Abstract
Purified bovine rhodopsin was reconstituted into vesicles consisting of 1-stearoyl-2-oleoyl phosphatidylcholine or 1-stearoyl-2-docosahexaenoyl phosphatidylcholine with and without 30 mol % cholesterol. Rhodopsin stability was examined using differential scanning calorimetry (DSC). The thermal unfolding transition temperature (Tm) of rhodopsin was scan rate-dependent, demonstrating the presence of a rate-limited component of denaturation. The activation energy of this kinetically controlled process (Ea) was determined from DSC thermograms by four separate methods. Both Tm and Ea varied with bilayer composition. Cholesterol increased the Tm both the presence and absence of docosahexaenoic acid acyl chains (DHA). In contrast, cholesterol lowered Ea in the absence of DHA, but raised Ea in the presence of 20 mol % DHA-containing phospholipid. The relative acyl chain packing order was determined from measurements of diphenylhexatriene fluorescence anisotropy decay. The Tm for thermal unfolding was inversely related to acyl chain packing order. Rhodopsin kinetic stability (Ea) was reduced in highly ordered or disordered membranes. Maximal kinetic stability was found within the range of acyl chain order found in native bovine rod outer segment disk membranes. The results demonstrate that membrane composition has distinct effects on the thermal versus kinetic stabilities of membrane proteins, and suggests that a balance between membrane constituents with opposite effects on acyl chain packing, such as DHA and cholesterol, may be required for maximum protein stability.
INTRODUCTION
An extensive literature, accumulated over the past 40 years, demonstrates that variation of essentially any aspect of membrane lipid composition leads to a modulation of the functional efficacy or oligomeric state of membrane proteins (1–3). The effects that membrane composition may also have on membrane protein-folding and structural stability have received somewhat less attention. Characterizing the fundamental forces that determine the thermodynamic stability of membrane proteins is an area of intense interest (4). The energetic contribution of lipid-protein interactions to membrane protein stability is likely to be as important for this class of proteins as the energetics of protein-solvent interactions is to the stability of soluble proteins. Determining the effects of membrane composition on the energy required for thermal denaturation provides a measure of the effects of the lipid bilayer on relative protein-folding energetics.
Rhodopsin, a G protein-coupled receptor found in the disk membrane of retinal rod cells, is responsible for vision under low-light conditions. Light-induced isomerization of the 11-cis retinal chromophore initiates a series of rapid conformational changes, resulting in the active metarhodopsin II state (MetaII), which is in quasistable equilibrium with an inactive conformation, MetaI (5). MetaII binds and activates the G protein transducin, Gt, initiating the visual-signal transduction cascade in retinal rod cells (6–9). The MetaI-MetaII conformational equilibrium is sensitive to membrane physical properties such as elastic curvature stress (10,11), acyl chain packing free volume (12), and hydrophobic thickness (13). The MetaII-MetaI equilibrium is also altered by aspects of membrane composition, including head group (14), acyl chain unsaturation (11,15–17), and cholesterol (18,19). The effects of cholesterol and acyl chain unsaturation are particularly relevant to receptor function in humans, because these two aspects of biological membrane composition are subject to alteration by dietary fat intake or changes in lipid metabolism. The formation of MetaII is particularly enabled by the presence of docosahexaenoic acid acyl chain (DHA) 22:6n3, which is present in abundance in the rod outer segment disk membrane (20). In contrast, increased cholesterol reduces the formation of MetaII in native disk membranes and in rhodopsin-containing proteoliposomes. Cholesterol is present in bovine disk membranes at 5–30 mol %, depending on the age of the disk (21,22). One of the interesting questions posed by the disk membrane-rhodopsin system involves how the contrasting forces exerted by high levels of 22:6n3 acyl chains and significant levels of cholesterol are balanced to provide optimal receptor function and structural stability.
The goal of this study was a detailed calorimetric analysis of the combined effects of 22:6n3 acyl chains and cholesterol on integral membrane protein stability. Protein stability is a term that requires explicit definition, because it can refer to several different characteristics of protein energetics. We refer to thermal stability as the temperature at which a protein denatures, defined here as the maximum heat capacity of a thermal unfolding transition (Tm). Using this terminology, more thermostable proteins possess higher denaturation temperatures. The thermal stability of a protein is determined by a complex balance of the number and energies of intraprotein hydrogen bonds and salt bridges (23), interactions with other molecules such as lipid and solvent, and the interplay between the entropy and enthalpy change of unfolding (24). The thermodynamic stability of a protein is defined as the difference in free energy between the native and denatured states (24). For proteins that undergo irreversible thermal unfolding transitions, thermodynamic stability in terms of free energy cannot be determined by calorimetry (25). The kinetic stability of a protein refers to the rate at which a denaturation process occurs, where kinetic destabilization results in an increase in the rate of denaturation. An integral feature of any kinetic process is the activation energy barrier (Ea), where large activation energy barriers result in low rates of denaturation. Thermal and kinetic stabilities are quite different properties, with thermal stability resulting from the energies of intramolecular and intermolecular interactions, whereas kinetic stability results from the time-dependence of the physical mechanics of the denaturation processes.
Receptor thermal stability may be measured directly from the thermogram obtained by differential scanning calorimetry (DSC). Kinetic stability is obtained via model-dependent analyses of DSC data, such as the widely used methods described by Sanchez-Ruiz et al. (26). A “kinetically controlled” denaturation process is a process whose rate is on the order of the timescale of the DSC measurement, such that alterations in the rate of measurement result in apparent changes in the kinetic process being observed. At high thermal scan rates, few of the rate-limited denaturation events have time to occur at a given temperature. At slower scan rates, more of the rate-limited events have time to occur at lower temperatures, causing a redistribution of denaturation events in the temperature regime, and a shift of the observed thermal transition curve to lower temperatures. This alters both the shape of the curve and the temperature of transition maximum, Tm. The four methods developed by Sanchez-Ruiz et al. (26) involve calculation of Ea from both the shape of the transition and the shift in Tm induced by variation in scan rate (26).
EXPERIMENTAL PROCEDURES
Materials
Concanavalin A sepharose was purchased from Amersham Biosciences (Piscataway, NJ). Phospholipids 18:0, 18:1PC, and 18:0,22:6n3PC were purchased from Avanti Polar Lipids (Alabaster, AL). The fluorescent probe diphenylhexatriene (DPH) was purchased from Molecular Probes (Eugene, OR). Cholesterol was purchased from Calbiochem (San Diego, CA). Bovine retinas were from James and Wanda Lawson (Lincoln, NE).
Sample preparation
Rod outer segments (ROS) were isolated from bovine retinas, using the modified Shake-ate method (27). The ROS were solubilized in 30 mM octyl-β-D-glucopyranoside (OG) and rhodopsin was purified using a concanavalin A affinity column (28). Rhodopsin was eluted with 150 mM mannoside, and fractions with an absorbance ratio (A280/A500) <1.8 were pooled and dialyzed against OG-containing buffer to remove mannoside. Purified rhodopsin was analyzed for residual native phospholipid by the method of Bartlett (29), and the residual phospholipid content was <1 phospholipid per 10 rhodopsins.
