Abstract
Fourier transform infrared spectroscopy was used to analyze the chromophore structure in the parent states Pr and Pfr of plant phytochrome phyA and the respective photoproducts lumi-R and lumi-F. The spectra were obtained from phyA adducts assembled with either uniformly or selectively isotope-labeled phytochromobilin and phycocyanobilin. The interpretation of the experimental spectra is based on the spectra of chromophore models calculated by density functional theory. Global 13C-labeling of the tetrapyrrole allows for the discrimination between chromophore and protein bands in the Fourier transform infrared difference spectra. All infrared difference spectra display a prominent difference band attributable to a stretching mode with large contributions from the methine bridge between the inner pyrrole rings (B-C stretching). Due to mode coupling, frequencies and isotopic shifts of this mode suggest that the Pr chromophore may adopt a distorted ZZZssa or ZZZasa geometry with a twisted A-B methine bridge. The transition to lumi-R is associated with only minor changes of the amide I bands indicating limited protein structural changes during the isomerization site of the C-D methine bridge. Major protein structural changes occur upon the transition to Pfr in which the chromophore adopts a ZZEssa or ZZEasa-like state. In addition, specific interactions with the protein alter the structure of the B-C methine bridge as concluded from the substantial downshift of the respective stretching mode. These interactions are removed during the photoreaction to lumi-F (ZZE→ZZZ), which involves only small protein structural changes.
INTRODUCTION
Plants sense light via several types of photoreceptors, among which the phytochrome family is responsible for many photomorphogenetic processes: seed germination, hypocotyl elongation, shade avoidance, and flowering (1). Phytochromes are large proteins (124 kDa) that possess an open-chain tetrapyrrole chromophore, phytochromobilin (PΦB), covalently bound via a thioether linkage in the N-terminal domain of the protein (2). Red-light irradiation converts the red-absorbing Pr state into the far-red absorbing Pfr state. This photoconversion is reversible, Pfr returning to Pr by absorption of far-red light or thermally by a very slow dark state reversion (3–5). The forward photoreaction proceeds via the intermediates lumi-R, meta-Ra, and meta-Rc, the backward photoreaction via the intermediates lumi-F and meta-F (in the order of their appearance). Plant phytochromes are members of a bigger family of phytochrome-like photoreceptors that are found in cyanobacteria (Cph1 and Cph2 from Synechocystis), CphA and CphB from Calothrix, other bacteria (BphP from Deinococcus radiodurans, Agrobacterium tumefaciens, and Rhodopseudomonas palustris), or fungi (FphA from Aspergillus nidulans) (see Karniol et al. (6) for an overview). All these proteins share the same features: the N-terminal half carrying the photosensory domain and the C-terminal part with the signaling domain. The N-terminal domain comprises highly conserved regions: the PAS, GAF, and PHY domains (7,8). The C-terminal region contains a histidine kinase-related domain in the plant phytochromes or a histidine kinase domain (e.g., in many bacterial phytochromes) with phosphotransferase activity (9–11). The signaling pathways are very complex and involve processes like dimerization, nuclear translocation, and interaction with many binding partners (12–18), and it was shown recently that signaling actually also involves the N-terminal region (19–21).
Because the assembly process between apoprotein and bilin chromophore is autocatalytic in phytochromes (22,23), heterologously expressed apoproteins can be reconstituted in vitro with naturally occurring bilins or chemically synthesized (modified) derivatives (24–27). As was shown recently, some prokaryotic phytochromes use covalently bound biliverdin as chromophore (10,28,29).
Despite numerous studies involving biophysical and spectroscopical techniques, the structure-function relationship or the photochemical process in phytochrome is not yet elucidated. Crystal structures have recently been resolved for the PAS-GAF domain of the Pr states of bacteriophytochrome DrBphP from Deinococcus radiodurans (30,31) and of bacteriophytochrome RpBphP3 from Rhodopseudomonas palustris (32), which both show the biliverdin chromophore in a ZZZssa configuration with twists around the AB and CD methine bridges, whereas the BC part is essentially planar. Important residues have been pinpointed, such as H260 for interaction with ring B propionate side chain, and D207 in conjugation with some water molecules for interaction with NH groups of rings A, B, C (amino acid numbering for Deinococcus). However, this aspartic acid does not appear as a classical counterion, as the interaction involves its main chain carbonyl oxygen instead of the side chain. In these respects, the available crystal structures constitute an important starting point for molecular phytochrome research. Specifically, these structures now allow development of hypotheses for the photoinduced reaction mechanism of phytochromes, which can then be tested by using advanced experimental and theoretical techniques.
Crucial issues to be addressed include the chromophore structures in the various states of phytochrome's photocycle for which no crystal structure data are available. Furthermore, it remains to be proven whether or not the lack of the photoinactive crystallized fragments lacking the PHY domain represent a good structural model for the chromophore binding domain in the native phytochromes. In addition, it is not yet clear if structural and mechanistic details are the same for bacterial and plant phytochromes. On the one hand, there is a far-reaching overall similarity with respect to the domain structure and amino acid sequences; specifically, key amino acid residues appear to be highly conserved (6). On the other hand, plant phytochrome binds PΦB via a Cys at a different position as biliverdin in bacterial phytochromes (10), and it has been speculated that these different binding sites may be associated with different chromophore structures in the Pr state (33).
In fact, spectroscopic data cannot readily be reconciled with a ZZZssa geometry in the Pr state of plant phytochrome phyA (34). In a recent combined resonance Raman (RR) spectroscopic and theoretical study on phyA a ZZZasa geometry has been proposed to be the chromophore configuration in the Pr state (35). This conclusion was based on the comparison of RR spectra of phyA adducts with PΦB and isotopically labeled phycocyanobilin (PCB) and spectra calculated for model chromophores using density functional theory (DFT). Conversely, the spectra calculated for the ZZZssa geometry failed to reproduce essential features of the experimental spectra. However, these calculations refer to isolated tetrapyrroles in vacuo. It can thus not be excluded that specific protein-chromophore interactions may perturb the structure of a hypothetical ZZZssa chromophore such that it affords a RR spectrum similar to that calculated for an ZZZasa geometry in vacuo (35).
The uncertainties in the structural assignment of the chromophore are even more severe for the remaining states of the Pr→Pfr phototransformation. There is a general agreement, however, on the nature of the primary photochemical process as a Z/E isomerization of the C-D methine bridge (36). In the case of bacteriophytochrome Agp1, thermal relaxation processes after the photoisomerization have been shown to include a transient deprotonation of the chromophore (37). Further relaxations of phytochromes presumably involve a (partial) rotation around the A-B methine bridge single bond (34,38,39). Even less unambiguous information is available for the chromophore structural during the Pfr→Pr back reaction. The date presented so far on the chromophore structure and its conformational changes upon light excitation show clearly our still insufficient understanding of this reaction pathway. A combined approach, including various spectroscopic techniques, biochemical techniques, and chemical synthesis, is a clear demand to investigate in greater detail the photochemistry of phytochromes.
Accordingly, we present studies on phyA adducts assembled with PΦB and PCB (isotopomers) by Fourier transform infrared (FTIR) spectroscopy, a method sensitive to conformational changes of the chromophore an the protein. The spectral changes observed during the photocycle of phytochrome were interpreted on the basis of spectra calculated for isolated model chromophores using DFT.
MATERIALS AND METHODS
PCB was extracted from freeze-dried Spirulina platensis cell powder (Bär Company, Bonn, Germany) following the published protocol (40).
Preparation of labeled chromophores
13C labeled chromophores
13C(10) labeled PΦB and 13C(5) labeled PCB were synthesized as described in (41).
Uniformly 15N- or 13C-labeled PCB
Fully nitrogen-labeled (15N4)-PCB was obtained from cyanobacterial cell cultures (Synechocystis PCC6803) that were grown in a Na15NO3 supplemented medium in an illumination growth chamber (Biomol, Hamburg, Germany). The same vessel was used for the uniform labeling with 13C, except that in this case gaseous 13CO2 was used as sole carbon source. Chlorosomes were isolated after a passage of a harvested cell culture through a French press, followed by a sucrose gradient centrifugation and methanolysis to release the phycocyanin-bound chromophore. PΦB was obtained from the simultaneously isolated phycoerythrobilin by addition of mercury chloride during the bilin extraction procedure (42). Purity and amount were checked by HPLC and mass spectrometry. Isotope content was routinely larger than 90%.
