Abstract
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) play a key role in membrane fusion in the secretory pathway. In vitro, SNAREs spontaneously assemble into helical SNARE complexes with the transmembrane domains at the C-terminal end. During fusion, SNAREs are thought to bridge the two membranes and assemble in a zipper-like fashion, pulling the membranes together and initiating fusion. However, it is not clear to what extent SNARE assembly contributes to membrane attachment and membrane fusion. Using the neuronal SNAREs synaptobrevin (VAMP), SNAP-25, and syntaxin as examples, we show here that liposomes containing synaptobrevin firmly attach to planar surfaces containing immobilized syntaxin. Attachment requires the formation of SNARE complexes because it is dependent on the presence of SNAP-25. Binding is competed for by soluble SNARE fragments, with noncognate SNAREs such as endobrevin (VAMP8), VAMP4, and VAMP7 (Ti-VAMP) being effective but less potent in some cases. Furthermore, although SNAP-23 is unable to substitute for SNAP-25 in the attachment assay, it forms complexes of comparable stability and is capable of substituting in liposome fusion assays. Vesicle attachment is initiated by SNARE assembly at the N-terminal end of the helix bundle. We conclude that SNAREs can indeed form stable trans-complexes that result in vesicle attachment if progression to fusion is prevented, further supporting the zipper model of SNARE function.
INTRODUCTION
Intracellular membrane fusion in the secretory pathway of eukaryotic cells is mediated by conserved protein families, indicating that the basic mechanisms of vesicle docking and fusion are similar in all trafficking steps (1). Among these proteins, the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) have emerged as key players in the final steps of fusion (2). SNARE proteins are characterized by a conserved stretch of 60–70 amino acids arranged in heptad repeats (referred to as a SNARE motif) that is usually connected by a short linker to a single C-terminal transmembrane domain. Some SNAREs, such as the neuronal SNARE SNAP-25, lack transmembrane domains and possess two SNARE motifs separated by a linker that may be palmitoylated and serve as a membrane anchor. Furthermore, some subfamilies contain independently folded N-terminal domains that probably exert a regulatory function (for review see Jahn and Scheller (2) and Rizo et al. (3).
SNAREs spontaneously associate into SNARE complexes of very high stability, and the concerted action of the AAA-ATPase NSF and SNAP-cofactors is required for disassembly. Association is mediated by the SNARE motifs. The crystal structures of SNARE complexes that are only distantly related to each other show a remarkable degree of structural conservation (4–7), allowing the deduction of basic features shared by all SNAREs. Each SNARE complex is represented by a heterooligomeric bundle of four parallel α-helices. The helices form a coiled coil that is stabilized by 16 stacked layers of interacting amino acid side chains. Although most of these side chains are hydrophobic, the central layer (“0” layer) is formed by one arginine (R) and three glutamine (Q) residues that are all highly conserved. Accordingly, SNAREs are classified into Q- and R-SNAREs, with the Q-SNAREs being further subdivided into Qa-, Qb-, and Qc-SNAREs (8,9). Sequence analysis has revealed that these four subfamilies are conserved and probably diverged very early in eukaryotic evolution, lending further support to the view that all SNARE complexes that function in membrane fusion have a QaQbQcR composition (2,10,11).
Fusion requires complementary SNAREs to be present on both membranes that are destined to fuse, and assembly of SNAREs into SNARE complexes is supposed to play a key role in fusion. According to the now widely accepted “zipper model,” the SNARE motifs initiate assembly at their N-terminal ends (“trans”-complex), bridging the membranes (12). Assembly then progresses toward the C-terminal membrane anchors, thus forming a tight connection between the membranes that may overcome the energy barrier for fusion. Although this model is supported by a large body of evidence, including both biochemical in vitro studies and analysis of SNARE mutants in living cells (for review see Sørensen [13]), the mechanistic details of the assembly pathway and the structure of the intermediates are less well understood. For instance, it was recently shown that, at least for the SNAREs that function in neuronal exocytosis, assembly is not the result of random collisions between four independent SNARE motifs, but rather proceeds via defined intermediates. Detailed studies of the SNAREs syntaxin 1 (Qa), SNAP-25 (Qbc), and synaptobrevin (VAMP) (R) have revealed that an unstable acceptor complex, consisting of the three Q-SNARE motifs, is a mandatory intermediate for SNARE assembly (14). In vitro, this complex readily recruits a second syntaxin that fills the position of the R-SNARE, resulting in a functionally inactive (and probably nonphysiological) QaQaQbc complex, which explains why SNARE assembly is very slow in vitro (15).