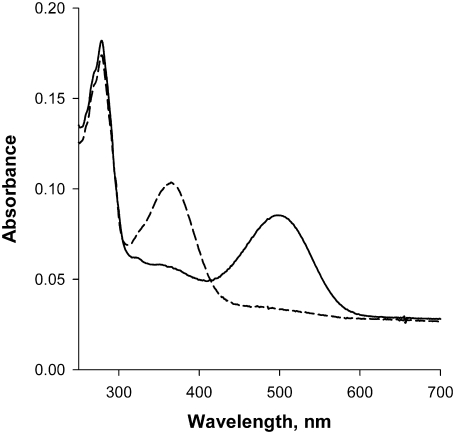
Rhodopsin was reconstituted into vesicles consisting of five different compositions, varying in acyl chain composition and cholesterol: 1), 18:0, 18:1 phosphatidylcholine (1-stearoyl-2-oleoyl-phosphatidyl choline, or SOPC); 2), SOPC combined with 30 mol % cholesterol (SOPC/chol); 3), 18:0, 22:6n3 phosphatidyl choline (1-stearoyl-2-docosahexaenoyl phosphatidyl choline, or SDPC); 4), SOPC + 20 mol % SDPC (SOPC/SDPC); and 5), SOPC/SDPC plus 30 mol % cholesterol (SOPC/SDPC/chol). Proteoliposomes were prepared using the rapid dilution method, as previously described (30). Briefly, lipids in chloroform were mixed at the desired ratio, lyophilized to complete dryness, dissolved in OG-containing buffer, and mixed with purified rhodopsin in OG buffer. This mixture was equilibrated at 4°C with gentle stirring for 4 h to ensure complete mixing of lipid-containing and rhodopsin-containing micelles. Suspensions of rhodopsin/lipid/OG mixed micelles were slowly dripped (∼1 drop/s) into rapidly stirred cold PIPES-buffered solution (PBS) (10 mM PIPES, 30 mM NaCl, 60 mM KCl, 2 mM MgCl2, 30 μM DTPA, pH 7.0) to reduce the OG concentration below 10 mM. The resulting dilute proteoliposome suspension was concentrated by spinning at 1000 × g in a Sorvall GSA rotor (Thermo Scientific, Waltham, MA) in Vivacell 70 10,000 MWCO spin filters (VivaScience, Sartorius Stedim Biotech, Goettingen, Germany) and dialyzed against PBS (100-fold excess, changed 3 times) to remove OG monomers. All final samples had a lipid/rhodopsin ratio of 100:1 ± 5%, as shown by analysis for rhodopsin content (ΔA500 nm, using an extinction coefficient of 40,600 cm−1 M−1 (31)) and phospholipid content (according to the method of Bartlett (29)). All sample preparations involving 22:6n3-containing phospholipids were conducted in an argon-filled glove box, and all buffers were thoroughly degassed with argon before use. The functionality of rhodopsin in liposomes was judged by the presence of the native 11-cis retinal absorption band at 500 nm, and by the complete conversion of this band to a band at ∼380 nm by exposure to light, as shown in Fig. 1.
FIGURE 1.
Example of dark-adapted rhodopsin (solid curve) and bleached rhodopsin (dashed curve) in SDPC proteoliposomes. The sold curve shows the broad absorption band at 500 nm of native, unbleached rhodopsin, whereas the dashed curve shows the characteristic shift to 380 nm of bleached rhodopsin.
Differential scanning calorimetry
Differential scanning calorimetry measurements were performed with a 6100 Nano-Scan II calorimeter equipped with capillary cells (Calorimetry Sciences, Provo, UT). Samples were degassed and loaded into the DSC under a stream of argon in complete darkness, using infrared night-vision goggles. Cells were sealed in the dark and maintained at a pressure of 4.0 atm to maintain a stable baseline. Samples were scanned at rates of 0.25, 0.5, 1.0, and 1.5 C/min from 45°C to 95°C. A second heating scan was repeated after the sample was cooled and equilibrated at 40°C, and this scan was used to correct for heat capacity changes not related to the protein unfolding transition. The DSC scans were normalized to excess molar heat capacity (Cp), using CpCalc 2.1 (Calorimetry Sciences), and the thermograms were analyzed for Tm and ΔH using an Origin 7 (OriginLab, Northampton, MA) baseline correction, as described elsewhere (32). Thermograms were fit using the equation Asym2Sig in Origin.
Determination of Ea from DSC data
The methods of determining Ea developed by Sanchez-Ruiz et al. (26) make use of either the shift in Tm with change in scan rate, or various aspects of the shape of the denaturation transition. Each of the methods of calculating the activation energy are based on a two-state model of the denaturation process, N → D, in which native molecules proceed irreversibly in a single step to the denatured state with a first-order rate constant k (26). The scan rate-independent methods utilize characteristics of the thermogram such as the total heat of the transition, Qt (obtained by integrating the peak), the heat evolved as a function of temperature, Q(T); the scan rate, ν; and the heat capacity as a function of temperature, Cp(T). The Asym2Sig equations describing the thermograms were integrated in Mathcad (Insightful, Inc., Seattle, WA) to obtain Qt − Q(T) and Qt/(Qt − Q(T)), which are required for two of the methods (described below).
Method A
In this method, the slope of an Arrhenius plot is used to calculate Ea. The denaturation rate constant, k, is calculated from the shape of a single thermogram according to k(T) = νCp(T)/(Qt − Q(T)). A plot of ln(k) vs. 1/T yields a line whose slope is −Ea/R (26), where R is the gas constant. Multiple thermograms collected at different scan rates should yield similar activation energies.
Method B
The unique feature of this method is explicit use of the dependence of Tm on scan rate. It is independent of thermogram shape, and utilizes thermograms collected at different scan rates to calculate the thermal denaturation activation energy according to 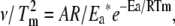 where A is the frequency factor or activation entropy (33). As in method A, the slope of the plot ln(
where A is the frequency factor or activation entropy (33). As in method A, the slope of the plot ln(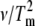 ) vs. 1/Tm is equal to −Ea/R (26).
) vs. 1/Tm is equal to −Ea/R (26).
Method C
This method strictly analyzes the shape of the transition, and does not make use of the scan rate. By plotting ln[ln(Qt/(Qt − Q(T))] vs. 1/T, the slope of the line is again equal to −Ea/R. The intercept of this method corresponds to the Tm of the transition, and should be equal to the thermogram Tm (26).
Method D
A unique feature of this method is the explicit dependence of Ea on the height of the transition peak, 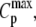 relative to the peak area Qt. This method analyzes the shape of the thermogram according to
relative to the peak area Qt. This method analyzes the shape of the thermogram according to 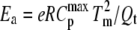 (26).
(26).
Time-resolved fluorescence
Vesicles were suspended in PBS buffer at a phospholipid concentration of 0.5 mM. The DPH was dissolved in tetrahydrofuran, and added at a phospholipid/DPH ratio of 300:1. Fluorescence lifetime and differential polarization measurements at 37°C were acquired with a K2 multifrequency cross-correlation phase fluorometer (ISS, Urbana, IL), as previously described (34). Fifteen modulation frequencies, logarithmically spaced from 5–150 MHz, were used for both lifetime and differential polarization measurements. All measurements were repeated a minimum of three times with each sample composition. Measured polarization-dependent differential phases and modulation ratios for each sample were combined with the measured total intensity decay to yield the anisotropy decay, r(t). All anisotropy decay data were analyzed in terms of the Brownian rotational diffusion model (34,35). The results of the Brownian rotational diffusion model-based analysis were interpreted in terms of an angular distribution function, which is symmetric about θ = π/2, f(θ). The extent to which the equilibrium orientational freedom of DPH is restricted by the phospholipid acyl chains was quantified using the disorder parameter, fv, which is proportional to the overlap of f(θ)sinθ, with the orientational distribution corresponding to randomly oriented DPH (34,36).
Meta I-Meta II equilibrium measurements
Spectra of the MI-MII equilibrium of photoactivated rhodopsin were collected and deconvolved according to Straume et al. (37). Briefly, rhodopsin-containing vesicles were diluted to 0.3 mg/mL in PBS buffer, pH 7.0, and equilibrated at 37°C in a thermally regulated sample holder. A set of four absorption spectra was collected sequentially in an Agilent 8453 diode array spectrophotometer (Agilent Technologies, Santa Clara CA). These included the spectra acquired 1), after the sample was equilibrated in the dark at 37°C; 2), 3 s after the sample was 15–20% bleached by a 520-nm flash; 3), 10 min after addition of 30 mM hydroxylamine to convert bleached rhodopsin to opsin and retinal oxime; and 4), after complete bleaching of the sample. Individual MI and MII spectra were deconvolved from their equilibrium mixture, using a nonlinear least-squares method, and the equilibrium constant, Keq, was calculated according to Keq = [MII]/[MI].