Preparation of recombinant 65 kDa phytochrome
The N-terminal domain of recombinant oat (plant) phytochrome (phyA), encompassing amino acids 1–595, was heterologously expressed in Hansenula polymorpha, essentially as described by Mozley, Remberg, and Gärtner (43). Holoprotein formation by the addition of chromophore was accomplished in the crude lysate of a cell culture (cleared by ultracentrifugation). The recombinant phytochrome was affinity-purified, making use of a C-terminally attached His6-tag and analyzed for its quality and purity by absorption difference spectroscopy (Pr–Pfr) and by SDS-PAGE. This fragment will be denoted as phyA for the sake of simplicity.
Sample preparation for FTIR spectroscopy
PhyA-13C(10)-PΦB was initially dissolved in a 10 mM Na-phosphate, 10 mM NaCl buffer (pH 8) containing 2 mM DTT, and 200 μM Pefabloc (Biomol, Hamburg, Germany). The stock protein concentration was determined by measuring the UV absorbance of a diluted solution at 280 nm, where the extinction coefficient is 132,000 M−1 cm−1 (44). The concentration was 1.378 g l−1 for phyA-13C(10)-PΦB, 2.2 g l−1 for phyA-13C(5)-PCB, 2.26 g l−1 for phyA-PCB, and 1.67 g l−1 for phyA-15N-PCB. The Pr→Pfr photoconversion was followed by irradiating the sample using fiber optics connected to a 150 W slide projector fitted with a band-pass filter with peak transmission at 633 nm (BP633). A long pass filter (RG695, Schott, Mainz, Germany) was used for the Pfr-Pr conversion. After the changes in the UV-Vis spectrum between 400 and 800 nm as a function of the illumination time, full Pr→Pfr photoconversion was obtained after 6 min. This duration was used in FTIR measurements for all intermediates. For the FTIR measurements, sandwich samples were prepared: ∼1 nmol of protein (60 μl) was dried, using a gentle stream of nitrogen, on the central surface (7-mm diameter) of a specially designed BaF2 round infrared window, separated from the 7–8 μm higher rim of the same window by a 1 mm wide round groove (45). The obtained homogeneous film was rehydrated by adding 6–8 μl buffer and sealed by placing a second plane BaF2 window on top of the first one, the higher rim serving as a spacer. The quality of the film was increased when the protein solution was first dialyzed against a 5 mM Tris, 10 mM NaCl buffer, containing 2 mM 2-mercaptoethanol, and 0.2% glycerol by volume. Glycerol is used to decrease aggregation, and 2-mercaptoethanol to retain photoreversibility after rehydration of the film (46,47).
FTIR measurements
The spectra were measured using a Bruker IFS 28 FTIR spectrophotometer equipped with an MCT detector, and a thermostated sample holder. Spectrophotometer and sample chamber were purged with dry air. For the cryogenic experiments, a home-built cryostat was used that allows measurements in the temperature range between −190 and 0°C (±0.5°C). Before insertion into the cryostat, the samples were converted either into the Pr state for measurements of the intermediates of the Pr→Pfr pathway or into the Pr/Pfr mixture for measurements of the reverse reaction, using the above described filters. The degree of rehydration of the protein film was checked using the broad absorption band of water ∼3300 cm−1. Sealing the sandwich windows with silicone on the edges prevented dehydration of the film in the cryostat. The intermediates were accumulated by irradiation of Pr or Pfr at the temperatures given in the figure captions. They were chosen according to Eilfeld and Rüdiger (48). The phototransformation back to the initial state had to be accomplished at ambient temperature. For each single-beam spectrum, 512 scans were accumulated with a resolution of 4 cm−1. For measurements in D2O, H/D exchange was accomplished by five successive additions and evaporation of D2O on the protein film, which was finally rehydrated using 6–8 μl of D2O.
Each FTIR difference spectrum displayed represents an average of many (∼10–30) single difference spectra corresponding to several samples and several experiments per sample. Each single difference spectrum was obtained from five single-beam spectra recorded before and three single-beam spectra recorded after illumination. First, a “time” baseline was measured as the average difference between successive spectra, before illumination. This time baseline represents the minor changes occurring during the time elapsed between two successive 512 scan single beam spectra (∼1 min). For each of the three photoproduct difference spectra, the correct amount of this time baseline had to be subtracted according to the elapsed time. Second, such a photoproduct difference spectrum had to be corrected also for heat effect due to the duration of irradiation by applying, after measuring the photoproduct spectra, a second irradiation of the same duration, and subtracting this effect. When necessary, a supplementary water vapor spectrum subtraction was applied.
The noise level can be deduced from the baseline above 1750 cm−1, where no bands are present. The band intensities of the spectra can be estimated from the strongest bands that extend from 0.1 milliabsorbance unit for the early photoproducts (lumi-R, meta-Ra) to 1 milliabsorbance unit for the Pfr-spectrum. The differences in spectral intensities are due to the gradually reduced photoconversion in the Pr:Pfr pathway at low temperatures. Based on infrared (IR) bands common to both transitions, the yield of lumi-R is <20% of that of Pfr.
As a general rule, we denote the difference spectrum for the Pr→Pfr transition as Pfr/Pr spectrum, and negative and positive bands are ascribed to the Pr and Pfr states, respectively. The difference spectrum for the Pr→lumi-R transition is named lumi-R/Pr spectrum, and negative and positive bands are ascribed to the Pr and lumi-R states, respectively. The backreaction from Pfr to lumi-F is denoted as lumi-F/Pfr, and the negative and positive bands are ascribed to Pfr and lumi-F, respectively.
Quantum chemical calculations
Quantum chemical calculations of the vibrational spectra and infrared intensities were carried out using DFT with the hybrid exchange functional B3LYP level (49) implemented in GAUSSIAN03 program package (50). The geometries and corresponding force field of the protonated PCB and PΦB molecules with ZZZasa and ZZZssa conformation and configuration of the methine bridges correspond to those computed previously (35) (Fig. 1). In these models the positive charge of the tetrapyrrole was compensated by a chloride ion placed between the two inner pyrrol rings. The sulfur of the thioether linkage was replaced by hydrogen. The geometries of the ZZEasa and ZZEssa conformations were built from the two structures mentioned above by rotating the C(15)=C ∼180° and optimized under tight convergence criteria. All calculations were carried out using the 6-31G* basis set except for the chlorine atom for which a 6-31+G* basis set was adopted. Further details of the computational method are given elsewhere (35).
FIGURE 1.
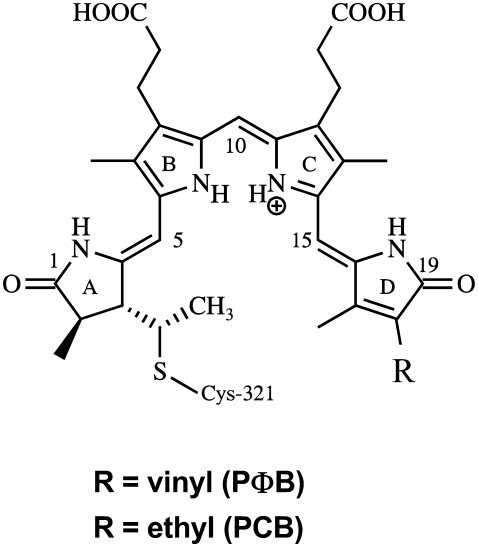
Structure of phytochromobilin (PΦB; R = vinyl) and phycocyanobilin (PCB; R = ethyl). Positions for 13C labeling are at C(5) and C(10). 15N labeling has been uniformly carried out at all nitrogens. In the text the individual methine bridges are denoted by A-B, B-C, and C-D.