One of the most controversial issues under discussion is whether SNARE assembly results primarily in membrane attachment before fusion with fusion being brought about by other factors, or whether it mediates fusion whereas membrane attachment involves other proteins. Reconstitution of SNAREs into liposomes results in spontaneous fusion that is dependent on the formation of SNARE complexes (16), which suggests that SNAREs function as fusion factors. Whereas this assay is widely used as a reduced in vitro model for fusion (see, e.g., Parlati et al. [17], Bacia et al. [18], and Tucker et al. [19]), other studies argue that fusion in such systems is partially relying on nonspecific fusogenic properties of the liposomes and thus does not represent a valid model for the fusion pathway in biological membranes (20,21) (see Rizo et al. [3] for a comprehensive discussion). Unfortunately, it is experimentally not easy to capture short-lived intermediates such as trans-SNARE complexes and to differentiate between docking and fusion. In neuronal exocytosis, indirect evidence indicates an interaction of synaptobrevin with syntaxin and SNAP-25 before exocytosis, such as differential sensitivity to cleavage by clostridial neurotoxins or to inhibition by antibodies (22,23). More direct evidence for trans intermediates was recently obtained for SNAREs involved in the fusion of yeast vacuoles, using an assay that is based on coprecipitation of tagged SNAREs from docked vesicles (24).
In liposome fusion assays, the frequency of collisions between liposomes is so high that docking is not rate-limiting (25), although in some systems the need for preincubation at low temperature seems to indicate that SNARE-mediated docking must precede fusion (16). Furthermore, recent studies have suggested that a binary interaction between synaptobrevin and syntaxin is sufficient for docking and subsequent fusion of artificial vesicles to planar lipid bilayers (26,27). However, fusion of liposomes requires the presence of all three SNAREs, and no stable binary interaction between syntaxin and synaptobrevin has been detected. Interestingly, two studies (21,28) showed that although docking of liposomes occurred in the presence of synaptobrevin and syntaxin, an interaction between these proteins as measured by fluorescence resonance energy transfer was only detectable in the presence of SNAP-25.
In this study we investigated whether SNAREs are capable of docking vesicles to a planar solid surface. To this end, we immobilized syntaxin on gold-coated glass in a functionally active form. Binding of synaptobrevin liposomes then occurs in a SNAP-25-dependent manner. Further characterization revealed that docking correlates well with the formation of SNARE complexes, as observed in both solution studies and liposome fusion assays, but differences in stability are uncovered when cognate SNARES are exchanged for noncognate SNAREs.
MATERIALS AND METHODS
Molecular cloning, expression, and purification of recombinant proteins
The following constructs of SNARE proteins from Rattus norvegicus, as described previously (14,15,29), were used in this study: synatobrevin full length (aa1-116), synaptobrevin cytosolic part (aa1-96), syntaxin 1a cytosolic part (aa1-262), syntaxin 1a H3 domain (aa180-262), N-terminally truncated syntaxin (aa183-288), SNAP23 full length (aa1-210), SNAP25a full length (aa1-206) mutant in which all cysteines were substituted for serines (30), SNAP25a C-terminal BotNT/A fragment (aa1-197), and SNAP25a C-terminal BoNT/E (aa1-180) fragment. Syntaxin 1a cytosolic part and H3 domain with N-terminal cysteine (aa1-262 263Cys and 180-262 263Cys, respectively) were generated using site-directed mutagenesis (31). Recombinant proteins were expressed as 6xHis fusion proteins in BL21DE Escherichia coli strain from pET28 vector and purified on nickel-nitrilotriacetic acid (Ni-NTA) agarose, followed by further purification with an Äkta system (GE Healthcare, Freiburg, Germany) on MonoS or MonoQ columns (GE Healthcare). Protein purification was carried out essentially as described previously (32). The 6xHis tag was removed by thrombin cleavage after the Ni-NTA purification step. For the purification of proteins containing the transmembrane domain, sodium cholate was used for membrane solubilization and was present at 1.5% in all buffers during the purification procedure.
Atomic force microscopy (AFM)
Coverslips were coated for 1 h with 1 mg/mL BSA (fraction V, protease-free; Sigma-Aldrich, Taufkirchen, Germany), extensively washed with phosphate-buffered saline (PBS; 40 mM sodium phosphate pH 7.3, 150 mM NaCl) and incubated for 30 min with 5 nm colloidal gold at room temperature. After the coverslips were washed with HPLC-grade water free from particulate matter (Merck, Darmstadt, Germany), they were dried and imaged on a NanoScope MultiMode IIIa scanning probe microscope (Veeco, Mannheim, Germany). The images (1 μm × 1μm) were recorded in air with an RTESP cantilever (Veeco Probes, Mannheim, Germany; tip radius < 10 nm) in tapping mode.