RESULTS
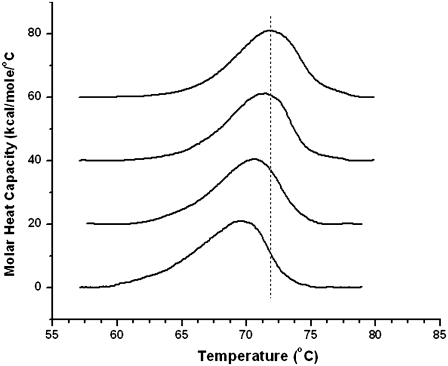
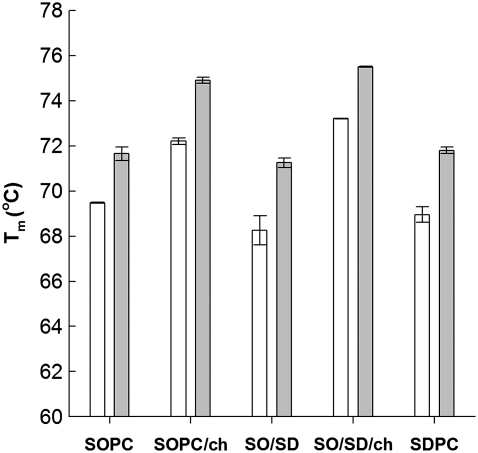
The DSC thermograms were collected by heating from 40°C to 95°C at four different scan rates for rhodopsin in all five bilayer compositions: SOPC, SOPC combined with 30 mol % cholesterol (SOPC/chol), SDPC, SOPC + 20 mol % SDPC (SOPC/SDPC), and SOPC/SDPC plus 30 mol % cholesterol (SOPC/SDPC/chol). Increasing the scan rate from 0.25°C/min to 1.5°C/min caused a progressive increase in the apparent Tm of thermal unfolding for rhodopsin in all five bilayer compositions, as shown for rhodopsin in SOPC in Fig. 2. The Tm for thermal denaturation of rhodopsin was also dependent on lipid composition (Fig. 3). At the fastest scan rate, the Tm of rhodopsin in SOPC increased by 3.3°C with the addition of 30 mol % cholesterol. In 80/20 SOPC/SDPC, cholesterol increased Tm by 4.25°C, suggesting that the presence of a polyunsaturated 22:6n3 acyl chain in the membrane amplifies the thermostabilizing effects of cholesterol, perhaps by enhancing the interaction between rhodopsin and cholesterol. At all scan rates, the essential features of the effects of cholesterol and 22:6n3 acyl on thermostability were qualitatively similar. Cholesterol raised Tm by 3–4°C, and the difference in Tm between rhodopsin in SOPC and SDPC was <0.5°C.
FIGURE 2.
Thermograms of rhodopsin in SOPC collected at scan rates of 1.5°C/min, 1.0°C/min, 0.5°C/min, and 0.25°C/min (from top to bottom, respectively). Dotted line corresponds to the Tm of rhodopsin in SOPC at 1.5°C/min, and is included to illustrate the shift in Tm with scan rate.
FIGURE 3.
Effects of lipid composition on thermal denaturation temperature and scan rate-induced Tm shift of rhodopsin. Gray bars correspond to transition temperature at the fastest scan rate (1.5°C/min, average of two scans), and open bars correspond to Tm at the slowest scan rate (0.25°C/min).
The Tm of rhodopsin was found to be scan rate-dependent for all compositions, consistent with a previous report for rhodopsin in ROS disk membranes (38). To facilitate comparisons with other studies, the scan rate-induced Tm shift (ΔTm) is normalized to the scan rate range, Δν. The value of ΔTm/Δν ranged from 1.7–2.4°C, and varied with membrane composition, as shown in Table 1. The addition of 30 mol % cholesterol produced opposite changes in Tm/Δν in SOPC and 80/20 SOPC/SDPC. The addition of 30% cholesterol to SOPC increased ΔTm/Δν by ∼0.5°C, but in SOPC/SDPC, 30 mol % cholesterol lowered ΔTm/Δν by 0.6°C. The addition of 20 mol % SDPC to an SOPC membrane raised ΔTm/Δν by ∼0.7°C, to a value approximately the same as that obtained in pure SDPC. This suggests that with respect to the rate-limited process, the destabilizing effects of SDPC may saturate at 20 mol %. Interestingly, the addition of both SDPC and cholesterol to an SOPC membrane resulted in a ΔTm/Δν close to what is observed in pure SOPC bilayers, suggesting that the individual destabilizing effects of cholesterol and SDPC may cancel each other out when both cholesterol and 22:6n3 acyl chains are present in the membrane.
TABLE 1.
Effects of membrane composition on rhodopsin thermal transition properties
| Sample | ΔTm/Δν | Tm (°C) | ΔHcal (kJ/mol) | 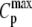 |
T1/2 (°C) |
|---|---|---|---|---|---|
| SOPC | 1.7 ± 0.30 | ||||
| 1.5°C/min | 71.65 ± 0.30 | 590 ± 21 | 20.4 ± 1.5 | 6.1 ± 0.2 | |
| 1.0 | 71.43 ± 0.04 | 536 ± 25 | 20.6 ± 0.7 | 5.7 ± 0.1 | |
| 0.5 | 70.73 ± 0.18 | 544 ± 17 | 20.6 ± 0.2 | 5.8 ± 0.1 | |
| 0.25 | 69.49 ± 0.02 | 578 ± 4 | 21.1 ± 0.2 | 6.2 ± 0.0 | |
| SOPC + cholesterol | 2.2 ± 0.20 | ||||
| 1.5°C/min | 74.90 ± 0.14 | 766 ± 8 | 25.1 ± 0.4 | 6.6 ± 0.0 | |
| 1.0 | 74.40 ± 0.14 | 766 ± 42 | 25.3 ± 0.5 | 6.5 ± 0.3 | |
| 0.5 | 73.50 ± 0.28 | 729 ± 38 | 26.4 ± 0.4 | 6.1 ± 0.7 | |
| 0.25 | 72.20 ± 0.14 | 846 ± 59 | 25.5 ± 3.2 | 7.1 ± 0.1 | |
| SOPC/SDPC | 2.4 ± 0.67 | ||||
| 1.5°C/min | 71.25 ± 0.21 | 846 ± 38 | 27.2 ± 1.4 | 7.0 ± 0.1 | |
| 1.0 | 70.95 ± 0.07 | 909 ± 92 | 30.0 ± 1.3 | 6.8 ± 0.1 | |
| 0.5 | 69.95 ± 0.49 | 1005 ± 50 | 29.8 ± 0.4 | 7.2 ± 0.4 | |
| 0.25 | 68.25 ± 0.64 | 1197 ± 301 | 38.3 ± 4.7 | 7.3 ± 1.0 | |
| SOPC/SDPC + cholesterol | 1.8 ± 0.00 | ||||
| 1.5°C/min | 75.50 ± 0.00 | 557 ± 13 | 21.2 ± 0.6 | 5.8 ± 0.0 | |
| 1.0 | 75.30 ± 0.28 | 758 ± 193 | 28.2 ± 8.5 | 5.8 ± 0.3 | |
| 0.5 | 74.00 ± 0.00 | 729 ± 155 | 24.0 ± 1.6 | 6.5 ± 0.9 | |
| 0.25 | 73.20 ± 0.00 | 582 ± 50 | 21.8 ± 1.6 | 6.0 ± 0.1 | |
| SDPC | 2.3 ± 0.38 | ||||
| 1.5°C/min | 71.80 ± 0.14 | 720 ± 46 | 25.8 ± 1.2 | 6.3 ± 0.0 | |
| 1.0 | 71.65 ± 0.07 | 716 ± 54 | 25.5 ± 2.9 | 6.3 ± 0.4 | |
| 0.5 | 70.35 ± 0.21 | 967 ± 100 | 29.2 ± 2.3 | 7.1 ± 0.1 | |
| 0.25 | 68.95 ± 0.35 | 1285 ± 130 | 31.2 ± 4.6 | 9.5 ± 0.2 |
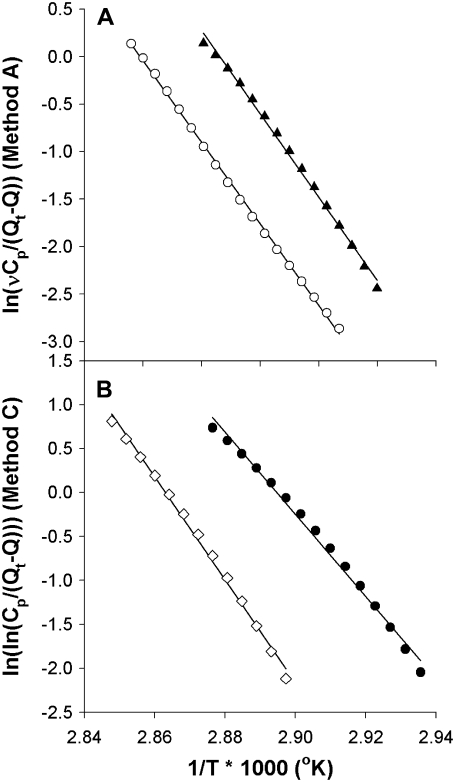
The dependence of Tm on scan rate and the shape of the denaturation transition were used to calculate the activation energy of the kinetically limited process (26), using each of the 4 methods developed by Sanchez-Ruiz et al. (26). Methods A and C use the shape of the thermogram, specifically νCp/(Qt− Q(T)) in method A and Qt/(Qt− Q(T)) in method C, as shown in Fig. 4. Values of Ea determined by methods A and C were in good agreement with each other for every thermogram, i.e., at all scan rates for each membrane composition. In method C, the x axis intercepts correspond to Tm, and these values were compared to the actual thermogram Tm to validate the calculation. This analysis typically yielded values of Tm that were <1°C different from the actual Tm, and data from analyses that exceeded a 1°C error in Tm were discarded.