RESULTS
Uniform 13C-labeling of the PΦB chromophore
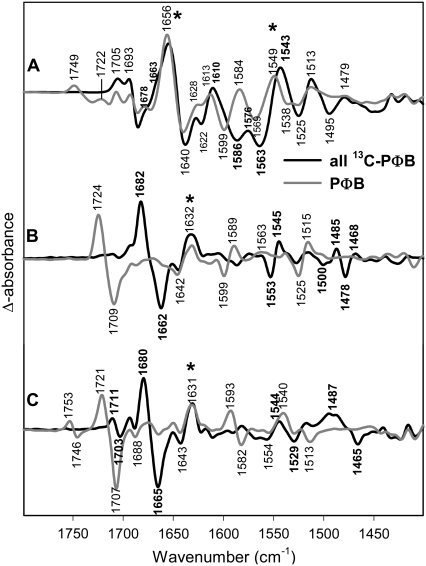
In infrared studies of photobiological systems, it is difficult to assign spectral contributions to the chromophore or to the protein, because both components add changes in the difference spectrum. In our earlier studies of phyA we could unequivocally identify the C=O stretch of ring A by 18O-labeling due to the different substitution pattern of rings A and D. Furthermore, the C=O stretch of ring D could be identified by comparing the spectra of phyA reconstituted with PΦB versus PCB (38). However, most other bands could not be assigned. Therefore, we reconstituted the apoprotein phyA with fully 13C-labeled PΦB. The Pfr/Pr, lumi-R/Pr, and lumi-F/Pfr spectra are shown in Fig. 2. (See Materials and Methods for the nomenclature of the spectra).
FIGURE 2.
Comparison of FTIR difference spectra of phyA reconstituted with nonlabeled PΦB and phyA reconstituted with uniformly 13C-labeled PΦB. The spectra of the adducts with nonlabeled and labeled PΦB are given by the gray and black lines, respectively. The spectra (from top to bottom) refer to the “Pfr” minus “Pr” (A), “lumi-R” minus “Pr” (B), and “lumi-F” minus “Pfr” (C) differences. Protein changes are denoted by an asterisk. Further details are given in the text (Materials and Methods).
In the lumi-R/Pr and lumi-F/Pfr spectra most of the bands display 13C/12C isotopic shifts indicating that these spectra are largely dominated by the contributions of the chromophore. In contrast, several bands in the Pfr/Pr difference spectrum are insensitive toward 13C/12C labeling. These are specifically the bands between 1620 and 1680 cm−1 that, hence, are assigned to amide I modes of the protein. This finding is consistent with the view that the primary photochemical process is not associated with significant protein structural changes such that the most reliable information on the chromophore structures can be derived from the Pr/lumi-R and Pfr/lumi-F difference spectra.
Quantum chemical calculations of the IR spectra
The quantum chemical calculations predict a variety of IR active modes in the spectral range between 1650 and 1500 cm−1 that is of particular interest for analyzing chromophore structural changes during the phytochrome photocycle (Supplementary Material, Data S1). Regardless of the methine bridge conformation and configuration, the highest IR intensity of protonated PΦB and PCB is predicted for a mode calculated to be between 1600 and 1590 cm−1. This mode is dominated by the B-C methine bridge stretching in geometries with anti, syn, anti conformations of the methine bridges whereas in geometries with syn, syn, anti conformation the contributions from stretchings of the A-B bridge and C=C bonds in ring B are larger than those of the B-C stretching. Notwithstanding the different compositions we will denote this mode as B-C stretching for the sake of simplicity. The modes that include major contributions from the A-B and C-D methine bridge stretchings have medium or low IR intensities but their frequencies depend more sensitively on the respective methine bridge geometries. These modes, however, are expected to be particularly strong in the RR spectrum. All geometries give rise to two modes with relatively large contributions from the N-H in-plane bending of the rings B and C. For both modes that are calculated at ∼1560–1570 cm−1 and ∼1510–1530 cm−1 a weak to medium IR intensity is expected. Strong IR intensities are associated with the ring A and ring D C=O stretching vibrations that are located above 1700 cm−1.
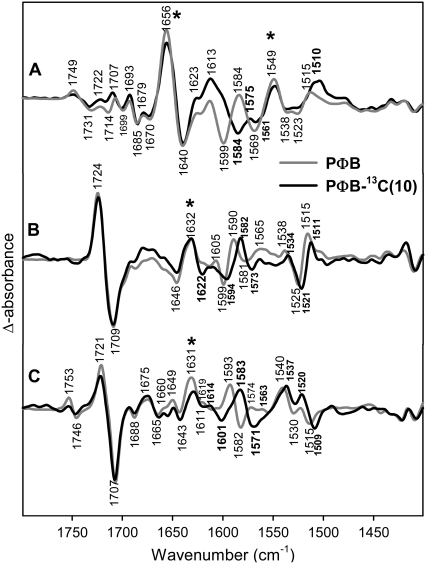
13C(10)-labeling of PΦB
In Fig. 3, the lumi-R/Pr difference spectra are shown for phyA reconstituted with unlabeled PΦB (gray) and 13C(10)-labeled PΦB (black). The large difference band at 1709(−)/1724(+) cm−1 has been assigned previously to the C=O stretch of ring D undergoing an environmental change upon the Z→E isomerization (38). Because the feature between 1646 and 1632 cm−1 is affected only a little, if at all, in the fully 13C-labeled chromophore (Fig. 2), we assign most of those signals to amide I band changes.
FIGURE 3.
Comparison of FTIR difference spectra of phyA reconstituted with nonlabeled PΦB and phyA reconstituted with 13C(10)-labeled PΦB. The spectra of the adducts with nonlabeled and labeled PΦB are given by the gray and black lines, respectively. The spectra (from top to bottom) refer to the “Pfr” minus “Pr” (A), “lumi-R” minus “Pr” (B), and “lumi-F” minus “Pfr” (C) differences. Protein changes are denoted by an asterisk. Further details are given in the text (Materials and Methods).
A clear isotopic shift can be seen for the difference band 1599(−)/1590(+) cm−1 that shifts to 1594(−)/1582(+). Thus, the corresponding modes of Pr and lumi-R must contain the B-C methine stretching coordinate. In addition, the difference spectrum displays a broad negative feature on the high-frequency side of the negative shifted band that is missing in the corresponding difference spectra obtained with 13C(10)-labeled PCB. Because both RR spectroscopy and normal mode analyses indicate that the vibrational spectra of both chromophores are essentially identical in this spectral range (35), this feature is ascribed to a baseline drift, which influences the position of the shifted stretching mode of the Pr state. Because the photoproduct yield for lumi-R is at least a factor of 5 smaller than for Pfr (see Material and Methods), the lumi-R spectra are especially prone to such distortions. We will show below that the real position of this shifted Pr band is around 1590 cm−1 instead of 1594 cm−1. Given the significant shift of ∼9 cm−1 induced upon 13C(10)-labeling, this band is assigned to a mode of largely B-C stretching character in agreement with normal mode analysis (vide supra).
Further effects induced by 13C(10)-labeling are the small difference band 1525(−)/1538(+) cm−1 and the larger difference band 1525(−)/1515(+) cm−1 that all shift down by ∼4 cm−1.
The corresponding B-C methine bridge stretching modes for lumi-F/Pfr give rise to the difference band at 1582(−)/1593(+) cm−1 that is downshifted by 10 cm−1 upon 13C(10)-labeling (Fig. 3). Interestingly, in Pfr this mode is at a frequency even lower than in lumi-R (Table 1). In addition, labeling leads to a new band at 1601 cm−1 in the Pfr state.
TABLE 1.
Experimental and calculated data of the B-C methine bridge stretching mode
| Adducts | n.a. [PED] | 13C(10) | H/D | 13C(5) | |
|---|---|---|---|---|---|
| Phytochromobilin | |||||
| ZZZ conformers | |||||
| Experiment | Pr | 1599 | 1590 | 1590 | |
| lumi-F | 1593 | 1583 | 1586 | ||
| Calculated | ZZZssa | 1597 [19% C=C (AB), 18% C–C (BC), 9% C–H rock (BC)] | 1585 | 1589 | |
| ZZZasa | 1599 [51% C–C (BC), 14% C–H rock (BC)] | 1578 | 1591 | ||
| ZZE conformers | |||||
| Experiment | Pfr | 1584 | 1575 | 1575 | |
| lumi-R | 1590 | 1582 | 1580 | ||
| Calculated | ZZEssa | 1594 [15% C=C (AB), 20% C–C (BC), 8% C–H rock (BC)] | 1582 | 1587 | |
| ZZEasa | 1598 [50% C–C(BC), 15% C–H rock (BC)] | 1577 | 1591 | ||
| Phycocyanobilin | |||||
| ZZZ conformers | |||||
| Experiment | Pr | 1602 | 1597 | ||
| lumi-F | 1598 | 1593 | |||
| Calculated | ZZZssa | 1596 [23% C=C (AB), 13% C–C (BC), 18% C=C (B)] | 1586 | ||
| ZZZasa | 1602 [49% C–C(BC), 18% C–H rock (BC)] | 1600 | |||
| ZZE conformers | |||||
| Experiment | Pfr | 1586 | 1581 | ||
| lumi-R | 1592 | 1590 | |||
| Calculated | ZZEssa | 1593 [18% C=C (AB), 17% C–C (BC), 20% C=C (B)] | 1584 | ||
| ZZEasa | 1603 [50% C–C(BC), 16% C–H rock (BC)] | 1601 | |||
Frequencies are given in cm−1 (experimental values are indicated in bold); calculated normal mode compositions (in brackets) are given in terms of potential energy distribution (PED). AB, A-B methine bridge; BC, B-C methine bridge; B, ring B; rock, rocking coordinate; C=C and C–C, stretching coordinates of C=C and C–C double and single bonds, respectively.
