Abstract
Enterohemorrhagic Escherichia coli O157:H7 and enteropathogenic E. coli cause a characteristic histopathology in intestinal cells known as attaching and effacing. The attaching and effacing lesion is encoded by the Locus of Enterocyte Effacement (LEE) pathogenicity island, which encodes a type III secretion system, the intimin intestinal colonization factor, and the translocated intimin receptor protein that is translocated from the bacterium to the host epithelial cells. Using lacZ reporter gene fusions, we show that expression of the LEE operons encoding the type III secretion system, translocated intimin receptor, and intimin is regulated by quorum sensing in both enterohemorrhagic E. coli and enteropathogenic E. coli. The luxS gene recently shown to be responsible for production of autoinducer in the Vibrio harveyi and E. coli quorum-sensing systems is responsible for regulation of the LEE operons, as shown by the mutation and complementation of the luxS gene. Regulation of intestinal colonization factors by quorum sensing could play an important role in the pathogenesis of disease caused by these organisms. These results suggest that intestinal colonization by E. coli O157:H7, which has an unusually low infectious dose, could be induced by quorum sensing of signals produced by nonpathogenic E. coli of the normal intestinal flora.
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) has caused numerous food and waterborne outbreaks of bloody diarrhea and hemolytic uremic syndrome throughout the world. The organism colonizes the large intestine and produces a potent toxin, Shiga toxin (Stx), which is responsible for the major symptoms of hemorrhagic colitis and hemolytic uremic syndrome (1, 2). Enteropathogenic E. coli (EPEC), a related pathogen that does not produce Stx, is a major cause of infant diarrhea in children in developing countries (2, 3). Both EHEC and EPEC cause a histopathological lesion on intestinal epithelial cells termed attaching and effacing (AE). This lesion is characterized by the destruction of the microvilli and the rearrangement of the cytoskeleton to form a pedestal-like structure that cups the bacteria individually (2, 4).
The genes involved in the formation of the AE lesion are encoded on a pathogenicity island named the Locus of Enterocyte Effacement (LEE) (5, 6). This region contains (i) sep and esc genes encoding a type III secretion system (7); (ii) the eae gene encoding an adhesin called intimin that is responsible for the intimate attachment of the bacteria to the epithelial cell (8); (iii) the espABD genes, which encode proteins secreted by the type III secretion system, including EspA, which forms a filamentous-like structure that facilitates translocation of EspB and the translocated intimin receptor (Tir) (9–12); (iv) the tir gene, which encodes the translocated intimin receptor, the receptor for the intimin (13); and (v) the ler gene (LEE-encoded regulator), which encodes a positive regulator of LEE genes (14).
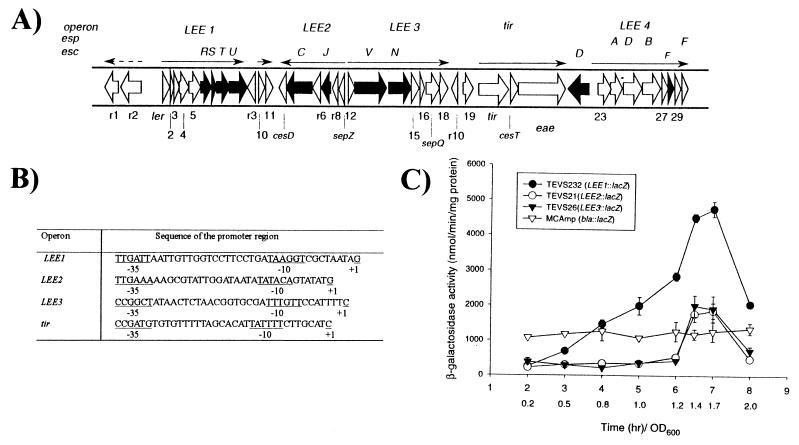
Sequence analysis of the LEEs for EPEC O127:H6 (6) and EHEC O157:H7 (15) reveals 41 ORFs that are highly conserved at the DNA and protein levels for the type III secretion genes but are more variable for the esp, eae, and tir genes. The majority of the LEE genes are in five major polycistronic operons named LEE1 through LEE4 and tir (Fig. 1A) (14, 16).
Figure 1.
(A) Map of the locus of enterocyte effacement (LEE) pathogenicity island showing operons LEE1, LEE2, LEE3, tir, and LEE4. Operons LEE1, LEE2, and LEE3 encode the type III secretion system; the tir operon encodes the translocated intimin receptor (Tir) and the adhesin intimin; the LEE4 operon encodes the EspA, EspB, and EspD secreted proteins. Black arrows indicate genes whose predicted proteins share homology with components of type III secretion systems present in other Gram-negative pathogens. (B) Predicted promoters of the EHEC LEE1, LEE2, LEE3, and tir operons. (C) β-galactosidase specific activity from LEE1∷lacZ, LEE2∷lacZ and LEE3∷ lacZ chromosomal fusions in a K-12 background throughout the bacterial growth curve.