Preparation of proteoliposomes
Proteoliposomes were prepared essentially as previously described (25). Briefly, lipids (Avanti Polar Lipids, Alabaster, AL) were mixed in chloroform to yield (molar ratios) phosphatidylcholine (5), phosphatidylethanolamine (2), phosphatidylserine (1), phosphatidylinositol (1), and cholesterol (1). The lipid mix for the liposomes used in the docking assay contained 2% (n/n) 1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N-lissamine Rhodamine B sulfonyl. For preparation of the NBD/Rhodamine-labeled liposomes used in the fusion assay, 1.5% (n/n) 1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N-lissamine Rhodamine B sulfonyl, 1.5% (n/n) 1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl), and 17% (n/n) phosphatidylethanolamine were used instead of 20% (n/n). After drying, the liposomes were resuspended in 20 mM Hepes/KOH pH 7.4, 100 mM KCl, 5 mM dithiothreitol, and 5% (w/v) sodium cholate at a total lipid concentration of 13.5 mM. SNARE proteins in 1.5% sodium cholate were added to yield a lipid/protein molar ratio of 160:1.
Liposomes were formed via detergent removal on a G-25 superfine Sephadex (GE Healthcare) column equilibrated either in PBS for the liposome docking assay or in 20 mM Hepes pH 7.4, 140 mM KCl for the liposome fusion assay by using a sample/column volume ratio of 1:30. Liposomes for the docking assay were diluted 1:20 in PBS and snap-frozen in liquid nitrogen. After thawing, they were kept on ice and not frozen again. Liposomes for the fusion assay were prepared fresh each time, kept on ice, and used undiluted.
Liposome fusion assay
Fusion of liposomes was measured by fluorescence dequenching at 30°C in a FluoroMax II fluorometer (HORIBA Jobin Yvon, Edison, NJ) essentially as previously described (25). Unlabeled syntaxin (aa183–288) liposomes were mixed with equal amounts of synaptobrevin-containing liposomes labeled with NBD/Rhodamine in a total volume of 30 μl (final buffer concentrations: 20 mM Hepes/KOH pH 7.4, 140 mM KCl). The final lipid concentration was ∼0.3 mM. Fluorescence dequenching was measured using 460 nm for excitation and 538 nm for emission. Fluorescence intensities were normalized to the initial fluorescence intensity.
Liposome binding assay
Glass coverslips (12 mm diameter) were sonicated for 10 min in 2% Hellmanex II (Helma, Müllheim, Germany) in an ultrasound bath, washed extensively, and sonicated in water for another 10 min. Then the water was exchanged with 100% methanol and the coverslips were sonicated again for 10 min to remove the remaining traces of Hellmanex detergent. Methanol was washed away with water and the coverslips were dried for 3 h at 80°C or overnight at 50°C. For coating, each coverslip was incubated with a 120 μl drop of 1% bovine serum albumin (BSA) in PBS, ensuring compete coverage of the surface. After 1 h at room temperature, the coverslips were rinsed extensively. Next, a 120 μl drop of colloidal gold suspension (5-nm particle size; BB International, Cardiff, UK) was placed on the coverslip and incubated for 30 min. To ensure binding of the gold particles, complete removal of nonadsorbed BSA in the preceding rinsing steps was essential. Then the coverslips were washed with PBS and incubated with 0.7 mg/mL syntaxin (H3 domain containing a C-terminal cysteine) at 37°C for 2 h (standard assay conditions) unless indicated otherwise. The coverslips were rinsed again with PBS and then used in the binding assay.
For liposome binding, the coverslips were placed upside down on a drop with synaptobrevin-containing liposomes diluted 1:20 in PBS containing 1 mg/mL and SNAP25 or its mutants/homologs at a final concentration of 1 mg/mL. Unless indicated otherwise, incubation was carried out for 15 min at room temperature (standard assay conditions). The coverslips were then washed with PBS, incubated for 5 min with green fluorescent beads (200 nm, 2% solids diluted 1:10,000 in PBS; Molecular Probes) as markers for adjusting the focal plane. Coverslips were imaged on a Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss, Oberkochen, Germany) with a 1.4 numerical aperture 63× objective and appropriate filter sets. For each experiment, two coverslips were analyzed in parallel, three images per coverslip were obtained, and the average of the six values was calculated.