The complex structure in the 1535–1505 cm−1 frequency range of the lumi-F spectra is also affected by labeling. In this case, we detect a 2 cm−1 downshift of the weak positive band located around 1520 cm−1. However, the position of these bands may be perturbed by the large and broad positive band at 1540 cm−1 shifted down by 3 cm−1. This broad band is a peculiarity of the lumi-F state and has no counterpart in the Pfr state. In addition, the negative band at 1515 cm−1, which is associated to a vibrational mode of the Pfr state, is downshifted by 6 cm−1, similarly to the bands described in the lumi-R spectrum for this spectral region.
The identification of the IR bands of the Pr and Pfr states is confirmed by the analysis the Pfr/Pr difference spectrum. In principle, it should be possible to observe the negative bands of the lumi-R/Pr and lumi-F/Pfr difference spectra also in the Pfr/Pr difference spectrum as negative (Pr) and positive bands (Pfr), respectively. However, one has to take into account that the Pr→Pfr transition is associated with larger protein structural changes such that protein IR bands may interfere with those of the chromophore in the Pfr/Pr difference spectrum. Nevertheless, the band attributable to the B-C methine stretching mode of Pr is observed at the same position (1599 cm−1) as in the lumi-R/Pr difference spectrum whereas for Pfr the apparent peak position is slightly higher (1584 cm−1) as compared to that in the lumi-F/Pfr spectrum (1582 cm−1). 13C(10)-labeling gives rise to a positive shoulder at ∼1600 cm−1, supporting the finding that labeling causes a new band of the Pfr state at this position. The 13C-downshift of the 1599-cm−1 band of Pr (B-C stretching) is significantly larger (−15 cm−1) than that in the lumi-R/Pr difference spectrum (−5 cm−1). This discrepancy is attributed to the interference with the positive shoulder in the Pfr/Pr and the baseline drift in the lumi-R/Pr spectrum (vide supra), which may cause an over- and underestimation of the shifts, respectively. The true shift may lie within these extreme values (∼−10 cm−1) and is likely to be similar to that of the B-C methine bridge stretching of lumi-R. The corresponding band of Pfr is shifted down to ∼1575 cm−1 where it overlaps with the nearby positive band at 1568 cm−1 such that the intensity of the peak is relative low. Altogether, the difference spectra indicate that the frequency of the B-C stretching decreases in the order Pr > lumi-F > lumi-R > Pfr (Table 1).
A new difference signal is observed in the Pr→Pfr difference spectrum at 1569(−)/1549(+) cm−1. Labeling at C(10) does not seem to shift this band. We will provide evidence that this band must be assigned to amide II spectral changes, although it seems to be downshifted by uniformly 13C labeling of the chromophore (Fig. 2). Finally, there is a positive band at 1515 cm−1 that is shifted down to ∼1510 cm−1. It corresponds to the Pfr band observed in the lumi-F spectrum at 1515 cm−1. Surprisingly, there is no corresponding negative Pr band as it is observed it in the lumi-R spectrum.
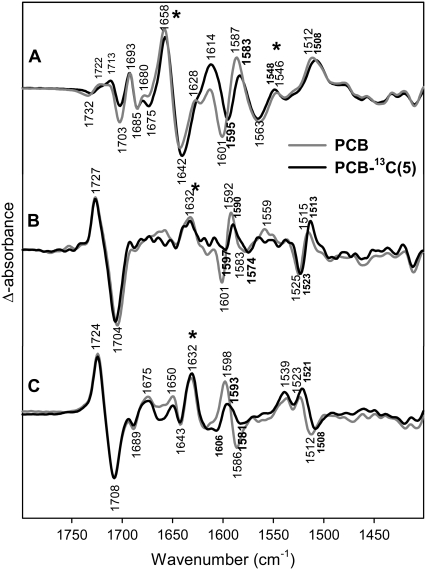
13C(5)-labeling of PCB
13C-labelling at position C(5) was carried out only for PCB that differs from PΦB by the ethyl substituent at ring D instead of a vinyl group. Both chromophores exhibit the same structure when incorporated in phyA (35,51) and the few spectral differences can be attributed readily to the different substituents at ring D. In the frequency range under consideration, the main spectral difference refers to the C-D stretching that increases in frequency by ∼15 cm−1 such that it overlaps with the A-B stretching at ∼1644 cm−1 as observed in the RR spectrum. As predicted by calculations (vide supra, Data S1), the C-D stretching exhibits low IR activity. Among the IR active modes, the B-C stretching is predicted to be between 1602 and 1594 cm−1 in the four geometries calculated for PCB and PΦB. Experimentally, a 2–3 cm−1 lower frequency is observed for all states of the phyA(PCB) adducts (Table 1). 13C-labelling at position C(5) causes a moderate downshift of this mode by ∼5 cm−1 in Pr, Pfr, and lumi-F and by 2 cm−1 in lumi-R (Fig. 4). For Pr, the mode that is dominated by the A-B stretching coordinate has been assigned to a band at ∼1644 cm−1 in the RR spectra of both phyA(PCB) and phyA(PΦB) adducts (35) that undergoes a ca. 15-cm−1 downshift upon 13C(5)-labeling. No difference signals are observed in this region of the lumi-R/Pr IR difference spectra, whereas in the Pfr/Pr difference spectrum relatively strong signals from the amide I bands prohibit the identification of any chromophore bands. Also in the lumi-F/Pfr difference spectrum, no bands shifts due to 13C(5)-labeling are detected between 1600 and 1650 cm−1 (Fig. 4).
FIGURE 4.
Comparison of FTIR difference spectra of phyA reconstituted with nonlabeled PΦB and phyA reconstituted with uniformly 13C(5)-labeled PCB. The spectra of the adducts with nonlabeled and labeled PΦB are given by the gray and black lines, respectively. The spectra (from top to bottom) refer to the “Pfr” minus “Pr” (A), “lumi-R” minus “Pr” (B), and “lumi-F” minus “Pfr” (C) differences. Protein changes are denoted by an asterisk. Further details are given in the text (Materials and Methods).
Below 1550 cm−1, the effects of 13C(5)-labeling are small and refer to relatively weak difference signals (Fig. 4). These are, for instance, the lumi-R/Pr difference bands at ∼1518 cm−1 (downshift 2 cm−1), the lumi-F band at 1523 cm−1 (downshift 2 cm−1) and the Pfr band at 1512 cm−1 (downshift 4 cm−1). This latter band is observed in both the lumi-F/Pfr and in the Pfr/Pr difference spectra whereas the corresponding Pr band is only detected in the lumi-R/Pr difference spectrum, in analogy to the spectra of the 13C(10)-labeled chromophore.