In addition to the shared chromosomal LEE genes, EPEC and EHEC also contain unique large plasmids. The 70-kb EPEC adherence factor plasmid encodes a regulator of virulence genes called Per (plasmid-encoded regulator) consisting of three ORFs, perA, perB, and perC. PerA is an AraC homologue (17) and regulates the expression of intimin and bfpA encoding the bundle-forming pilus structural subunit (17, 18). We recently reported that Per also regulates the expression of ler, the first gene in the LEE1 operon whose product activates expression of the LEE2, LEE3, tir, and LEE4 operons in EPEC in a regulatory cascade (14). EHEC lacks Per, and little is known about the regulation of virulence genes in this organism.
In this study we demonstrate that a regulatory mechanism known as quorum sensing is involved in the regulation of the expression of the type III secretion system in both EHEC and EPEC. Quorum sensing is a mechanism of cell-to-cell signaling via the production of compounds known as autoinducers that allow a bacterium to “sense” its own population as well as the population of other bacteria in a given environment. Quorum sensing was first described as being involved in the regulation of luminescence in Vibrio fischrei (19) and since then has been shown to be a widespread gene regulation mechanism present in both Gram-negative and Gram-positive bacteria. Our results suggest a novel mechanism for the regulation of type III secretion systems and may help explain the low infectious dose of EHEC.
Materials and Methods
Strains and Plasmids.
EHEC strain 86-24h11 is a derivative of wild-type strain 86-24 containing a mutation in the stxA2 gene that inactivates Stx2 (Ann Hull, personal communication). All E. coli strains were grown aerobically at 37°C in LB broth unless otherwise noted. All Vibrio harveyi strains were grown aerobically in Autoinducer Bioassay (AB) medium at 25°C (20). For details on strains, clones, and primers see Tables 4 and 5, which are published as supplemental data on the PNAS web site, www.pnas.org.
Molecular Cloning and Sequencing Procedures.
Plasmid pVS33 was constructed by amplifying the rpoS gene from EHEC O157:H7 strain 86-24 with Pwo polymerase (Boehringer Mannheim) by using primers K1193 and K1020 and was cloned into the Psp AI site of plasmid pBAD33 (21). The cloned rpoS gene in pVS33 was induced by addition of 0.2% arabinose to the growth medium. Plasmid pVS68 was constructed by amplifying the luxS gene from EHEC O157:H7 strain 86-24 with Pwo using primers K1663 and K1664 and was cloned into the EcoRV site of pBluescript II SK(−) (Stratagene).
To mutate the luxS gene in EPEC strain E2348/69, we cloned a tetracycline cassette derived from pBR322 into an EcoRV site in the middle of luxS. This construct was then cloned into the suicide vector pCVD442 (22), yielding pVS72, which was used to generate an EPEC luxS mutant strain named VS81 by allelic exchange. Strain VS82 is VS81-complemented with pVS68.
Primer Extension.
Primer extension analysis on E. coli O157:H7 strain 86-24 used RNA extracted from cells grown until late exponential phase in DMEM at 37°C following standard procedures (23). Primers used to determine transcriptional start sites were designed from sequences within the first gene of each operon and included K983 (LEE1); K870 (LEE2); K985 (LEE3); and K1490 (tir). The start sites were confirmed by using two additional primers. Primer extension reactions were run in parallel with DNA sequencing reactions of the corresponding genes to determine the transcriptional start site.
lacZ Fusions.
lacZ operon fusions were constructed by amplifying the promoter regions of the operons using Pwo and cloning into plasmid pRS551, which contains a promoterless lac operon (24). Primers used to clone the promoter regions were K981 and K982 (LEE1); K1083 and K1084 (LEE2); K887 and K888 (LEE3); K979 and K980 (tir); K1641 and K1642 (LEE4); K1428 and K1429 (stx2); K1547 and K1549 (perA). To generate single-copy chromosomal lacZ fusions, the resulting plasmids were linearized with XhoI and were transformed into strain TE2680 (MC4100 recD∷Tn10) as described by Elliott (25). The operon∷lacZ fusions were confirmed by DNA sequencing and the chromosomal insertions were analyzed by Southern blotting to confirm that they were in single copy. The resulting plasmids containing the lacZ fusions and strains containing the chromosomal fusions were designated pVS232Z and TEVS232 (LEE1 fusion), pVS21 and TEVS21 (LEE2), pVS26 and TEVS26 (LEE3), pVS24 and TEVS24 (tir), pVS76 and TEVS76 (LEE4), pSS206 and SS575 (perA), and pVS44 and TEVS44 (stx). A β-lactamase control fusion (bla∷lacZ) was generated by using promoter sequences from pBR322, which were cloned into pRS551 to generate pVSAPR. This plasmid was transformed into E. coli MC4100, and λRS45 phage lysates were prepared (24) and were used to transduce the fusion into the chromosome of MC4100.
