteins and analyze the subtle characteristics that underlie the force activation hypothesis. When visually comparing the TTN-1 model to the twitchin and titin crystal structures, TTN-1 seems most structurally related to twitchin (Fig. 2, A and B), however there does exist a few varying regions that seem to be upheld only between the TTN-1 model and the structure of human titin. The topology and the bond-structure that holds the regulatory domain in intimate contact with the catalytic core seem closely related for all four kinases. It seems likely that, if the force activation hypothesis is true, all of these enzymes could adhere to such an on/off mechanism.
In conclusion, we have provided evidence that in response to mechanical force, two giant titin-like protein kinase domains unfold in a step-wise manner. The first step is likely to be the movement of the autoinhibitory domain from the catalytic pocket, without complete unfolding of the domain. With the application of increased force to the protein, the kinase domain will rupture, in a biphasic manner. These data bolster the hypothesis that the autoinhibited giant kinases in muscle cells may be activated in response to the forces generated during each contraction/relaxation cycle. Further experiments are underway to test if applying small forces to these enzymes, forces sufficient only to remove the autoinhibitory region (Fig. 6), would indeed result in the activation of these enzymes.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We are grateful to Henry Epstein for suggesting the collaboration between the Benian and Oberhauser labs.
This work was funded by National Institutes of Health grants R01DK073394 (to A.F.O.), RO1AR051466 (to G.M.B.), the John Sealy Memorial Endowment Fund for Biomedical Research (to A.F.O.) by the Polycystic Kidney Foundation (grant 116a2r, A.F.O.) and a training fellowship from the Keck Center for Computational and Structural Biology of the Gulf Coast Consortia (National Library of Medicine grant 5T15LM07093 to T.G.).
Dina N. Greene and Tzintzuni Garcia contributed equally to this work.
Editor: Jane Clarke.
References
- 1.Lange, S., F. Xiang, A. Yakovenko, A. Vihola, P. Hackman, E. Rostkova, J. Kristensen, B. Brandmeier, G. Franzen, B. Hedberg, L. G. Gunnarsson, S. M. Hughes, S. Marchand, T. Sejersen, I. Richard, L. Edstrom, E. Ehler, B. Udd, and M. Gautel. 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 308:1599–1603. [DOI] [PubMed] [Google Scholar]
- 2.Witt, C. C., S. H. Witt, S. Lerche, D. Labeit, W. Back, and S. Labeit. 2008. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 27:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotthardt, M., R. E. Hammer, N. Hubner, J. Monti, C. C. Witt, M. McNabb, J. A. Richardson, H. Granzier, S. Labeit, and J. Herz. 2003. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J. Biol. Chem. 278:6059–6065. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara, T. M., D. B. Flaherty, and G. M. Benian. 2005. Titin/connectin-related proteins in C. elegans: a review and new findings. J. Muscle Res. Cell Motil. 26:435–447. [DOI] [PubMed] [Google Scholar]
- 5.Benian, G. M., J. E. Kiff, N. Neckelmann, D. G. Moerman, and R. H. Waterston. 1989. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature. 342:45–50. [DOI] [PubMed] [Google Scholar]
- 6.Benian, G. M., S. W. L'Hernault, and M. E. Morris. 1993. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein, twitchin, of Caenorhabditis elegans. Genetics. 134:1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Probst, W. C., E. C. Cropper, J. Heierhorst, S. L. Hooper, H. Jaffe, F. Vilim, S. Beushausen, I. Kupfermann, and K. R. Weiss. 1994. cAMP-dependent phosphorylation of Aplysia twitchin may mediate modulation of muscle contractions by neuropeptide cotransmitters. Proc. Natl. Acad. Sci. USA. 91:8487–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegman, M. J., D. Funabara, S. Kinoshita, S. Watabe, D. J. Hartshorne, and T. M. Butler. 1998. Phosphorylation of a twitchin-related protein controls catch and calcium sensitivity of force production in invertebrate smooth muscle. Proc. Natl. Acad. Sci. USA. 95:5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty, D. B., K. M. Gernert, N. Shmeleva, X. Tang, K. B. Mercer, M. Borodovsky, and G. M. Benian. 2002. Titins in C. elegans with unusual features: coiled-coil domains, novel regulation of kinase activity and two new possible elastic regions. J. Mol. Biol. 323:533–549. [DOI] [PubMed] [Google Scholar]
- 10.Lei, J., X. Tang, T. C. Chambers, J. Pohl, and G. M. Benian. 1994. Protein kinase domain of twitchin has protein kinase activity and an autoinhibitory region. J. Biol. Chem. 269:21078–21085. [PubMed] [Google Scholar]
- 11.Hu, S. H., M. W. Parker, J. Y. Lei, M. C. Wilce, G. M. Benian, and B. E. Kemp. 1994. Insights into autoregulation from the crystal structure of twitchin kinase. Nature. 369:581–584. [DOI] [PubMed] [Google Scholar]
- 12.Kobe, B., J. Heierhorst, S. C. Feil, M. W. Parker, G. M. Benian, K. R. Weiss, and B. E. Kemp. 1996. Giant protein kinases: domain interactions and structural basis of autoregulation. EMBO J. 15:6810–6821. [PMC free article] [PubMed] [Google Scholar]
- 13.Wilmann, M., M. Gautel, and O. Mayans. 2000. Activation of calcium/calmodulin regulated kinases. Cell Mol. Biol. 46:883–894. [PubMed] [Google Scholar]