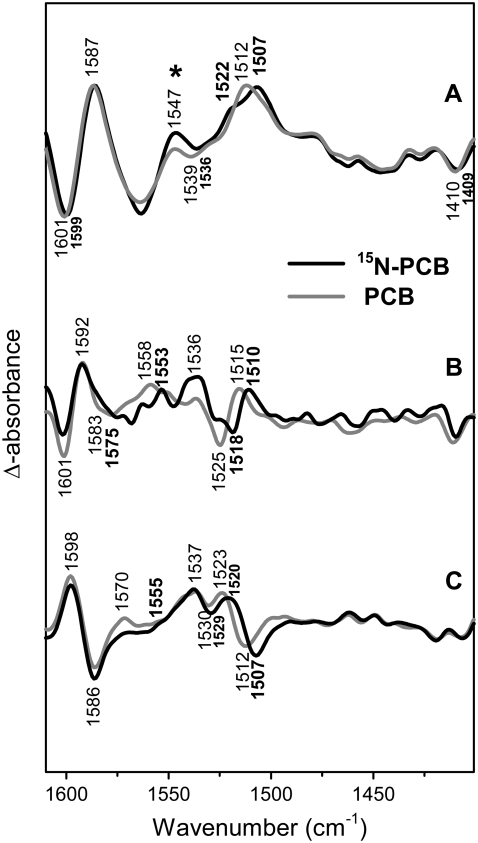
Uniform 15N-labeling of the PCB chromophore
Difference spectra of phyA adducts with uniformly 15N-labeled PCB show no effect on the B-C stretching mode for any of the phytochrome states (Fig. 5). Isotopic shifts are restricted to bands between 1535 and 1500 cm−1. These are the bands at 1512 (Pfr), 1515 (lumi-R), 1523 (lumi-F), and 1525 cm−1 (Pr) that all display a 5–7-cm−1 downshift in the adduct carrying the labeled chromophore. In addition, the Pr band seems to split by the labeling, causing the shoulder ∼1528 cm−1. These bands are attributed to a mode involving the N-H in-plane bending coordinates of ring B and C as predicted by the calculations (Data S1).
FIGURE 5.
Comparison of FTIR difference spectra of phyA reconstituted with nonlabeled PΦB and phyA reconstituted with uniformly 15N-labeled PCB. The spectra of the adducts with nonlabeled and labeled PΦB are given by the gray and black lines, respectively. The spectra (from top to bottom) refer to the “Pfr” minus “Pr” (A), “lumi-R” minus “Pr” (B), and “lumi-F” minus “Pfr” (C) differences. Protein changes are denoted by an asterisk. Further details are given in the text (Materials and Methods).
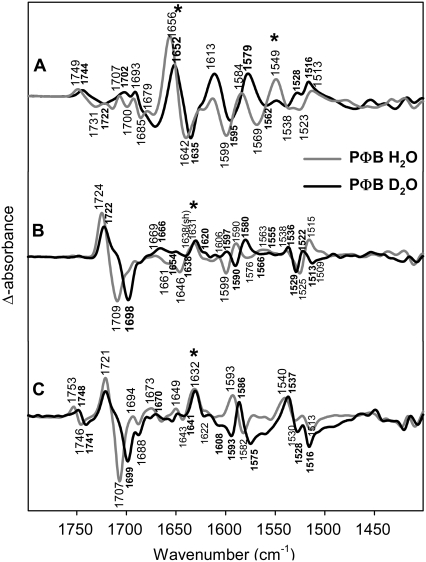
H/D exchange in phyA(PΦB) adducts
In the frequency range under consideration, isotopic effects produced upon H/D exchange may lead to difference bands originating from the amide I (>1620 cm−1) and amide II modes (1570–1540 cm−1) and from those chromophore modes that include N-H in-plane bending coordinate (Fig. 6). Bands attributable to the amide I observed for lumi-R and lumi-F at ∼1630 cm−1 show only a small downshift (−1 cm−1), which, at first, may be interpreted in terms of limited solvent exposure for those peptide groups undergoing a conformational transition during the primary photochemical processes. An alternative and more likely interpretation is based on the assignment of the 1630-cm−1 band to an amide I of a β-sheet structure that displays very small (<2 cm−1) H/D shifts (52,53). This would imply that the Pr→lumi-R transition is associated with a structural distortion of a peptide bond in a β-sheet structure. A similar amide I band is also observed in the transition from Pfr to lumi-F. Conversely, the amide I bands in the Pfr/Pr difference spectrum at 1656 (Pfr) and 1642 cm−1 (Pr) shift down by ∼6 cm−1 in D2O indicating that the conformational changes associated with the Pr/Pfr transition involve additional parts of the protein having random-coil and/or α-helical structures.
FIGURE 6.
Comparison of FTIR difference spectra of phyA reconstituted with nonlabeled PΦB and phyA reconstituted with PΦB in H2O (gray) and D2O (black). The spectra (from top to bottom) refer to the “Pfr” minus “Pr” (A), “lumi-R” minus “Pr” (B), and “lumi-F” minus “Pfr” (C) differences. Protein changes are denoted by an asterisk. Further details are given in the text (Materials and Methods).
Among the chromophore bands, we note distinct H/D effects for the B-C stretching modes that exhibit frequencies lowered by ∼7–9 cm−1 in all four states (Table 1). Furthermore, deuteration induces a new Pfr band around 1600 cm−1, which can be seen both in the lumi-F/Pfr and in the Pfr/Pr difference spectra. The change of the band structure around 1520 cm−1 is quite complicated and cannot be described by simple H/D induced shifts. This region includes the 15N/14N-sensitive N-H in-plane bending mode of ring B and C and, therefore, the composition of this mode and the adjacent modes will be altered drastically upon H/D exchange. In Pr the N-H in-plane bending at 1525 cm−1 is replaced by two bands at higher (1529 cm−1) and lower frequency (1513 cm−1), whereas in Pfr the 1513 cm−1 band in H2O has to be compared with a band at 1516 cm−1 in D2O. H/D-dependent changes in the vicinity of the 1523 cm−1 band in the lumi-F spectrum can hardly be determined due to the adjacent relatively strong band at 1540 cm−1. In lumi-R, the 1515-cm−1 band disappears and it is not clear whether the feature at 1522 cm−1 represents a true band of the chromophore.
The large difference band at 1569(−)/1549(+) cm−1 in the Pfr/Pr spectrum disappears in D2O. As discussed above, uniform 13C-labeling seems to shift this band down by 6 cm−1 (Fig. 2). However, the intensity of this signal is considerably increased, suggesting that two difference bands are superimposed. In fact, taking into account the 13C-downshift of the B-C methine stretching mode of Pr to 1553 cm−1 and assuming a comparably large downshift of the corresponding Pfr mode to ∼1535 cm−1, the observed difference band in the Pfr spectrum of the 13C-labeled species may be rationalized in terms of a superposition of the 13C-insensitive 1568(–)/1549(+) cm−1 difference band and the downshifted signal from the B-C methine stretching modes. Further support for this interpretation is obtained by measurements in D2O of phyA with the uniformly 13C-labeled chromophore. Here, the 1569(−)/1549(+) cm−1 difference band disappears, whereas the signal due to the B-C methine bridge stretching is further downshifted (not shown). Thus we assign the 1568(−)/1549(+) difference signal to changes of amide II modes, accompanying the significant amide I changes in the Pfr/Pr difference spectrum described above. In agreement with the deuteration-induced shift of the amide I band, the amide II band disappears in D2O.
DISCUSSION
We will focus on those modes that can be assigned safely on the basis of isotopic shifts and DFT calculations. Together with RR data reported previously, the changes of these in the IR differences spectra allows for a description of the chromophore and protein structural changes during the photocycle of phyA.
The chromophore structure in Pr
The chromophore structure in the parent state Pr of plant phytochrome phyA is a matter of controversial debate. On the basis of the crystal structures of the chromophore binding domains of bacterial phytochromes a ZZZssa configuration has been suggested (30,32). Such a geometry in which the rings A, B, and C are, however, essentially coplanar cannot be reconciled with the analysis of the RR spectra (35). On the basis of phyA assembled with isotopically labeled PCB, the A-B and C-D methine bridge stretching modes have been unambiguously assigned to the bands at 1644 and 1637 cm−1, respectively. This band pattern as well as the isotopic shifts for the C(15)D- and 13C(5)-labeled PCB adducts are well described by the spectra calculated for the ZZZasa geometry whereas the spectra obtained for the ZZZssa geometry do not reproduce the experimental data. However, this conclusion is based on spectra calculated for chromophore geometries optimized in vacuo, i.e., in the absence of interactions with the surrounding protein matrix. It can, therefore, not be excluded that protein-chromophore interactions affect the spectra of a hypothetical ZZZssa geometry to produce a band pattern and isotopic effects that are characteristic of the ZZZasa geometry in vacuo (35).