β-Galactosidase Assays.
Resuspended cell pellets were diluted in Z buffer (0.06 M Na2HPO4⋅7H2O/0.04 M Na2HPO4⋅H2O/0.01 M KCl/0.001 M MgSO4⋅7H2O/0.05 M β-mercaptoethanol, pH 7.0) and were assayed for β-galactosidase activity by using o-nitrophenyl-β-d-galactopyranoside as substrate as previously described (26). Specific activity (nM/min/mg protein) was measured by using protein concentrations determined by the Lowry assay (27).
Preconditioned Media Assays.
An initial inoculum grown shaking in LB for 18 hours at 37°C was diluted 1:100 in fresh LB and was grown until an OD600 of 0.2, when it was once again diluted 1:100 and grown until an OD600 of ≈1.2. After centrifugation (12,000 × g, 4 min, 25°C) and filtration (0.45 μM filter), media nutrients were corrected by adding a solution of 20× LB to a final concentration of 0.5×, and the pH was adjusted to 7.2 as described (28). The promoter-lacZ fusions were grown in this preconditioned medium until OD600 < 0.2 and was assayed for β-galactosidase activity. These tests were performed at both 30°C and 37°C with LB, LB with 0.5% glucose, LB with 0.4M NaCl, DMEM, and DMEM with 10% FBS.
Vibrio harveyi Luminescence Assay.
The presence of autoinducer 2 (AI-2) in the preconditioned media was assayed by using the V. harveyi BB170 (luxN∷Tn5) reporter strain, which responds only to AI-2 (29). The luminescence assays were performed as described (29), and the assays were read in a Wallack 1420 multilable counter.
Secreted Proteins.
Secreted proteins from EPEC VS81 and VS82 strains were prepared as described (7) after growing these strains to an OD600 of 1.0 in DMEM at 37°C.
Results
Regulation by Quorum Sensing of the EHEC LEE Genes.
We investigated the regulation of genes involved in the formation of the AE lesion in EHEC O157:H7 strain 86-24h11. The five major polycistronic operons responsible for the AE lesion include the LEE1, LEE2, and LEE3 operons, which encode a type III secretion system, the tir operon, which encodes Tir, CesT (a chaperone for Tir), and intimin, and the LEE4 operon, which encodes the secreted Esp proteins (6) (Fig. 1A). The transcriptional start sites for these operons were determined by using primer extension analysis (Fig. 1B), and the predicted promoters derived from these data were used to construct lacZ reporter fusions. The promoter sequences for the LEE4 operon are virtually identical for EPEC and EHEC, and an EHEC LEE4∷lacZ fusion was constructed based on the transcription start site reported for EPEC LEE4 operon (14). The operon fusions were initially constructed in multicopy plasmids and then were inserted into the chromosome of E. coli K-12 strain TE2680 to create single copy operon fusions. The LEE1, LEE2, and LEE3 promoters were expressed during the transition from late exponential to stationary phase, and their expression decreased in stationary phase (Fig. 1C).
The maximum expression of the three LEE operons during the transition from late exponential to stationary phase plus a recent report that E. coli and Salmonella possess quorum sensing activity (29) prompted us to examine the possibility that these operons were regulated by a quorum sensing mechanism. We tested for quorum sensing activation of the LEE1, LEE2, LEE3, LEE4, and tir operons in media preconditioned by growing E. coli O157:H7 strain 86-24h11 in LB broth at 37°C until an OD600 of ≈1.2. Culture supernatants of the preconditioned (PC) medium were prepared as described (28) and were added to cultures containing the operon fusions, and β-galactosidase activities were measured. Addition of the PC medium resulted in a three-fold increase in the expression of the LEE1 and LEE2 operons (Table 1) relative to expression in nonpreconditioned medium. The same PC medium had no effect on the LEE3, tir (Table 1), and LEE4 (data not shown) operons or on β-lactamase (bla∷lacZ) or Stx2 (stx2∷lacZ) promoters (Table 1). These results indicate that E. coli O157:H7 secretes a substance that induces transcription of the LEE1 and LEE2 operons encoding components of the type III secretion system.
Table 1.