FIGURE 4.
Methods for determining Ea, based on shape of the thermogram. (A) Representative plots of ln(k) vs. 1/T (method A): rhodopsin in SOPC (solid triangles), rhodopsin in SOPC + 30% cholesterol (open circles). Activation energy is determined from the slope of the line. (B) Method C analysis of rhodopsin in SOPC + 20% SDPC (solid circles) and SOPC/SDPC/cholesterol (open diamonds). Activation energy is determined from the slope of the line.
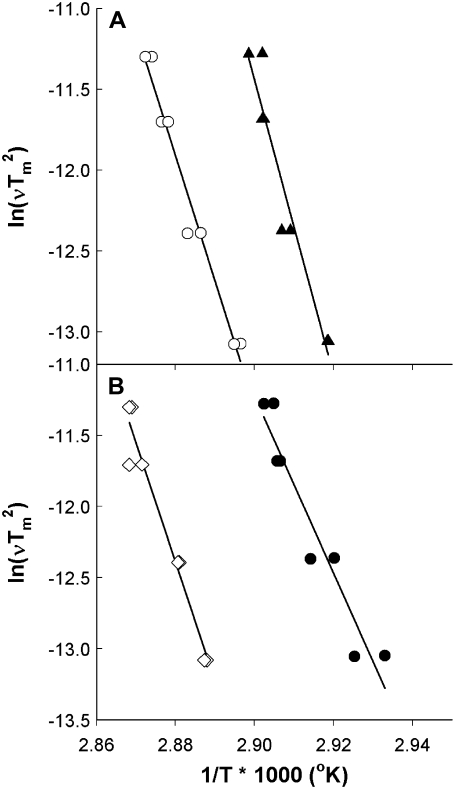
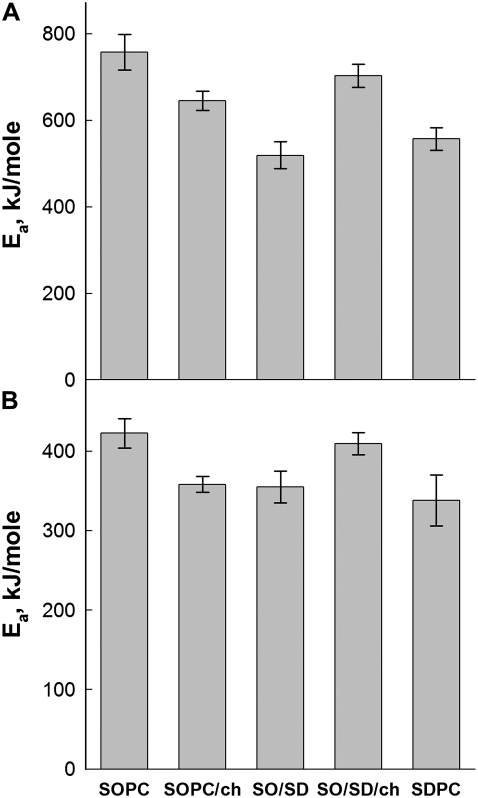
Method B is based on the scan rate-induced temperature shift, and does not depend on the shape of the thermogram. Fig. 5 shows representative plots used to determine activation energy by this method. The shallower slope in the plot of SOPC/cholesterol in Fig. 4 A, reflects the lower activation energy in this sample compared with rhodopsin in pure SOPC. In contrast, the steeper slope of the plot for SOPC/SDPC/cholestrol in Fig. 4 B shows that the addition of cholesterol to an SOPC/SDPC bilayer raises the activation energy of denaturation. The values of Ea derived via method B were approximately twice the values produced by the other 3 methods (Table 2). Method B is based solely on the shift in Tm with scan rate, whereas the other methods are derived from the detailed shape and height of the thermogram. All 4 analytical methods produced values of Ea that varied with membrane composition in an essentially identical manner, as shown in Fig. 6. Because of their similar theoretical basis, the values of Ea determined by methods A, C, and D were combined in Fig. 5 B.
FIGURE 5.
Scan rate-dependent method of determining denaturation activation energies. Activation energies are determined from the slopes of the lines. (A) Application of the scan rate-dependent method to rhodopsin in SOPC (solid triangles) and in SOPC + 30% cholesterol (open circles). (B) Rhodopsin in SOPC + 20% SDPC (solid circles) and in SOPC/SDPC/cholesterol (open diamonds).
TABLE 2.
Values of Ea (kJ/mol) determined by the four methods of Sanchez-Ruiz et al. (26)
| Sample | Method A | Method B | Method C | Method D | Keq | fv |
|---|---|---|---|---|---|---|
| SOPC | 394 ± 14 | 758 ± 41 | 440 ± 10 | 414 ± 19 | 2.00 ± 0.13 | 0.129 ± 0.009 |
| SOPC/cholesterol | 352 ± 23 | 645 ± 22 | 352 ± 31 | 377 ± 31 | 0.94 ± 0.09 | 0.050 ± 0.004 |
| SOPC/SDPC | 335 ± 33 | 519 ± 31 | 406 ± 53 | 356 ± 27 | 2.65 ± 0.21 | 0.139 ± 0.004 |
| SOPC/SDPC/cholesterol | 398 ± 23 | 703 ± 27 | 444 ± 51 | 419 ± 32 | 1.14 ± 0.07 | 0.058 ± 0.003 |
| SDPC | 301 ± 42 | 557 ± 26 | 385 ± 51 | 348 ± 56 | 3.20 ± 0.20 | 0.158 ± 0.005 |
FIGURE 6.