The presented FTIR data provide complementary information for determining the chromophore structure, particularly based on the B-C stretching mode, which due to its high IR intensity can be analyzed in terms of its isotopic shifts and compared with the calculated spectra. This mode can be reliable identified in all states. The calculated frequencies of this mode for PΦB and PCB of both the ZZZasa and the ZZZssa geometry agree very well (Δν ≤ 4 cm−1) with the experimental values. However, there are distinct discrepancies in the predicted and experimental isotopic shifts between the experimental data and the frequencies calculated for both geometries (Table 1). The experimental 13C(5) isotopic shift of −5 cm−1 in the phyA(PCB) adduct is overestimated and underestimated in the spectra calculated for the ZZZssa (−10 cm−1) and ZZZasa (−2 cm−1), respectively. Conversely, the calculated 13C(10)-isotopic shift for the phyA(PΦB) adduct is much higher for the ZZZasa (−21 cm−1) than for the ZZZssa geometry (−12 cm−1) that compares much better with the experimental shift of −9 cm−1.
These differences in the calculated spectra can be attributed readily to the different compositions of this mode. It possesses ∼50% B-C stretching character in the ZZZasa geometry, thereby accounting for the large 13C(10)-isotopic shift in the calculated spectra, whereas the contribution of this mode does not exceed 20% in the ZZZssa geometry, for which, in turn, a much higher A-B stretching character is predicted as reflected by the larger 13C(5) isotopic shift. We have, therefore, to ask what kind of chromophore structural changes might be induced by the interactions with the protein that alter the mode composition such that it is somehow in between the limiting cases of the in vacuo ZZZasa and ZZZssa geometries.
The unique structural feature of the ZZZssa geometry are the interactions of the ring A, B, and C N-H groups with the common counterion (Cl− in these calculations) (Data S1) that cause a near coplanarity of the three rings. This is most likely the origin for the substantial admixture of the A-B stretching coordinate to the “B-C” stretching mode. The interactions with the ring A N-H group are removed in the ZZZasa configuration and the ring A moves out of the plane spanned by the rings B and C. This geometric change and the alterations in the electrostatic interactions may cause a redistribution of the normal mode compositions such that the B-C content dominates. One may therefore imagine that in the true chromophore geometry of Pr the electrostatic interactions with a common hydrogen bond acceptor are weakened via a partial rotation around the A-B methine bridge single bond. Such a distorted ZZZssa structure may account for a mixing of the A-B and B-C stretching coordinates that is smaller than in the relaxed ZZZssa but larger than in the ZZZasa geometry, thereby providing a qualitative explanation for the isotopic shifts of the B-C stretching mode discussed above. An additional consequence of a twist around A-B methine single bond is that the π-electron delocalization is reduced and the A-B stretching approaches the character of an isolated C=C stretching mode, i.e., the frequency should be distinctly higher than that of a fully coplanar (relaxed ZZZssa) system. In fact, the RR spectra unambiguously identify the A-B stretching at a frequency that is higher by more than 20 cm−1 than that calculated for the relaxed ZZZssa geometry (35). The high frequency of this mode and the isotopic shifts produced by 13C-labelling at C(5) and C(15) have been well reproduced by the spectrum calculated for the ZZZasa geometry but they may well be also consistent with a distorted ZZZssa structure.
Also the C-D methine bridge is likely to be twisted taking into account the crystal structure data of the phytochrome fragment from Deinococcus radiodurans that indicate a ∼45° rotation of ring D out of the plane of the central pyrrole rings (30). This may be also the case in phyA as indicated by the relatively strong RR activity of the C(15)-H out-of-plane mode (51).
The photoconversion to lumi-R
The photoinduced transition from the Pr to the lumi-R state is generally assumed to be a Z→E isomerization of the C-D methine bridge double bond (36). The corresponding lumi-R/Pr difference spectrum is dominated by vibrational bands of the chromophore as identified by global labeling of PΦB. The number and intensities of IR difference bands attributable to the protein are small. In the amide I band region, there is only a relatively weak signal at 1631 cm−1 (Fig. 2). Thus, we conclude that the primary photoreaction of Pr is associated only with small adjustments of the peptide bonds that most likely are localized in the immediate vicinity of the chromophore, i.e., of the C-D entity. These subtle protein structural changes may be localized in β-sheet structure segments that are found in the immediate vicinity of the chromophore (30).
In the lumi-R state, the B-C methine bridge stretching shifts down by 10 cm−1 but the isotopic shifts are similar to that in Pr. In general, the ZZEssa geometry provides a better description of most of the isotopic shifts of the B-C stretching (Table 1) and predicts an intense and slightly 13C(10) sensitive band at 1546 cm−1 (−3 cm−1) that may account for the experimental band at 1538 cm−1 (−4 cm−1) (Fig. 3). However, the ZZEssa geometry overestimates the 13C(5) downshift (−9 cm−1 calculated vs. −2 cm−1 experiment; Table 1). Conversely, this shift is well reproduced by the spectrum for the ZZEasa geometry. In addition, for the ZZEasa, but not for the ZZEssa geometry, the calculations predict the high position of the A-B methine stretching mode as found for lumi-R by RR experiments (∼1648 cm−1) (34,51). Because this band is at about the same position as in the Pr state, it is not detected in the lumi-R/Pr IR difference spectrum, despite a medium IR intensity predicted by the calculations.
Evidently, the origin for these discrepancies between the calculated and the experimental spectra is the same as in the Pr state, i.e., a dihedral distortion of the A-B methine bridge leading to an admixture of the A-B stretching coordinate to the B-C stretching mode up to an extent that is higher and lower than in the relaxed ZZEasa and ZZEssa geometry, respectively. This would imply that the A-B methine bridge geometry remains unchanged during the Pr→lumi-R transition that is consistent with the view that chromophore structural changes in the primary photoreaction are restricted to the isomerization site, i.e., to the C-D methine bridge. This isomerization does not lead to a coplanar C-D entity as indicated by the strong C(15)-H out-of-plane mode in the RR spectrum (51). A complete rotation around the C-D methine bridge is likely to be prohibited due to steric constraints in the chromophore binding pocket as suggested by crystal structure data (30). Thus, it may well be that in the Z→E isomerization of the C-D methine bridge represents a ∼90° rotation of the double bond, from a twisted Z configuration in Pr to a twisted E configuration in lumi-R.
The PFR state
The thermal relaxation from lumi-R via the Meta-R states to Pfr is associated with major protein structural changes as already shown previously for Agrobacterium Agp1 (54). This data on the amide I band changes in the Pfr/Pr difference confirm this finding also for phyA. Furthermore, the significant H/D shifts of the amide I IR difference signals (Fig. 6) imply that the underlying protein structural changes include segments that readily exchange the amide protein, an indication for largely solvent-exposed peptide bonds.
In the Pfr state the B-C stretching shifts down further by 6 cm−1 whereas the isotopic shifts are not different from those in Pr and lumi-R. This finding suggests that also the normal mode composition is not distinctly altered such that the considerable downshift of this mode is attributed to changes of the geometries or the force constants of the coordinates involved rather than to a major change of the mode composition. These coordinates are mainly the B-C and A-B stretchings as well as the N-H in-plane bending of rings B and C.
It has been suggested that the transition to Pfr is associated with a (partial) rotation of the A-B methine bridge (34). This is reflected by a substantial downshift of the A-B stretching from ∼1645 cm−1 (lumi-R) to ∼1618 cm−1 in the RR spectrum (34,51) that is well reproduced by the spectrum calculated for the ZZEasa geometry. On the other hand, this calculated spectrum cannot account for the downshift of the B-C stretching (IR; Table 1). Previous RR experiments have shown that the Pfr state is characterized by a very strong C(15)-H out-of-plane mode, indicating a strongly twisted C-D methine bridge (51), and it could be argued that this twist causes the downshift. However, as shown by DFT calculations comparing the spectra obtained for PCB conformers that exhibit different C-D conformations in vacuo, the increase of the C-D methine bridge torsion does not affect the B-C methine stretching mode (55) Thus we conclude that the low frequencies of the B-C stretching in the IR spectrum and of the C-D stretching in the RR spectrum must reflect a structural change localized at the B-C methine bridge that may be induced by altering the interactions with the rings B and C. Model calculations on PCB and PΦB show that omitting the Cl− counterion causes a substantial frequency downshift of the C-D and B-C stretching modes. These frequency shifts may either be due to a lowering of the respective force constants or the substantial decrease of the (ring B)-C(10)-(ring C) bond angle or the increase of the B-C methine bridge dihedral angles. Unfortunately, no structure is available for the chromophore binding domain of plant phyA, which would be required to take in to account the chromophore-protein interaction in a quantum-mechanical/molecular-mechanics approach. The steric interactions and the interaction of the positively charged B-C part would be of special importance. As already mentioned, the structures of the bacterial phytochromes do not show a localized counterion (30,32).