Activation of LEE operons by preconditioned media
| lacZ fusions | β-galactosidase activity LB | β-galactosidase activity LB-PC* |
|---|---|---|
| LEE1† | 178 ± 62.12‡ | 523 ± 80 |
| LEE2† | 155 ± 25 | 455 ± 23 |
| LEE3† | 76 ± 17 | 100 ± 27 |
| LEE3 (pSE1093)† | 285 ± 14 | 603 ± 32 |
| tir† | 32 ± 3.4 | 42 ± 3.6 |
| tir (pSE1093)† | 62 ± 3 | 212 ± 79 |
| stx† | 30 ± 7 | 45 ± 6 |
| bla† | 368 ± 48 | 361 ± 37 |
| LEE1 (MC4100)§ | 860 ± 276 | 4,118 ± 230 |
| LEE1 (86-24h11)§ | 1,681 ± 276 | 5,266 ± 230 |
| LEE2 (MC4100)§ | 1,563 ± 153 | 3,113 ± 116 |
| LEE2 (86-24h11)§ | 1,597 ± 239 | 6,525 ± 507 |
| LEE3 (MC4100)§ | 1,078 ± 25 | 1,066 ± 50 |
| LEE3 (86-24h11)§ | 1,074 ± 21 | 2,200 ± 237 |
LB-PC, LB medium preconditioned with EHEC strain 86-24h11.
† Single copy lacZ fusions in K-12 strain TE2680; pSE1093 contains EHEC ler gene.
‡ β-galactosidase activity in Miller units.
§ Multicopy lacZ fusions in K-12 strain MC4100 or EHEC strain 86-24h11.
In EPEC expression of the LEE2, LEE3, and LEE4 operons is regulated in a cascade fashion by the product of the first gene in the LEE1 operon, the transcriptional activator Ler (14). In EHEC, Ler also activates LEE2, LEE3, and tir operons, but not LEE4, which appears to be constitutively expressed at a high level in EHEC (V.S. and J.B.K., unpublished data). We tested the hypothesis that quorum sensing acts through this regulatory cascade by examining expression of the LEE3 and tir operons in the presence of the ler gene cloned from EHEC (pSE1093). Although in the absence of ler these operons were not activated by PC medium, in the presence of ler the expression of both operons by PC medium was increased (Table 1). The LEE1 and LEE2 operons are induced by quorum sensing in both K-12 (MC4100) and EHEC (86-24h11) backgrounds (Table 1), and LEE2 is induced to an even higher level in an EHEC background (Table 1) because this operon is also up-regulated by Ler. The LEE3 operon is only induced by quorum sensing in an EHEC background (Table 1) in which Ler is present, consistent with the activation of LEE3 in a K-12 background only when ler is added (Table 1). The LEE1 and LEE2 promoters appear to be typical σ70 promoters whereas the LEE3 and tir promoters more closely resemble promoter sequences recognized by the alternative σ factor RpoS (σ38) (Fig. 1B). Decreased expression of LEE3∷lacZ in an rpoS mutant (RH90) (30), which was restored by complementation with the cloned rpoS gene (Table 2), indicates that the LEE3 is transcribed by RpoS. Together, these results indicate that the LEE1 and LEE2 operons are directly regulated by quorum sensing in a K-12 background whereas the LEE3 and tir operons are regulated by quorum sensing indirectly via Ler, a product of the LEE1 operon. The LEE3 operon (and presumably tir) is transcribed via RpoS in addition to being regulated by quorum sensing. The fact that the LEE3 operon is also regulated by RpoS probably accounts for the data in Fig. 1C showing that this operon is also up-regulated in the transition of late exponential to stationary phase in a K-12 background. Although this pattern of expression for the LEE1 and LEE2 promoters is attributable to quorum sensing, for the LEE3, it is probably attributable to the induction by RpoS. Other promoters transcribed by RpoS have previously been reported to be regulated by quorum sensing (31, 32).
Table 2.
Effect of RpoS on type III promoters from EHEC strain 86-24
| lacZ fusions* | β-galactosidase activity, nmol/min/mg protein
|
||
|---|---|---|---|
| MC4100 | RH90† | RH90 (pVS33) | |
| LEE1 | 46,540 ± 6,938 | 40,310 ± 3,020 | nt |
| LEE2 | 47,396 ± 8,870 | 41,212 ± 4,615 | nt |
| LEE3 | 36,250 ± 884 | 14,250 ± 204 | 34,724 ± 754 |
| bla | 33,783 ± 7,250 | 34,073 ± 6,215 | nt |
nt, not tested.
*Multicopy lacZ fusions in plasmid pRS551 in MC4100.
† RH90 is MC4100 (rpoS∷Tn10); pVS33; rpoS in pBAD33 (induced with 0.2% Arabinose).