Comparison of activation energies determined by different methods. (A) Activation energies determined by method B. (B) Ea values obtained by averaging methods A, C, and D. In all four methods, the addition of either 22:6 acyl chains or cholesterol separately reduces the activation energy of rhodopsin denaturation. All four methods also suggest that the presence of both cholesterol and 22:6 acyl chains essentially nullifies the destabilizing properties that each exhibits when added alone.
The most striking effect of variation in membrane composition on Ea involves the differential effects of 30 mol % cholesterol. The addition of 30% cholesterol to an SOPC membrane results in a decrease in the Ea for rhodopsin denaturation. The addition of this amount of cholesterol to an 80/20 SOPC/SDPC membrane raises Ea, and both of these effects are statistically significant (Fig. 6). A second unexpected result is that the addition of 20% SDPC to an SOPC membrane also results in a decrease in Ea. All 4 methods indicate that the simultaneous incorporation of SDPC and cholesterol into an SOPC bilayer results in a rhodopsin denaturation activation energy that is nearly the same as in pure SOPC bilayers. All 4 methods show that Ea for rhodopsin thermal denaturation is significantly lower in SDPC than in SOPC. Method B indicates that the Ea of rhodopsin in SDPC is slightly larger than in 80/20 SOPC/SDPC, whereas the average of methods A, C, and D indicates that it is slightly less. The differences are not significant, suggesting that the kinetic acceleration of rhodopsin thermal denaturation by SDPC is saturated at 20 mol % SDPC.
The compositional parameters that varied in this study (cholesterol and DHA content) were selected because they are aspects of biological membrane composition that may be altered by diet. The principal membrane property that is altered by an adjustment of both of these compositional variables is acyl chain packing, or fluidity. To quantify the changes in acyl chain packing because of variations in membrane composition, we measured DPH fluorescence anisotropy decays for all rhodopsin-containing proteoliposomes at 37°C. The effects of membrane composition on acyl chain packing were quantified in terms of the disorder parameter fv (34,36). The values of fv obtained for SOPC/cholesterol and SOPC/SDPC/cholesterol are <0.06, indicating a narrow orientational probability distribution for DPH which is interpreted as the result of relatively condensed acyl chain packing. In the cholesterol-free bilayers, fv ranged from 0.129–0.158, indicating that acyl chain packing in these three bilayers is much less constrained, or more fluid, than in cholesterol-containing bilayers.
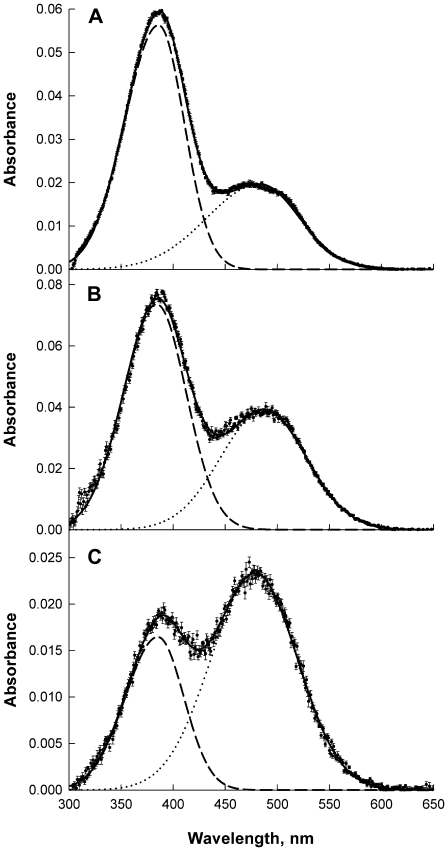
The ability of photo-activated rhodopsin to undergo the MetaI-to-MetaII conformation change was assessed from corrected difference spectra of equilibrium MetaI-MetaII mixtures acquired at 37°C, as shown in Fig. 7. The MetaI/MetaII equilibrium constant ranged from 0.94 for SOPC/cholesterol to 3.2 for rhodopsin in SDPC (Table 2). Both a reduction in Keq with the addition of cholesterol (19,39), and an increase in Keq with the addition of DHA, are consistent with previous reports (11,12,17).
FIGURE 7.
Examples of spectra of MI-MII equilibrium mixtures obtained at 37°C, used for determining Keq: rhodopsin in SDPC (A) SOPC (B) and SOPC/30 mol % cholesterol (C). A spectrum acquired in the dark was subtracted from a spectrum acquired immediately after a brief (∼1 ms) green flash that bleached 20–25% of the sample. The band corresponding to unbleached rhodopsin in the resulting difference spectrum was subtracted after analytical determination of the shape and height of the rhodopsin absorption band.
DISCUSSION
The thermal and kinetic stabilities of rhodopsin are affected by the composition of the surrounding lipid membrane. The values of Tm for thermal denaturation of rhodopsin in Fig. 3 show that the thermostability of rhodopsin in SOPC bilayers is increased by ∼3°C by the addition of 30% cholesterol, and modestly decreased by ∼0.5°C by the addition of 20% SDPC. The thermal stabilization of rhodopsin by cholesterol, and destabilization by polyunsaturated acyl chains, was previously observed (40,41). We report here that the presence of both 30 mol % cholesterol and 20% SDPC in an SOPC bilayer results in rhodopsin that is ∼4.4°C more thermostable than in 80/20 SOPC/SDPC, and ∼0.6°C more thermostable than rhodopsin in SOPC/cholesterol. This suggests that the presence of SDPC in an SOPC bilayer containing cholesterol amplifies the thermostabilizing effect of cholesterol on rhodopsin.
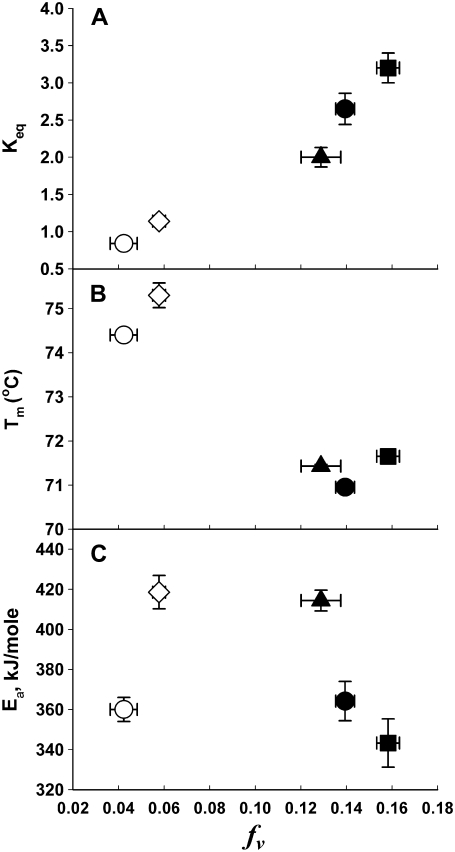
We considered a possible relationship between acyl chain packing and rhodopsin stability by comparing the parameters that characterize the thermal denaturation of rhodopsin with acyl chain packing, as summarized by fv (Fig. 8, B and C). This comparison was motivated by the linear relationship between fv and the MetaI-to MetaII conformation change, summarized by Keq, reported previously for rhodopsin in ROS disk membranes (19) and in reconstituted membranes consisting of a wide range of phospholipid compositions (12,18). For the bilayer compositions examined in this study, a comparison of these two parameters at 37°C showed this same linear relationship, as seen in Fig. 8 A. However, Fig. 8, B and C show that there is not a simple relationship between acyl chain packing and the thermal or kinetic stability of rhodopsin.
FIGURE 8.
Relationship between acyl chain packing, fv, and parameters that summarize rhodopsin conformation change (Keq) or thermal denaturation for all 5 bilayer compositions: SOPC/cholesterol (open circles), SOPC/SDPC/cholesterol (open diamonds), SOPC (solid triangles), SOPC/SDPC (solid circles), and SDPC (solid squares). (A) The MetaI-MetaII equilibrium constant, Keq, has a linear relationship with fv. (B) Rhodopsin thermostability, Tm, at a scan rate of 1.5°C/min. (C) Denaturation activation energies, Ea, determined from the average of methods A, C, and D.