The photoreaction of PFR
In lumi-F, the composition of the B-C stretching mode does not appear to be altered as concluded from the isotopic shifts. However, the frequency has increased to a value between those of the Pr and lumi-R states implying that the suggested interactions between the protein and rings B and C in Pfr (vide supra) are removed in lumi-F. The IR difference spectrum indicates only small protein structural changes that most likely refer to the same part of the protein as in the Pr to lumi-R transition (vide supra). Thus, we conclude that, similar to the Pr→lumi-R transition, the structural changes associated with the photoreaction of Pfr are largely restricted to the isomerization site. The conclusion also implies that the A-B methine bridge geometry remains essentially unchanged, which is consistent with the fact that no IR difference band at ∼1620 cm−1 attributable to A-B stretching is observed. The changes of the C=O stretch of ring A as indicated by Figs. 2, 3, and 6 in the transition from Pfr to lumi-F (see also Foerstendorf et al. (38)). do not necessarily contradict this conclusion because they could reflect a very small reorientation of ring A.
CONCLUSIONS
The combined approach presented here, using isotope labeled chromophores, vibrational spectroscopy, and DFT calculations, adds significant information on the photochemistry of the plant photoreceptor phytochrome. However, the inherent problems also become evident, including the constraints for the theoretical calculations. Yet, one can unambiguously assign amide I and amide II bands in the Pfr spectrum, and amide I bands in the lumi-R and lumi-F spectra by the incorporation of the fully 13C labeled chromophore. Similarly, the selectively labeled chromophores allow for the interpretation of the IR bands in terms of internal coordinates contributing to a normal mode. Whereas in the lumi intermediates the structural changes are restricted to the chromophore and its immediate environment (probably including small changes of a β-sheet structure), later intermediates and the Pfr state show large protein conformational changes of α-helical and random coil parts. These observations are of importance for the signaling function of phytochrome.
Comparing the three-dimensional structures of the chromophore-bearing domains with the results obtained by the theoretical analysis of the isotope-edited IR spectra, problems are evident. It can be stated clearly that simple E-Z-isomerizations do not describe sufficiently the molecular changes of the chromophore and the compensatory movements after the photoisomerization. Even if double and single bond rotations are identified as ZZZ(E)ssa, or -asa, one has to propose a distorted conformation of the chromophore. The current state of interpretation of RR and the IR data favors a twisted A-B methine bridge either in the ssa, but also in the asa geometry. The former would be in agreement with the structural data of bacterial phytochromes. Furthermore, the B-C stretching mode appears especially sensitive to changes of the chromophore-protein interaction involving rings B and C that are especially pronounced in the transition to the Pfr state.
In future studies, the focus will be on improvements of the vibrational analysis by inclusion of the protein in DFT QM/MM calculations, provided a structure of the chromophore-binding domain becomes available. In further experiments it will be interesting to study by IR and RR spectroscopy the impact of mutations that are thought, based on available structural data, to impose constraints on the chromophore or to alter polar interactions with the NH group of the four pyrrole rings. For these future investigations, the normal modes identified here by isotopic labeling will be an important basis.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft grants to M.A.M. and P.H. (SFB 498), W.G. (GA 377/13-1,2), and F.S. and P.S. (SI 278/25-1,2).
Editor: Brian R. Dyer.
References
- 1.Schäfer, E., and F. Nagy. 2006. Photomorphogenesis in plants and bacteria. Springer, Dordrecht, The Netherlands.
- 2.Hershey, H. P., R. F. Barker, K. B. Idler, J. L. Lissemore, and P. H. Quail. 1985. Analysis of cloned cDNA and genomic sequences for phytochrome: complete amino acid sequences for two gene products expressed in etiolated Avena. Nucleic Acids Res. 13:8543–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sineshchekov, V. A. 1995. Photobiophysics and photobiochemistry of the heterogeneous phytochrome system. Biochim. Biophys. Acta. 1228:125–164. [Google Scholar]
- 4.Ruddat, A., P. Schmidt, C. Gatz, S. E. Braslavsky, W. Gärtner, and K. Schaffner. 1997. Recombinant type A and B phytochromes from potato. Transient absorption spectroscopy. Biochemistry. 36:103–111. [DOI] [PubMed] [Google Scholar]
- 5.Gärtner, W., and S. Braslavsky. 2004. The phytochromes: spectroscopy and function. In Photoreceptors and Light Signaling. A. Batschauer, editor. Royal Society of Chemistry, Cambridge, UK. 136–180.
- 6.Karniol, B., J. R. Wagner, J. M. Walker, and R. D. Vierstra. 2005. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montgomery, B. L., and J. C. Lagarias. 2002. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 7:357–366. [DOI] [PubMed] [Google Scholar]
- 8.Hughes, J., and T. Lamparter. 1999. Prokaryotes and phytochrome. The connection to chromophores and signaling. Plant Physiol. 121:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh, K. C., and J. C. Lagarias. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA. 95:13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhoo, S. H., S. J. Davis, J. Walker, B. Karniol, and R. D. Vierstra. 2001. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 414:776–779. [DOI] [PubMed] [Google Scholar]
- 11.Hübschmann, T., H. J. M. M. Jorissen, T. Börner, W. Gärtner, and N. T. de Marsac. 2001. Phosphorylation of proteins in the light-dependent signaling pathway of a filamentous cyanobacterium. Eur. J. Biochem. 268:3383–3389. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser, C., K. C. Yeh, J. C. Lagarias, H. Zhang, T. D. Elich, and J. Chory. 1999. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 284:1539–1541. [DOI] [PubMed] [Google Scholar]
- 13.Choi, G., H. Yi, J. Lee, Y. K. Kwon, M. S. Soh, B. Shin, Z. Luka, T. R. Hahn, and P. S. Song. 1999. Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature. 401:610–613. [DOI] [PubMed] [Google Scholar]
- 14.Ni, M., J. M. Tepperman, and P. H. Quail. 1998. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 95:657–667. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Garcia, J. F., E. Huq, and P. H. Quail. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 288:859–863. [DOI] [PubMed] [Google Scholar]
- 16.Casal, J. J., L. G. Luccioni, K. A. Oliverio, and H. E. Boccalandro. 2003. Light, phytochrome signaling and photomorphogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2:625–636. [DOI] [PubMed] [Google Scholar]
- 17.Chen, M., R. Schwab, and J. Chory. 2003. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA. 100:14493–14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagatani, A. 2004. Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7:708–711. [DOI] [PubMed] [Google Scholar]
- 19.Sweere, U., K. Eichenberg, J. Lohrmann, V. Mira-Rodado, I. Bäurle, J. Kudla, F. Nagy, E. Schäfer, and K. Harter. 2001. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science. 294:1108–1111. [DOI] [PubMed] [Google Scholar]
- 20.Oka, Y., T. Matsushita, N. Mochizuki, T. Suzuki, S. Tokutomi, and A. Nagatani. 2004. Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell. 16:2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mateos, J. L., J. P. Luppi, O. B. Ogorodnikova, V. A. Sineshchekov, M. J. Yanovsky, S. E. Braslavsky, W. Gärtner, and J. J. Casal. 2006. Functional and biochemical analysis of the N-terminal domain of phytochrome A. J. Biol. Chem. 281:34421–34429. [DOI] [PubMed] [Google Scholar]