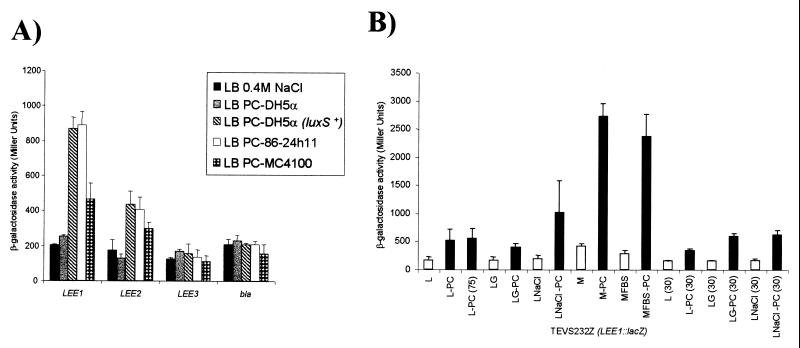
The E. coli quorum sensing mechanism described by Surette and Bassler (29) was active in all E. coli K-12 and O157:H7 strains tested by these investigators except for K-12 DH5α. We found that our LEE1∷lacZ and LEE2∷lacZ gene fusions were activated by quorum sensing through media preconditioned with either EHEC O157:H7 strain 86-24h11 or K-12 strain MC4100 but not with strain DH5α (Fig. 2A). These investigators also reported that, of the two quorum sensing systems present in V. harveyi, E. coli synthesized an autoinducer that activated the Autoinducer 2 (AI-2) system; the exact nature of the AI-2 compound has not been elucidated except that it is a small (Mr < 1,000), polar, organic compound that is heat resistant to 80°C (29). Using the V. harveyi AI-2 indicator strain BB170 (29), we confirmed that E. coli O157:H7 strain 86-24h11 produced a compound that activated the AI-2 system (Table 3). We prepared PC medium from 86-24h11 grown in a variety of conditions and found that autoinducer activity, measured by using the LEE1∷lacZ fusion, was stimulated by high osmolarity and growth at 37°C vs. 30°C and was resistant to 75°C for 10 min (Fig. 2B), conditions similar to those reported previously (33). We also found that growth in DMEM tissue culture media gave much higher levels of autoinducer activity than did growth in LB (Fig. 2B). Media preconditioned by growth of 86-24h11 in DMEM at 37°C activated the LEE1 operon by at least six-fold. Interestingly, these conditions were previously reported to produce optimal secretion of proteins via the Type III secretion system of EHEC (7, 34, 35).
Figure 2.
(A) Regulation of the LEE1, LEE2, and LEE3 EHEC operons in single copy lacZ fusions in TE2680 by media preconditioned (PC) with different E. coli strains. (B) Activation of EHEC LEE1∷lacZ fusion by PC-media (black bars) compared with activation by identical media that were not preconditioned (white bars). All PC media were prepared by growth of EHEC strain 86-24h11 under different conditions as indicated. L, LB broth; M, MEM; FBS, fetal bovine serum; G, 0.5% glucose added; NaCl, 0.4 M NaCl LB broth. Growth conditions included 37°C excepted for those experiments in which 30°C was used. Error bars represent SDs of at least three independent experiments.
Table 3.
Induction of luminescence in V. harveyi BB170 by preconditioned media
| Strains used to precondition media | Fold induction of luminescence in V. harveyi BB170 |
|---|---|
| EHEC 86-24h11 | 320 |
| MC4100 | 421 |
| DH5α | 1 |
| DH5α (pVS68)* | 113 |
| E2348/69 | 101 |
| VS81 (E2348/69 luxS−) | 1 |
| VS82 (VS81 with pVS68) | 180 |
| BB152 (AI-1−, AI-2+) | 251 |
Fold induction represents the average of ≥3 readings of induction of luminescence in V. harveyi reporter strain BB170 (sensor 1−, sensor 2+) by media preconditioned with indicated bacteria in relation to luminescence with LB media alone; pVS68 contains luxS.
A new family of genes responsible for production of AI-2 of E. coli, Salmonella typhimurium, and V. harveyi was recently cloned by Surette et al. (36) and named luxS. The luxS gene from E. coli K-12 corresponded to an ORF of previously unknown function, and analysis of the DH5α luxS gene revealed a frameshift mutation resulting in an inactive LuxS protein. We cloned the luxS gene from strain 86-24h11, generating plasmid pVS68, and tested it for the ability to activate the LEE∷lacZ fusions. As shown in Fig. 2A and Table 3, PC medium prepared from K-12 strain DH5-α containing the cloned luxS gene produces AI-2 and directly activated the LEE1 and LEE2 operons whereas medium preconditioned by growth of DH5α alone did not activate these operons, thus confirming that the luxS gene product is responsible for quorum sensing of the type III secretion system.