A striking feature of Fig. 8 B is that the values of Tm for rhodopsin in the 2 cholesterol-containing membranes (open symbols) vary by ∼0.9°C, and are ∼4°C higher than Tm in the 3 membranes without cholesterol (solid symbols), which vary by only ∼0.7°C. One clear difference between these 2 groups is the hydrophobic thickness of the bilayer. Cholesterol at 30 mol % increases the phosphate-to-phosphate spacing of SOPC by ∼5 Å (42), whereas an SDPC bilayer is <1 Å thicker than an SOPC bilayer (43). This suggests that the 3 cholesterol-free bilayers have essentially the same hydrophobic thickness, whereas the 2 cholesterol-containing bilayers are ∼5 Å thicker. Previous studies demonstrated that the thermal and thermodynamic stabilities of membrane-spanning proteins are altered by changes in the hydrophobic thickness of the surrounding membrane. In di-saturated lipids, the ΔGo for urea-induced reversible unfolding of OmpA increases ∼8 kJ mol−1, for a 5-Å increase in hydrophobic thickness (44), and cytochrome c oxidase is 6°C more thermostable in di-18:1n9(trans) phosphatidylcholine (PC) bilayers than in di-14:0 PC bilayers (45). Thus, the variation in Tm with bilayer composition in Fig. 8 B may reflect differences in membrane thickness. The lack of correlation between acyl chain packing and Tm is consistent with a previous study in which fv was varied by changing the lipid/protein ratio in SDPC membranes, but there was no variation in Tm (46).
Fig. 8 C shows the relationship between acyl chain packing and the activation energy barrier to thermal denaturation, Ea, and summarizes the complex relationship between bilayer composition and Ea. In the cholesterol-free bilayers, SOPC (solid triangles) produced the highest value of Ea (420 kJ/mol), SDPC (solid squares) produced the lowest value of Ea (345 kJ/mol), and 80/20 SOPC/SDPC (solid circles) had an intermediate value (365 kJ/mol). This large variation indicates that bilayer thickness is unlikely to determine Ea, because these 3 bilayers have essentially the same hydrophobic thickness, as discussed above. The addition of 30 mol % cholesterol produced opposite effects, depending on the host phospholipid matrix; it lowered Ea by 60 kJ mol in SOPC (open circles), and raised Ea by 55 kJ/mol in the SOPC/SDPC mixture (open diamonds).
It is particularly informative to consider the similar values of Ea obtained in SOPC and in the ternary lipid mixture SOPC + 20% SDPC + 30% cholesterol. The addition of either cholesterol or SDPC to SOPC lowers Ea, so the lack of a destabilizing effect in the ternary mixture is surprising and suggests several possibilities. One possibility is that rhodopsin denaturation activation energy is controlled by acyl chain packing, regardless of bilayer composition. Fig. 8 C, suggests that the kinetic stability of rhodopsin may be maximal within an intermediate range of acyl chain packing. We note that the apparent maximal value of Ea occurs within the range of free volume reported for native disk membranes (fv ∼0.12) (36). A biphasic effect of acyl chain packing on rhodopsin kinetic stability is consistent with the observations of Albert and Boesze-Battaglia (47), who found that Ea for rhodopsin thermal denaturation was increased by cholesterol up to 14 mol %, and was decreased by higher levels of cholesterol. Nonlinear effects of cholesterol were also observed in functional studies of membrane proteins (48). Several proteins, including the Na/Ca exchanger (49), Na/K ATPase (50), glutamate transporter (51), and GABA transporter (51), exhibit an increase in activity with increasing cholesterol concentrations, followed by a decrease in activity at higher cholesterol concentrations. The comparison between acyl chain packing and Ea in Fig. 8 C suggests that membranes may modulate the kinetics of protein thermal denaturation.
A second possibility is that specific rhodopsin-lipid interactions play a role in the modulation of denaturation kinetics. Evidence for specific GPCR-cholesterol and GPCR-DHA interactions was presented in a number of reports. Albert et al. reported fluorescence resonance energy transfer between rhodopsin and a fluorescent cholesterol analog which was specifically quenched by cholesterol, but not ergosterol, and suggested there may be a specific binding site on rhodopsin for cholesterol (52). The recently reported crystal structure of the β2 adrenergic receptor includes ordered cholesterol molecules in close proximity to the protein (53). Soubias et al. reported magnetization transfer between rhodopsin and both DHA and oleic acid acyl chains, and proposed that there are distinct interaction sites on the surface of rhodopsin for these two acyl chains (54). In addition, molecular dynamics simulations provide support for rhodopsin-DHA interactions (55,56). Further experiments involving the incremental titration of cholesterol and DHA-containing phospholipid and variations in rhodopsin/lipid ratio will be required to unambiguously the possible specific roles of interactions between rhodopsin DHA acyl chains and cholesterol in determining rhodopsin kinetic stability.
A third possibility is that lipid segregation in the ternary mixture prevents cholesterol or DHA from interacting with rhodopsin as they do in the binary mixtures, leaving the denaturation activation energy unchanged. In these pure lipid bilayers, cholesterol interacts with both SOPC and SDPC via association with the saturated 18:0 acyl chain (57). Thus, it seems unlikely that that the ternary SOPC/SDPC/cholesterol mixture is not uniformly mixed. However, the possibility that rhodopsin may induce de-mixing of this ternary mixture has not been explored.
Recent reports demonstrate that rhodopsin reconstituted in phospholipid bilayers does not exist strictly as monomers (13,58). In particular, Botelho et al. showed that the extent of rhodopsin monodispersion or oligomerization varies with bilayer thickness over the thickness range spanned by di-14:1 PC and di20:1 PC, for a lipid/rhodopsin ratio of 100 (13). They also showed that rhodopsin dispersion varies substantially with lipid/protein ratio. Their results indicate that at the 100:1 lipid/rhodopsin ratio employed in this study, the relative monodispersion of rhodopsin is quite high. Variation in rhodopsin monodispersion/oligomerization with bilayer thickness suggests that rhodopsin may be dispersed differently in cholesterol-containing and cholesterol-free bilayers. However, the large variation in Ea within each of these groups indicates that the relative oligomeric state does not play a role in determining the kinetic stability of rhodopsin thermal denaturation.
Previous studies that used all four methods of Sanchez-Ruiz et al. (26) reported discrepancies between the values of Ea, as summarized in Table 3. Values of Ea determined by method B often differed significantly from the values obtained via the methods based on the detailed shape of the DSC thermogram (methods A, C, and D). Various ranges of scan rates were used in the studies summarized in Table 3, and thus the ratio of Tm shift to scan rate range, ΔTm/Δν, is provided to facilitate comparisons of the magnitude of the scan rate-induced Tm shift. Values of ΔTm/Δν are weakly correlated with the extent of agreement between the four methods. For proteins with relatively large Tm shifts, such as concanavalin A (59) and thermolysin (26), the values of Ea calculated by method B are within 10% of the average values calculated by the other three methods. The other five proteins in Table 3 yielded values of ΔTm/Δν similar to the value obtained in this study. Comparing the method B value to the average of the other three methods for these five proteins shows that for glutathione-s-transferase and annexin V, the method B value is high, and for nitrite reductase and subtilisin BPN′, the method B value is low. However, for hemocyanin, the method B value is consistent with the values from the other methods. Consideration of structural fold, protein size, or oligomerization state provided no correlation with the extent of agreement of the method B value and the values obtained the other 3 methods. In our study, the discrepancy between Ea determined by method B and the values from the other 3 methods is 40–80%, much higher than the differences for the proteins summarized in Table 3. We speculate that this may be because ∼50% of the mass of rhodopsin is embedded in the membrane, whereas the proteins in Table 3 are soluble or bind to the membrane surface.
TABLE 3.