RESULTS
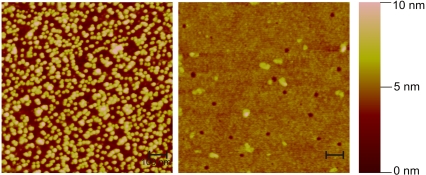
Exocytosis of synaptic vesicles involves the SNARE synaptobrevin (VAMP) located in the membrane of the synaptic vesicle, and the SNAREs syntaxin 1 and SNAP-25 located in the presynaptic plasma membrane. To study the role of these SNAREs in vesicle docking in a reduced in vitro system, we developed an assay that enables one to monitor the binding of liposomes reconstituted with purified synaptobrevin to a planar surface containing immobilized syntaxin. The planar surfaces to be used in such assays need to meet several requirements, including low nonspecific binding of liposomes and a cross-linking chemistry for immobilization that leaves the SNARE motif free to interact with the partner SNAREs. After screening a variety of approaches, we resorted to coverslips coated with colloidal gold as solid support. The surface of gold particles is negatively charged, and proteins can be firmly attached to gold via cysteine side chains; however, noncovalent hydrophobic and ionic interaction may also contribute to binding (33). For deposition of colloidal gold on glass surface, the coverslips were first coated with BSA (see Materials and Methods). Analysis of the resulting surface with AFM revealed a dense monolayer of individual, mostly nonaggregated gold particles (Fig. 1).
FIGURE 1.
Imaging by AFM of glass coverslips coated with BSA alone (right panel) or with BSA and 5 nm colloidal gold (left panel). The images were recorded in tapping mode and represent the height range in false colors with maximal height of 10 nm (brightness scale on the right). Notice that gold particles appear slightly bigger than their actual size of 5 nm due to the resolution limits of the cantilever. Scale bar = 100 nm.
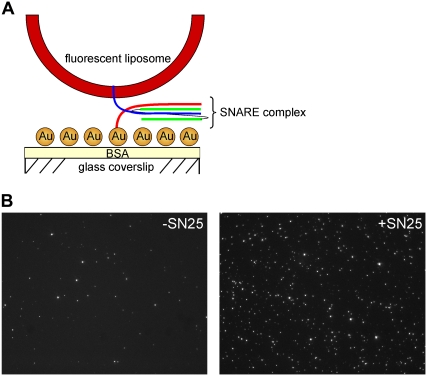
The principle for the vesicle binding assay is depicted in Fig. 2 A. Coated coverslips were placed on top of a drop of salt solution containing synaptobrevin liposomes that were labeled with the fluorescent dye Rhodamine. At the end of the reaction, the solution was washed off and the amount of liposomes bound per area was quantitated by fluorescence microscopy. As shown in Fig. 2 B, liposomes (visible as individual fluorescent dots) were randomly scattered on the surface. When SNAP-25 was present during the incubation, the number of bound liposomes was significantly increased, indicating that efficient binding requires the presence of a full complement of SNAREs and thus involves the formation of SNARE complexes.
FIGURE 2.
Docking of liposomes reconstituted with synaptobrevin to immobilized syntaxin is dependent on the presence of SNAP-25. (A) Cartoon showing the experimental setup of the SNARE-mediated docking assay used in this study. (B) Typical fluorescent image of synaptobrevin liposomes docked to syntaxin-coated coverslips in the absence (left panel) or presence (right panel) of SNAP-25. The experiment was carried out under standard assay conditions (see Materials and Methods for details).