Regulation of EPEC Virulence Genes by Quorum Sensing.
PC-media prepared with EPEC strain E2348/69 also produces AI-2 in the V. harveyi reporter assay (Table 3) and activates transcription of the EHEC LEE1, EPEC LEE1, and LEE2 operons, but not the LEE3 and LEE4 operons in a K-12 background (Fig. 3A). This pattern of gene regulation is very similar to the one observed in EHEC. We then generated a luxS mutant in EPEC strain E2348/69 (VS81) (Table 3). (We have been unable to generate a luxS mutant of EHEC strain 86-24h11 for unknown reasons.) Media preconditioned with strain VS81 (E2348/69 luxS−) did not activate the transcription of EHEC LEE1 operon or EPEC LEE1 and LEE2 operons. However addition of the cloned luxS gene to strain VS81, resulting in production of AI-2, and PC media prepared with this strain could activate transcription of EHEC LEE1 and EPEC LEE1 and LEE2 operons (Fig. 3A; Table 3).
Figure 3.
(A) Effect of media preconditioned (PC) by growth with EPEC strain E2348/69, VS81 (E2348/69 luxS−) or VS82 (VS81 luxS+) on EHEC and EPEC promoters. LEE1eh denotes LEE1∷lacZ fusion with the LEE1 operon of EHEC strain 86-24h11; all other fusions were with EPEC strain E2348/69 operons. All fusions were tested in MC4100. (B) SDS/PAGE (12%) analysis of secreted proteins from luxS mutant. Equal volumes of supernatant preparations were added to each lane: E2348/69; VS81 (E2348/69 luxS−); VS82 (VS81 complemented with cloned luxS).
The first gene of the LEE1 operon is ler, which Mellies and coworkers (14) recently demonstrated is activated by Per in EPEC. Because quorum sensing also regulates ler in EPEC, we investigated the possibility that the regulation by Per was linked to the quorum sensing regulation. Media preconditioned by EPEC strain E2348/69 had little, if any, effect on the expression of a perA∷lacZ fusion in a K-12 background (perA∷lacZ in LB produces 119 ± 29.5 Miller units; in PC media, 161 ± 7). However, when the perA∷lacZ fusion was in an EPEC background, addition of PC media increased perA expression two-fold (413 ± 2 Miller units vs. 793 ± 44). These results indicate that per is also subjected to regulation by quorum sensing in an indirect fashion through some unknown factor present in an EPEC but not in a K-12 background.
We also observed that the EPEC luxS mutant (VS81) secreted considerably lower amounts of protein than the wild-type strain E2348/69 and that this phenotype could be complemented by addition of the cloned luxS gene (pVS68) to the mutant (Fig. 3B). The decreased levels of secreted proteins in the luxS mutant is consistent with our gene fusion data showing that quorum sensing activates the type III secretion system at the transcriptional level.
Discussion
Although enteropathogenic E. coli (EPEC) was first recognized as a human pathogen more than 50 years ago, enterohemorrhagic E. coli (EHEC) was only recognized as a human pathogen in the 1980s (1). The most important difference in the pathogenesis of these organisms is the expression of a potent cytotoxin, Shiga toxin, by EHEC but not by EPEC. Despite this difference in toxigenicity, both pathogens interact with intestinal epithelial cells in a characteristic manner known as attaching and effacing (AE) (2, 4).
The AE phenotype is encoded by the LEE pathogenicity island (5), which encodes a type III secretion system, the intimin adhesin, and the translocated intimin receptor (Tir), which is translocated into the epithelial cells through the type III secretion system (reviewed in ref. 37). Knutton and coworkers (12) proposed a model of infection in which the first step would be the expression of the type III secretion system, including the secretion of EspA, which then forms a filamentous structure that makes the initial contact with epithelial cells. In a second step, Tir is translocated through the EspA filament inside the epithelial cells, where it functions as a receptor for intimin. Understanding the regulation and function of the LEE genes should provide important insights into the first step in the pathogenesis of disease attributable to EPEC and EHEC, intestinal colonization.
To study LEE gene regulation, we constructed lacZ operon fusions for the major operons of the EPEC and EHEC LEE. We recently reported that transcription of the EPEC and EHEC LEE2, LEE3, and tir operons are regulated by the product of the first ORF in the LEE1 operon, Ler (ref. 14; S. Elliott, V.S., J.L.M., J. A. Giron, S. Hutcheson, and J.B.K., unpublished work). We now report an additional level of virulence gene regulation for EPEC and EHEC whereby the majority of genes encoded on the LEE are also regulated by quorum sensing.