Denaturation activation energies for other proteins
| Protein | Molecular mass (kDa) | Structural fold (60), oligomeric state | Methods used to determine Ea | ΔTm/Δν* | Ea (kJ/mol) | Reference |
|---|---|---|---|---|---|---|
| Concanavalin A | 26.5 | All-β Tetramer | A | 21 | 126 | (59) |
| B | 138 | |||||
| C | 129 | |||||
| Thermolysin | 36.4 | α + β Monomer | A | 6 | 275 | (26) |
| B | 269 | |||||
| C | 296 | |||||
| D | 287 | |||||
| Hemocyanin | 400.0 | α + β Didecamer | A | 2 | 582 | (61) |
| B | 594 | |||||
| C | 626 | |||||
| D | 587 | |||||
| Glutathione-s-transferase | 26.0 | α + β Dimer | A | 3 | 398 | (62) |
| B | 456 | |||||
| C | 431 | |||||
| D | 402 | |||||
| Annexin V E17G | 35.7 | All-α Monomer (cytosol), polymeric (membrane) | A | 2 | 601 | (63) |
| B | 713 | |||||
| C | 546 | |||||
| D | 611 | |||||
| Nitrite reductase | 37.0 | All-β Trimer | A | 3 | 574 | (64) |
| B | 435 | |||||
| C | 523 | |||||
| D | 556 | (65) | ||||
| Subtilisin BPN′ | 27.5 | α + β Monomer | A | 3 | 255 | |
| B | 218 | |||||
| C | 280 | |||||
| D | 251 |
Some values of ΔTm/Δν were estimated from published thermograms.
In conclusion, we examined the role of lipid composition in the modulation of the thermal and kinetic stability of rhodopsin. The thermal stability, Tm, of rhodopsin in SOPC reconstituted membranes is increased by the addition of 30 mol % cholesterol, and decreased by the addition of 20% SDPC. The simultaneous addition of cholesterol and SDPC further enhances the thermostability of rhodopsin beyond the effects of cholesterol alone, which may suggest the promotion of rhodopsin-cholesterol interactions by SDPC. The highest value of thermal denaturation activation energy was also obtained in a bilayer containing both cholesterol and DHA acyl chains. These results imply that one important function of the combination of DHA acyl chains and cholesterol found in the rod outer segment disk membrane is to provide maximal thermal and kinetic stability to rhodopsin.
Editor: Lukas K. Tamm.
References
- 1.Lee, A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666:62–87. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerberg, J., and K. Gawrisch. 2006. The physical chemistry of biological membranes. Nat. Chem. Biol. 2:564–567. [DOI] [PubMed] [Google Scholar]
- 3.Marsh, D. 2003. Lipid interactions with transmembrane proteins. Cell. Mol. Life Sci. 60:1575–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White, S. H., A. S. Ladokhin, S. Jayasinghe, and K. Hristova. 2001. How membranes shape protein structure. J. Biol. Chem. 276:32395–32398. [DOI] [PubMed] [Google Scholar]
- 5.Matthews, R. G., R. Hubbard, P. K. Brown, and G. Wald. 1963. Tautomeric forms of metarhodopsin. J. Gen. Physiol. 47:215–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn, H. 1980. Light- and GTP-regulated interaction of GTPase and other proteins with bovine photoreceptor membranes. Nature. 283:587–589. [DOI] [PubMed] [Google Scholar]
- 7.Kwok-Keung Fung, B., and L. Stryer. 1980. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. Proc. Natl. Acad. Sci. USA. 77:2500–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung, B. K., J. B. Hurley, and L. Stryer. 1981. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc. Natl. Acad. Sci. USA. 78:152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylor, D. 1996. How photons start vision. Proc. Natl. Acad. Sci. USA. 93:560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber, T., A. V. Botelho, K. Beyer, and M. F. Brown. 2004. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys. J. 86:2078–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, M. F. 1994. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 73:159–180. [DOI] [PubMed] [Google Scholar]
- 12.Litman, B. J., and D. C. Mitchell. 1996. A role for phospholipid polyunsaturation in modulating membrane protein function. Lipids. 31:S193–S197. [DOI] [PubMed] [Google Scholar]
- 13.Botelho, A. V., T. Huber, T. P. Sakmar, and M. F. Brown. 2006. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 91:4464–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, N. J., and M. F. Brown. 1993. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 32:2438–2454. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell, D. C., S. L. Niu, and B. J. Litman. 2003. Enhancement of G protein-coupled signaling by DHA phospholipids. Lipids. 38:437–443. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien, D. F., L. F. Costa, and R. A. Ott. 1977. Photochemical functionality of rhodopsin-phospholipid recombinant membranes. Biochemistry. 16:1295–1303. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell, D. C., M. Straume, and B. J. Litman. 1992. Role of sn-1-saturated, sn-2-polyunsaturated phospholipids in control of membrane-receptor conformational equilibrium—effects of cholesterol and acyl chain unsaturation on the metarhodopsin-I-metarhodopsin-II equilibrium. Biochemistry. 31:662–670. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell, D. C., M. Straume, J. L. Miller, and B. J. Litman. 1990. Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry. 29:9143–9149. [DOI] [PubMed] [Google Scholar]
- 19.Niu, S. L., D. C. Mitchell, and B. J. Litman. 2002. Manipulation of cholesterol levels in rod disk membranes by methyl-beta-cyclodextrin. Effects on receptor activation. J. Biol. Chem. 277:20139–20145. [DOI] [PubMed] [Google Scholar]
- 20.Stinson, A. M., R. D. Wiegand, and R. E. Anderson. 1991. Fatty acid and molecular species compositions of phospholipids and diacylglycerols from rat retinal membranes. Exp. Eye Res. 52:213–218. [DOI] [PubMed] [Google Scholar]
- 21.Boesze-Battaglia, K., T. Hennessey, and A. D. Albert. 1989. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J. Biol. Chem. 264:8151–8155. [PMC free article] [PubMed] [Google Scholar]
- 22.Boesze-Battaglia, K., D. T. Organisciak, and A. D. Albert. 1994. RCS rat retinal rod outer segment membranes exhibit different cholesterol distributions than those of normal rats. Exp. Eye Res. 58:293–300. [DOI] [PubMed] [Google Scholar]
- 23.Vogt, G., S. Woell, and P. Argos. 1997. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 269:631–643. [DOI] [PubMed] [Google Scholar]
- 24.Becktel, W. J., and J. A. Schellman. 1987. Protein stability curves. Biopolymers. 26:1859–1877. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Ruiz, J. M. 1995. Differential scanning calorimetry of proteins. Subcell. Biochem. 24:133–176. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Ruiz, J. M., J. L. Lopez-Lacomba, M. Cortijo, and P. L. Mateo. 1988. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry. 27:1648–1652. [DOI] [PubMed] [Google Scholar]
- 27.McDowell, J. H., and H. Kuhn. 1977. Light-induced phosphorylation of rhodopsin in cattle photoreceptor membranes: substrate activation and inactivation. Biochemistry. 16:4054–4060. [DOI] [PubMed] [Google Scholar]
- 28.Litman, B. J. 1982. Purification of rhodopsin by concanavalin-A affinity-chromatography. Methods Enzymol. 81:150–153. [DOI] [PubMed] [Google Scholar]
- 29.Bartlett, G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234:466–468. [PubMed] [Google Scholar]
- 30.Jackson, M. L., and B. J. Litman. 1985. Rhodopsin-egg phosphatidylcholine reconstitution by an octyl glucoside dilution procedure. Biochim. Biophys. Acta. 812:369–376. [DOI] [PubMed] [Google Scholar]
- 31.Wald, G., and P. K. Brown. 1953. The molar extinction of rhodopsin. J. Gen. Physiol. 37:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, K., and J. M. Sturtevant. 1981. Thermal denaturation of streptomyces subtilisin inhibitor, subtilisin BPN′, and the inhibitor-subtilisin complex. Biochemistry. 20:6185–6190. [DOI] [PubMed] [Google Scholar]
- 33.Bischof, J. C., and X. He. 2005. Thermal stability of proteins. Ann. N. Y. Acad. Sci. 1066:12–33. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, D. C., K. Gawrisch, B. J. Litman, and N. Salem. 1998. Why is docosahexaenoic acid essential for nervous system function? Biochem. Soc. Trans. 26:365–370. [DOI] [PubMed] [Google Scholar]
- 35.Levine, Y. K., G. van Ginkel, G. R. Luckhurst, and C. A. Veracini. 1994. Molecular dynamics in liquid-crystalline systems studied by fluorescence depolarization techniques. In The Molecular Dynamic of Liquid Crystals. G. R. Luckhurst and C. A. Veracini, editors. Kluwer Academic, Amsterdam. 537–571.