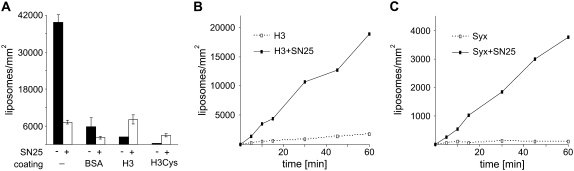
As discussed above, it is essential to differentiate between specific and nonspecific binding in such assays. Therefore, we investigated both surface treatment and incubation conditions in a systematic manner. When the gold surface was left untreated, a high degree of nonspecific binding was observed (Fig. 3 A). This may be due to an ionic interaction between synaptobrevin (exhibiting an overall positive charge) with the negatively charged colloidal gold, since syntaxin-containing liposomes do not show such pronounced unspecific binding (data not shown). Treatment of the surface with BSA, which is known to bind to colloidal gold, reduced the unspecific binding. Furthermore, nonspecific binding was reduced in the presence of SNAP-25, probably due to additional surface shielding. When the gold surface was first reacted with a fragment of syntaxin encompassing the SNARE motif, binding was dependent on the presence of SNAP-25. It is interesting that SNAP-25-dependent binding was also observed when a cysteine-free variant of syntaxin was used, which suggests that adsorption of syntaxin by noncovalent interactions does not interfere with its ability to engage in SNARE complexes. Under these conditions, however, background binding was higher (Fig. 3 A). Binding increased over time (Fig. 3 B). Similarly, a SNAP-25-dependent increase in bound liposomes was observed when the entire cytoplasmic domain was used instead of the H3 domain, but the degree of binding was considerably lower (Fig. 3 C), probably because of the well-known inhibitory effect of the N-terminal Habc domain on SNARE complex formation (34). In all subsequent experiments, for immobilization we used the fragment of syntaxin that contained the SNARE motif plus C-terminally attached cysteine (H3-Cys), and incubation with liposomes was carried out for 15 min (standard assay conditions).
FIGURE 3.
(A) Binding of synaptobrevin liposomes to colloidal gold-coated coverslips that were preincubated as indicated (standard assay conditions, see Materials and Methods for details). (H3) syntaxin fragment encompassing the SNARE motif (syx180-262); (H3-Cys) as before but with an additional cysteine in position 263 (n = 2, bars indicate range of values). (B and C) Time dependence of the binding of synaptobrevin liposomes to surfaces coated with either SNARE domain only (B, H3) or with full-length soluble syntaxin (C, syx) in the absence or presence of SNAP-25 (SN25).
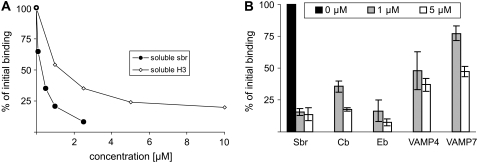
To confirm that docking depends on interaction between SNARE proteins, we performed competition experiments with soluble variants of syntaxin 1 and synaptobrevin 2 (Fig. 4 A). Micromolar amounts of either syntaxin or synaptobrevin were sufficient to decrease the binding to background levels. Moreover, soluble fragments of noncognate synaptobrevin homologs, such as cellubrevin, endobrevin, VAMP4, and VAMP7, were capable of competition at concentrations comparable to that of synaptobrevin but with different potencies, with VAMP7 being the weakest binding inhibitor and endobrevin the strongest (Fig. 4 B). VAMP7 is the most distantly related synaptobrevin homolog of the four R-SNAREs tested, suggesting a certain degree of preference for cognate SNAREs.
FIGURE 4.
Competition by soluble SNAREs of binding of synaptobrevin liposomes. (A) Binding is inhibited by increasing concentrations of a soluble syntaxin fragment containing the SNARE motif (H3, syx180-262) and synaptobrevin (sbr1-96). (B) Binding is inhibited by other R-SNAREs but with different potency. (Sbr) synaptobrevin; (Cb) cellubrevin; (Eb) endobrevin/VAMP8; (VAMP4) vesicle associated membrane protein 4; (VAMP7) vesicle associated membrane protein 7 (n = 2, bars indicate range of values).
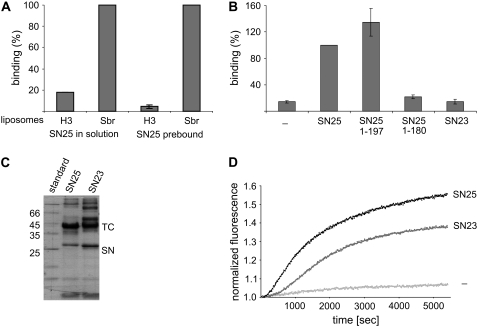
As discussed in the Introduction, syntaxin 1 and SNAP-25 are capable of forming a binary complex at a 2:1 stoichiometry. This complex is also represented by a four-helix bundle, which is structurally similar to the fully assembled SNARE complex (35,36) but significantly less stable (37), and is not capable of fusing liposomes (C. Schuette, personal communication, 2003). Thus, we sought to determine whether this complex is capable of mediating liposome docking. For this purpose, we incorporated syntaxin-H3 containing its transmembrane domain into liposomes and measured binding in parallel to synaptobrevin-containing liposomes. As shown in Fig. 5 A, no binding was observed over background, suggesting that the binary complex is not sufficient to achieve stable liposome docking. Similarly, no binding was observed when the coverslips were preincubated with excess SNAP-25, and SNAP-25 was left out during the subsequent liposome incubation. These results, together with the fact that SNAP-25 was used in excess in the first experiment, show that the lack of binding cannot be explained by a saturation of binding sites on the liposomes due to the formation of inactive 2:1 complexes between syntaxin and SNAP-25.