- 14.Heierhorst, J., X. Tang, J. Lei, W. C. Probst, K. R. Weiss, B. E. Kemp, and G. M. Benian. 1996. Substrate specificity and inhibitor sensitivity of Ca2+/S100-dependent twitchin kinases. Eur. J. Biochem. 242:454–459. [DOI] [PubMed] [Google Scholar]
- 15.Mayans, O., P. F. van der Ven, M. Wilm, A. Mues, P. Young, D. O. Furst, M. Wilmanns, and M. Gautel. 1998. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 395:863–869. [DOI] [PubMed] [Google Scholar]
- 16.Grater, F., J. Shen, H. Jiang, M. Gautel, and H. Grubmuller. 2005. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 88:790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullard, B., W. A. Linke, and K. Leonard. 2002. Varieties of elastic protein in invertebrate muscles. J. Muscle Res. Cell Motil. 23:435–447. [DOI] [PubMed] [Google Scholar]
- 18.Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, E., T. Garcia, S. Hultgren, and A. F. Oberhauser. 2006. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 91:3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberhauser, A. F., C. Badilla-Fernandez, M. Carrion-Vazquez, and J. M. Fernandez. 2002. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319:433–447. [DOI] [PubMed] [Google Scholar]
- 21.Oberhauser, A. F., P. K. Hansma, M. Carrion-Vazquez, and J. M. Fernandez. 2001. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 98:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- 23.Florin, E.-L., M. Rief, H. Lehmann, M. Ludwig, C. Dornmair, V. T. Moy, and H. E. Gaub. 1995. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 10:895–901. [Google Scholar]
- 24.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of λ-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 25.Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- 26.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 27.Lu, H., B. Isralewitz, A. Krammer, V. Vogel, and K. Schulten. 1998. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips, J. C., B. Rosemary, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., W. A. Linke, A. F. Oberhauser, M. Carrion-Vazquez, J. G. Kerkvliet, H. Lu, P. E. Marszalek, and J. M. Fernandez. 2002. Reverse engineering of the giant muscle protein titin. Nature. 418:998–1002. [DOI] [PubMed] [Google Scholar]
- 30.Bullard, B., T. Garcia, V. Benes, M. C. Leake, W. A. Linke, and A. F. Oberhauser. 2006. The molecular elasticity of the insect flight muscle proteins projectin and kettin. Proc. Natl. Acad. Sci. USA. 103:4451–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Best, R. B., S. B. Fowler, J. L. Toca-Herrera, and J. Clarke. 2002. A simple method for probing the mechanical unfolding pathway of proteins in detail. Proc. Natl. Acad. Sci. USA. 99:12143–12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma, D., O. Perisic, Q. Peng, Y. Cao, C. Lam, H. Lu, and H. Li. 2007. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. USA. 104:9278–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furst, D. O., M. Osborn, R. Nave, and K. Weber. 1988. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 106:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tskhovrebova, L., J. Trinick, J. A. Sleep, and R. M. Simmons. 1997. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 387:308–312. [DOI] [PubMed] [Google Scholar]
- 35.Obermann, W. M., M. Gautel, F. Steiner, P. F. van der Ven, K. Weber, and D. O. Furst. 1996. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J. Cell Biol. 134:1441–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moerman, D. G., G. M. Benian, R. J. Barstead, L. A. Schriefer, and R. H. Waterston. 1988. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 2:93–105. [DOI] [PubMed] [Google Scholar]
- 37.Carrion-Vazquez, M., H. Li, H. Lu, P. E. Marszalek, A. F. Oberhauser, and J. M. Fernandez. 2003. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10:738–743. [DOI] [PubMed] [Google Scholar]
- 38.Ainavarapu, S. R., L. Li, C. L. Badilla, and J. M. Fernandez. 2005. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 89:3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Best, R. B., B. Li, A. Steward, V. Daggett, and J. Clarke. 2001. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 81:2344–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, L., S. Wetzel, A. Pluckthun, and J. M. Fernandez. 2006. Stepwise unfolding of ankyrin repeats in a single protein revealed by atomic force microscopy. Biophys. J. 90:L30–L32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberhauser, A. F., and M. Carrion-Vazquez. 2008. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 283:6617–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, H., and K. Schulten. 1999. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins. 35:453–463. [PubMed] [Google Scholar]
- 43.Sotomayor, M., and K. Schulten. 2007. Single-molecule experiments in vitro and in silico. Science. 316:1144–1148. [DOI] [PubMed] [Google Scholar]
- 44.Carrion-Vazquez, M., A. F. Oberhauser, T. E. Fisher, P. E. Marszalek, H. Li, and J. M. Fernandez. 2000. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 74:63–91. [DOI] [PubMed] [Google Scholar]
- 45.Labeit, S., M. Gautel, A. Lakey, and J. Trinick. 1992. Towards a molecular understanding of titin. EMBO J. 11:1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.