- 36.Straume, M., and B. J. Litman. 1987. Equilibrium and dynamic structure of large, unilamellar, unsaturated acyl chain phosphatidylcholine vesicles—higher-order analysis of 1,6-diphenyl-1,3,5-hexatriene and 1-[4-(trimethylammonio)phenyl]-6-phenyl-1,3,5-hexatriene anisotropy decay. Biochemistry. 26:5113–5120. [DOI] [PubMed] [Google Scholar]
- 37.Straume, M., D. C. Mitchell, J. L. Miller, and B. J. Litman. 1990. Interconversion of metarhodopsin-I and metarhodopsin-II—a branched photointermediate decay model. Biochemistry. 29:9135–9142. [DOI] [PubMed] [Google Scholar]
- 38.Landin, J. S., M. Katragadda, and A. D. Albert. 2001. Thermal destabilization of rhodopsin and opsin by proteolytic cleavage in bovine rod outer segment disk membranes. Biochemistry. 40:11176–11183. [DOI] [PubMed] [Google Scholar]
- 39.Litman, B. J. and D. C. Mitchell. 1996. A role for phospholipid polyunsaturation in modulating membrane protein function. Lipids. 31:S193–S197. [DOI] [PubMed] [Google Scholar]
- 40.Albert, A. D., K. Boesze-Battaglia, Z. Paw, A. Watts, and R. M. Epand. 1996. Effect of cholesterol on rhodopsin stability in disk membranes. Biochim. Biophys. Acta. 1297:77–82. [DOI] [PubMed] [Google Scholar]
- 41.Polozova, A., and B. J. Litman. 2000. Cholesterol dependent recruitment of di22: 6-PC by a G protein-coupled receptor into lateral domains. Biophys. J. 79:2632–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung, W. C., M. T. Lee, F. Y. Chen, and H. W. Huang. 2007. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 92:3960–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Separovic, F., and K. Gawrisch. 1996. Effect of unsaturation on the chain order of phosphatidylcholines in a dioleoylphosphatidylethanolamine matrix. Biophys. J. 71:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong, H. and L. K. Tamm. 2004. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Nat. Acad. Sci. (USA) 101:4065–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morin, P. E., D. Diggs, and E. Freire. 1990. Thermal stability of membrane-reconstituted yeast cytochrome c oxidase. Biochemistry. 29:781–788. [DOI] [PubMed] [Google Scholar]
- 46.Niu, S. L., and D. C. Mitchell. 2005. Effect of packing density on rhodopsin stability and function in polyunsaturated membranes. Biophys. J. 89:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert, A. D., and K. Boesze-Battaglia. 2005. The role of cholesterol in rod outer segment membranes. Prog. Lipid Res. 44:99–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeagle, P. L. 1991. Modulation of membrane function by cholesterol. Biochimie. 73:1303–1310. [DOI] [PubMed] [Google Scholar]
- 49.Vemuri, R., and K. D. Philipson. 1989. Influence of sterols and phospholipids on sarcolemmal and sarcoplasmic reticular cation transporters. J. Biol. Chem. 264:8680–8685. [PubMed] [Google Scholar]
- 50.Yeagle, P. L., J. Young, and D. Rice. 1988. Effects of cholesterol on (Na+,K+)-ATPase ATP hydrolyzing activity in bovine kidney. Biochemistry. 27:6449–6452. [DOI] [PubMed] [Google Scholar]
- 51.Shouffani, A., and B. I. Kanner. 1990. Cholesterol is required for the reconstruction of the sodium- and chloride-coupled, gamma-aminobutyric acid transporter from rat brain. J. Biol. Chem. 265:6002–6008. [PubMed] [Google Scholar]
- 52.Albert, A. D., J. E. Young, and P. L. Yeagle. 1996. Rhodopsin-cholesterol interactions in bovine rod outer segment disk membranes. Biochim. Biophys. Acta. 1285:47–55. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen, S. G., H. J. Choi, D. M. Rosenbaum, T. S. Kobilka, F. S. Thian, P. C. Edwards, M. Burghammer, V. R. Ratnala, R. Sanishvili, R. F. Fischetti, G. F. Schertler, W. I. Weis, and B. K. Kobilka. 2007. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 450:383–387. [DOI] [PubMed] [Google Scholar]
- 54.Soubias, O., W. E. Teague, and K. Gawrisch. 2006. Evidence for specificity in lipid-rhodopsin interactions. J. Biol. Chem. 281:33233–33241. [DOI] [PubMed] [Google Scholar]
- 55.Feller, S. E., and K. Gawrisch. 2005. Properties of docosahexaenoic-acid-containing lipids and their influence on the function of rhodopsin. Curr. Opin. Struct. Biol. 15:416–422. [DOI] [PubMed] [Google Scholar]
- 56.Grossfield, A., S. E. Feller, and M. C. Pitman. 2006. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 103:4888–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huster, D., K. Arnold, and K. Gawrisch. 1998. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry. 37:17299–17308. [DOI] [PubMed] [Google Scholar]
- 58.Mansoor, S. E., K. Palczewski, and D. L. Farrens. 2006. Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. USA. 103:3060–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerjee, T., and N. Kishore. 2005. 2,2,2-trifluoroethanol-induced molten globule state of concanavalin A and energetics of 8-anilinonaphthalene sulfonate binding: calorimetric and spectroscopic investigation. J. Phys. Chem. 109:22655–22662. [DOI] [PubMed] [Google Scholar]
- 60.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536–540. [DOI] [PubMed] [Google Scholar]
- 61.Idakieva, K., K. Parvanova, and S. Todinova. 2005. Differential scanning calorimetry of the irreversible denaturation of Rapana thomasiana (marine snail, gastropod) hemocyanin. Biochim. Biophys. Acta. 1748:50–56. [DOI] [PubMed] [Google Scholar]
- 62.Quesada-Soriano, I., F. Garcia-Maroto, and L. Garcia-Fuentes. 2006. Kinetic study on the irreversible thermal denaturation of Schistosoma japonicum glutathione s-transferase. Biochim. Biophys. Acta. 1764:979–984. [DOI] [PubMed] [Google Scholar]
- 63.Vogl, T., C. Jatzke, H. J. Hinz, J. Benz, and R. Huber. 1997. Thermodynamic stability of annexin V E17G: equilibrium parameters from an irreversible unfolding reaction. Biochemistry. 36:1657–1668. [DOI] [PubMed] [Google Scholar]
- 64.Stirpe, A., R. Guzzi, H. Wijma, M. P. Verbeet, G. W. Canters, and L. Sportelli. 2005. Calorimetric and spectroscopic investigations of the thermal denaturation of wild type nitrite reductase. Biochim. Biophys. Acta. 1752:47–55. [DOI] [PubMed] [Google Scholar]
- 65.Arroyo-Reyna, A., S. R. Tello-Solis, and A. Rojo-Dominguez. 2004. Stability parameters for one-step mechanism of irreversible protein denaturation: a method based on nonlinear regression of calorimetric peaks with nonzero deltaCp. Anal. Biochem. 328:123–130. [DOI] [PubMed] [Google Scholar]