- 22.Elich, T. D., and J. C. Lagarias. 1989. Formation of a photoreversible phycocyanobilin-apophytochrome adduct in vitro. J. Biol. Chem. 264:12902–12908. [PubMed] [Google Scholar]
- 23.Terry, M. J., J. A. Wahleithner, and J. C. Lagarias. 1993. Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306:1–15. [DOI] [PubMed] [Google Scholar]
- 24.Lindner, I., B. Knipp, S. Braslavsky, W. Gärtner, and K. Schaffner. 1998. A novel chromophore selectivity modifies the spectral properties of one of the two stable states of the plant photoreceptor phytochrome. Angew. Chem. Int. Ed. Engl. 37:1843–1846. [Google Scholar]
- 25.Hanzawa, H., K. Inomata, H. Kinoshita, T. Kakiuchi, K. P. Jayasundera, D. Sawamoto, A. Ohta, K. Uchida, K. Wada, and M. Furuya. 2001. In vitro assembly of phytochrome B apoprotein with synthetic analogs of the phytochrome chromophore. Proc. Natl. Acad. Sci. USA. 98:3612–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robben, U., I. Lindner, W. Gärtner, and K. Schaffner. 2001. Analysis of the topology of the chromophore binding pocket of phytochromes by variation of the chromophore substitution pattern. Angew. Chem. Int. Ed. Engl. 40:1048–1050. [DOI] [PubMed] [Google Scholar]
- 27.Hanzawa, H., T. Shinomura, K. Inomata, T. Kakiuchi, H. Kinoshita, K. Wada, and M. Furuya. 2002. Structural requirement of bilin chromophore for the photosensory specificity of phytochromes A and B. Proc. Natl. Acad. Sci. USA. 99:4725–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamparter, T., N. Michael, O. Caspani, T. Miyata, K. Shirai, and K. Inomata. 2003. Biliverdin binds covalently to agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 278:33786–33792. [DOI] [PubMed] [Google Scholar]
- 29.Quest, B., T. Hübschmann, S. Sharda, N. Tandeau de Marsac, and W. Gärtner. 2007. Homologous expression of a bacterial phytochrome. The cyanobacterium Fremyella diplosiphon incorporates biliverdin as a genuine, functional chromophore. FEBS J. 274:2088–2098. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, J. R., J. S. Brunzelle, K. T. Forest, and R. D. Vierstra. 2005. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 438:325–331. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, J. R., J. Zhang, J. S. Brunzelle, R. D. Vierstra, and K. T. Forest. 2007. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282:12298–12309. [DOI] [PubMed] [Google Scholar]
- 32.Yang, X., E. A. Stojkovic, J. Kuk, and K. Moffat. 2007. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc. Natl. Acad. Sci. USA. 104:12571–12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mroginski, M. A., D. H. Murgida, and P. Hildebrandt. 2007. The chromophore structural changes during the photocycle of phytochrome: a combined resonance Raman and quantum chemical approach. Acc. Chem. Res. 40:258–266. [DOI] [PubMed] [Google Scholar]
- 34.Mroginski, M. A., D. H. Murgida, D. von Stetten, C. Kneip, F. Mark, and P. Hildebrandt. 2004. Determination of the chromophore structures in the photoinduced reaction cycle of phytochrome. J. Am. Chem. Soc. 126:16734–16735. [DOI] [PubMed] [Google Scholar]
- 35.Murgida, D. H., D. von Stetten, P. Hildebrandt, P. Schwinte, F. Siebert, S. Sharda, W. Gärtner, and M. A. Mroginski. 2007. The chromophore structures of the Pr states in plant and bacterial phytochromes. Biophys. J. 93:2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rüdiger, W. 1992. Events in the phytochrome molecule after irradiation. Photochem. Photobiol. 56:803–809. [DOI] [PubMed] [Google Scholar]
- 37.Borucki, B., D. von Stetten, S. Seibeck, T. Lamparter, N. Michael, M. A. Mroginski, H. Otto, D. H. Murgida, M. P. Heyn, and P. Hildebrandt. 2005. Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J. Biol. Chem. 280:34358–34364. [DOI] [PubMed] [Google Scholar]
- 38.Foerstendorf, H., C. Benda, W. Gärtner, M. Storf, H. Scheer, and F. Siebert. 2001. FTIR studies of phytochrome photoreactions reveal the C=O bands of the chromophore: consequences for its protonation states, conformation, and protein interaction. Biochemistry. 40:14952–14959. [DOI] [PubMed] [Google Scholar]
- 39.Rohmer, T., H. Strauss, J. Hughes, H. de Groot, W. Gärtner, P. Schmieder, and J. Matysik. 2006. (15)N MAS NMR studies of cph1 phytochrome: chromophore dynamics and intramolecular signal transduction. J. Phys. Chem. B. 110:20580–20585. [DOI] [PubMed] [Google Scholar]
- 40.Kufer, W. and H. Scheer. 1979. Chemical modification of biliprotein chromophores. Z. Naturforsch. 34:776–781. [DOI] [PubMed] [Google Scholar]
- 41.Makhynya, Y., Z. Hussain, T. Bauschlicher, P. Schwinté, F. Siebert, and W. Gärtner. 2007. Synthesis of selectively 13C-labelled bilin compounds. J. Org. Chem. 8:1287–1293. [Google Scholar]
- 42.Cornejo, J., S. I. Beale, M. J. Terry, and J. C. Lagarias. 1992. Phytochrome assembly. The structure and biological activity of 2(R),3(E)-phytochromobilin derived from phycobiliproteins. J. Biol. Chem. 267:14790–14798. [PubMed] [Google Scholar]
- 43.Mozley, D., A. Remberg, and W. Gärtner. 1997. Large-scale generation of affinity-purified recombinant phytochrome chromopeptide. Photochem. Photobiol. 66:710–715. [DOI] [PubMed] [Google Scholar]
- 44.Lagarias, J. C., J. M. Kelly, K. L. Cyr, and W. O. Smith, Jr. 1987. Comparative photochemical analysis of highly purified 124 kilodalton oat and rye phytochromes. Photochem. Photobiol. 46:5–13. [Google Scholar]
- 45.Vogel, R., and F. Siebert. 2003. Fourier transform IR spectroscopy study for new insights into molecular properties and activation mechanisms of visual pigment rhodopsin. Biopolymers. 72:133–148. [DOI] [PubMed] [Google Scholar]
- 46.Siebert, F., R. Grimm, W. Rüdiger, G. Schmidt, and H. Scheer. 1990. Infrared spectroscopy of phytochrome and model pigments. Eur. J. Biochem. 194:921–928. [DOI] [PubMed] [Google Scholar]
- 47.Foerstendorf, H., E. Mummert, E. Schäfer, H. Scheer, and F. Siebert. 1996. Fourier-transform infrared spectroscopy of phytochrome: difference spectra of the intermediates of the photoreactions. Biochemistry. 35:10793–10799. [DOI] [PubMed] [Google Scholar]
- 48.Eilfeld, P., W. Rüdiger. 1985. Absorption spectra of phytochrome intermediates. Z. Naturforsch. 40c:109–114. [Google Scholar]
- 49.Becke, A. D. 1993. Density-functional thermochemistry 3. the role of exact exchange. J. Chem. Phys. 98:5648–5652. [Google Scholar]
- 50.Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery, Jr., T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomai, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyouta, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, V. Bakken, C. Adamo, J. Jaramilo, R. Gomperts, R. E. Stratman, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez, and J. A. Pople. 2004. Gaussian 03, Revision C02. Gaussian, Inc., Wallingford, CT.
- 51.Kneip, C., P. Hildebrandt, W. Schlamann, S. E. Braslavsky, F. Mark, and K. Schaffner. 1999. Protonation state and structural changes of the tetrapyrrole chromophore during the Pr→Pfr phototransformation of phytochrome: a resonance Raman spectroscopic study. Biochemistry. 38:15185–15192. [DOI] [PubMed] [Google Scholar]
- 52.Goormaghtigh, E., V. Cabiaux, and J.-M. Ruysschaert. 1994. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. III. Secondary structures. In Subcellular Biochemistry, Vol. 23. H. J. Hilderson and G. B. Ralston, editors. Plenum Press, New York. 405–450. [DOI] [PubMed]
- 53.Arrondo, J. L. R., A. Muga, J. Castresana, and F. M. Goñi. 1993. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 59:23–56. [DOI] [PubMed] [Google Scholar]
- 54.Noack, S., N. Michael, R. Rosen, and T. Lamparter. 2007. Protein conformational changes of Agrobacterium phytochrome Agp1 during chromophore assembly and photoconversion. Biochemistry. 46:4164–4176. [DOI] [PubMed] [Google Scholar]
- 55.Mroginski, M. A., F. Mark, W. Thiel, and P. Hildebrandt. 2007. Quantum mechanics/molecular mechanics calculations of the Raman spectra of the phycocyanobilin chromophore in α-phycocyanin. Biophys. J. 93:1885–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.