Addition of media preconditioned (PC) by growth of EHEC, EPEC, or E. coli K-12 strains activated transcription of the lacZ reporter gene fused to LEE operons whereas fresh, nonpreconditioned media had no effect. Examination of single copy operon fusions inserted into the K-12 chromosome revealed that the LEE1 and LEE2 operons were directly activated by quorum sensing whereas the LEE3 and tir operons were indirectly activated via the Ler regulator (Table 1). The quorum sensing mechanism responsible for this activation is the same as that recently described by Surrette and Bassler (29, 33, 36) for E. coli, Salmonella, and V. harveyi as demonstrated by the lack of activation of the LEE1 and LEE2 operons by media preconditioned by growth of E. coli K-12 strain DH5α. This strain lacks a functional luxS gene that is responsible for synthesis of the AI-2 autoinducer. Addition of media prepared by growth of DH5α containing the cloned, proficient luxS gene restored activation of these operons (Fig. 2A). Similarly, media prepared with a luxS mutant of EPEC strain E2348/48 (VS81) did not activate expression of LEE1 and LEE2, and addition of the cloned luxS gene to VS81 restored the ability to activate these operons (Fig. 3A).
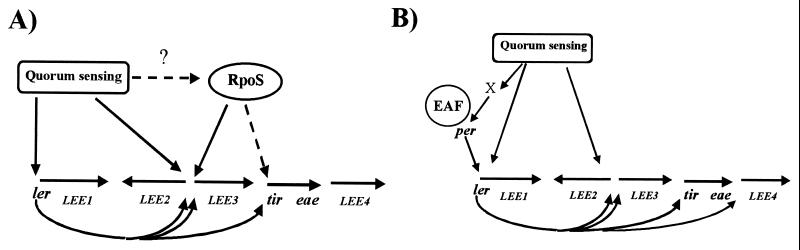
In addition to the quorum sensing regulation, in EHEC but not EPEC, LEE3 and probably tir operons are also regulated by rpoS (Fig. 1B; Table 2). Other operons transcribed by RpoS have previously been reported to be regulated by quorum sensing (31, 32, 38). EPEC but not EHEC contains the Per regulator, which positively regulates expression of ler and the LEE1 operon (14). Expression of a perA∷lacZ fusion was also observed to be regulated by quorum sensing in our studies. Also, the EPEC but not EHEC LEE4 operon is regulated by Ler and therefore indirectly by quorum sensing. Fig. 4 summarizes these observations and presents proposed models of regulation for the EPEC and EHEC LEE genes.
Figure 4.
Proposed model of LEE gene regulation in EHEC O157:H7 and EPEC O127:H6. (A) In EHEC, quorum sensing directly activates expression of the LEE1 and LEE2 operons and indirectly activates expression of LEE3 and tir operons via Ler, encoded in the LEE1 operon. The LEE3 and tir operons are regulated by the alternate σ factor RpoS, whose expression may also be regulated by quorum sensing. (Dashed lines indicate that no direct experimental evidence has yet been gathered for these steps.) (B) In EPEC, quorum sensing directly activates expression of the LEE1 and LEE2 operons and indirectly activates expression of LEE3, tir, and LEE4 operons via Ler. The Per regulator encoded on the EPEC adherence factor plasmid regulates expression of the LEE1 operon, and per is also regulated indirectly by quorum sensing, through some unknown factor (X) present only in EPEC.
Although Surrette and Bassler (29) reported that both pathogenic and nonpathogenic E. coli strains produced a substance that could activate quorum sensing-regulated genes in V. harveyi, no E. coli genes regulated by this mechanism have heretofore been identified. We have now shown that genes encoding the type III secretion system and the Tir and intimin intestinal colonization factors of EPEC and EHEC are regulated by quorum sensing, although it is likely that additional genes are regulated by this mechanism given the widespread distribution of the luxS gene.
The identification of density-regulated genes in EHEC O157:H7 may seem inconsistent with the unusually low infectious dose of EHEC required to cause disease, estimated at fewer than 50 organisms in one outbreak (39). The very low infectious dose suggests that such “self-signaling” may not occur in vivo, at least during the initial stages of infection. Instead, we propose that the low infectious dose of this pathogen could be explained, at least in part, by activation of the type III system by autoinducers synthesized by normal flora E. coli resident in the large intestine. The presence of autoinducers in the large intestine could activate the EHEC LEE genes, thereby allowing intestinal colonization mediated by intimin to proceed. In EPEC, the situation is more complicated because of the presence of the Per regulator, for which an EHEC counterpart has not been described. However, EPEC colonizes the small intestine, where the number of E. coli and other coliforms is much lower than in the large intestine (up to 107/g in the small intestine and up to 1010/g in the large intestine) (40). The Per regulator could help compensate for the lower amounts of autoinducer predicted to be present in the small intestine. In addition, EPEC, but not EHEC, produces a type IV pilus (BFP) that has been shown to mediate interbacterial adherence on intestinal epithelial cells (41), thereby leading to microcolony formation and locally higher densities.
The discovery of quorum sensing mechanisms in a variety of pathogens has stimulated a search for therapeutic approaches that would inhibit intercellular communication among the bacteria and decrease in vivo expression of virulence factors. Such an approach may also be possible for treatment or prophylaxis of disease caused by EPEC and EHEC once the autoinducer molecule has been identified and characterized. Our findings link several common themes of bacterial pathogenesis recognized in recent years, including quorum sensing, pathogenicity islands, regulatory cascades of virulence genes, and type III secretion systems. Given the widespread distribution of type III secretion systems and quorum sensing mechanisms in numerous bacterial pathogens, the regulation of such systems by quorum sensing may prove to be a paradigm of bacterial pathogenesis that can be exploited for novel therapeutic and prophylaxis approaches.
Supplementary Material
Acknowledgments
We thank Anne Hull for the EHEC stx− strain 86-24h11; Simon Elliott for the plasmid pSE1093 containing the cloned EHEC ler; and Bonnie Bassler for V. harveyi strains BB152 and BB170. This work was supported by Grants AI41325 and AI21657 from the National Institutes of Health. Vanessa Sperandio is a Pew Latin American Fellow.
Abbreviations
- EHEC
enterohemorrhagic Escherichia coli O157:H7
- Stx
Shiga toxin
- EPEC
enteropathogenic E. coli
- AE lesion
attaching and effacing lesion
- LEE
Locus of Enterocyte Effacement
- Tir
translocated intimin receptor
- Per
plasmid-encoded regulator
- AI-2
autoinducer 2
- PC medium
preconditioned medium
References
- 1.Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Washington, DC: Am. Soc. Microbiol.; 1998. [Google Scholar]
- 2.Nataro J P, Kaper J B. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine M M, Edelman R. Epidemiol Rev. 1984;6:31–51. doi: 10.1093/oxfordjournals.epirev.a036274. [DOI] [PubMed] [Google Scholar]
- 4.Kaper J B, Elliott S J, Sperandio V, Perna N T, Mayhew G F, Blattner F R. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 163–182. [Google Scholar]
- 5.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott S, Wainwright L A, McDaniel T, MacNamara B, Donnenberg M, Kaper J B. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 7.Jarvis K G, Girón J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerse A E, Kaper J B. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenny B, Lai L, Finlay B B, Donnenberg M S. Mol Microbiol. 1996;20:313–324. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Yu J, Kaper J B. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai L-C, Wainwright L A, Stone K D, Donnenberg M S. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 14.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 15.Perna N T, Mayhew G F, Pósfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott S J, Hutcheson S W, Dubois M S, Mellies J L, Wainwright L A, Batchelor M, Frankel G, Knutton S, Kaper J B. Mol Microbiol. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Duarte O G, Kaper J B. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 19.Nealson K H, Platt T, Hastings J W. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg E P, Hastings J W, Ulitzur S. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 21.Guzman L-M, Belin D, Carson M, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Simons R W, Houman F, Kleckner N. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 25.Elliott T. J Bacteriol. 1992;174:245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H, editor. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 27.Lowry O H, Rosenbrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 28.Baca-Delancey R R, South M M T, Ding X, Rather P N. Proc Natl Acad Sci USA. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surette M G, Bassler B L. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange R, Hengge-Aronis R. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 31.Sitnikov D M, Schineller J B, Baldwin T O. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huisman G W, Kolter R. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 33.Surette M G, Bassler B L. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 34.Jarvis K G, Kaper J B. Infect Immun. 1996;64:4826–2829. doi: 10.1128/iai.64.11.4826-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haigh R, Baldwin T, Knutton S, Williams P H. FEMS Microbiol Lett. 1995;129:63–68. doi: 10.1016/0378-1097(95)00136-S. [DOI] [PubMed] [Google Scholar]
- 36.Surette M G, Miller M B, Bassler B L. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 38.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 39.Tilden J, Young W, McNamara A, Custer C, Boesel B, Lambert-Fair M, Majkowski J, Vugia D, Werner S B, Hollingsworth J, Morris J G. Am J Public Health. 1996;86:1142–1145. doi: 10.2105/ajph.86.8_pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon G L, Gorbach S L. In: Infections of the Gastrointestinal Tract. Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. New York: Raven; 1995. pp. 53–69. [Google Scholar]
- 41.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.