FIGURE 5.
Binary complexes between SNAP-25 and syntaxin, or substitution of SNAP-25 with SNAP-23, are unable to dock liposomes. (A) Docking of liposomes containing N-terminally truncated syntaxin (syx183-288) in comparison with liposomes containing synaptobrevin. SNAP-25 was either added in solution (left), or the coverslips were preincubated with SNAP-25 for 30 min, followed by washing. (B) Docking of synaptobrevin liposomes in the presence of full-length SNAP-25, the BoNT/A fragment of SNAP-25 (SN251-197), the BoNT/E-fragment of SNAP-25 (SN251-180), or SNAP-23. (For A and B, n = 2 and bars indicate range of values.) (C) Formation of ternary SNARE complexes (TC) between SNAP-25 and SNAP23 (SN) in solution, monitored by the appearance of high-molecular-weight bands in SDS-PAGE, indicating SDS-resistance typical of SNARE complexes. (D) Fusion of liposomes containing synaptobrevin and a truncated variant of syntaxin 1 (syx183-288) mediated by SNAP-25 or SNAP-23. Fusion was monitored using a standard dequenching assay (see Materials and Methods). All binding experiments were normalized to the binding of synaptobrevin-containing liposomes in the presence of SNAP-25 under standard assay conditions.)
The light chains of botulinum neurotoxins are metalloendoproteases that specifically cleave synaptic SNAREs. These toxins cannot cleave SNAREs when they are fully assembled. Two of them, botulinum neurotoxin (BoNT) A and E, act on SNAP-25. BoNT A cleaves between positions 197 and 198, resulting in the deletion of nine amino acids involved in the formation of the two C-terminal layers in the SNARE complex. BoNT E cleaves between residues 180 and 181 next to the +2 layer (38). In intact synapses, poisoning with BoNT A can be partially rescued by elevation of intracellular calcium concentration (39,40), showing that the function of the SNARE complex in fusion is impaired but not abolished. Correspondingly, the SNAP-25 fragment arising from cleavage with BoNT A forms a stable SNARE complex. In contrast, BoNT/E fragments are no longer capable of forming stable complexes, and BoNT/E poisoning cannot be rescued (41). However, it is unclear whether these fragments are still capable of interacting with their partner SNAREs in a manner that may stabilize docking but may be insufficient for fusion. We therefore expressed and purified SNAP-25 fragments corresponding to the BoNT/A and BoNT/E cleavage products and tested them for their ability to support liposome docking. Only the BoNT/A fragment (SNAP-251-197) (and not the BoNT/E fragment [SNAP-251-180]) was able to promote liposome docking (Fig. 5 B). We also tested whether light chains of neurotoxins cleaving synaptobrevin (BoNT/D and tetanus toxin) were able to detach bound liposomes. These toxins are known to cleave free synaptobrevin and probably also loose trans-complexes (42). Accordingly, preincubation of liposomes with either of the toxins prevents binding. However, no dissociation of bound liposomes was observed even after extended toxin treatment (not shown).
We then tested whether SNAP-25 can be substituted by SNAP-23, a ubiquitously expressed SNAP-25 homolog involved in constitutive nonregulated fusion. In chromaffin cells, it has been shown that SNAP-23 can substitute for SNAP-25 in calcium-triggered exocytosis, but with reduced efficiency (29). As shown in Fig. 5 B, no docking above background was observed under these conditions. Neither a prolonged incubation time of 1 h nor preincubation of syntaxin-coated coverslips with SNAP-23 resulted in binding significantly higher then background (not shown). Contrary to this, incubation of SNAP-23 with synaptobrevin 2 and syntaxin 1 in solution yielded an SDS-resistant SNARE complex that formed with kinetics similar to that formed in the presence of SNAP-25 (Fig. 5 C, and data not shown). Additionally, SNAP-23 supported fusion between synaptobrevin and H3-containing liposome, although with a slower kinetics than SNAP-25 (Fig. 5 D). Together, these experiments suggest that the reduced efficiency of SNAP-23 is primarily due to an impaired nucleation or to a reduced stability of the trans-complex that forms before the fusion reaction is carried out (see Discussion).
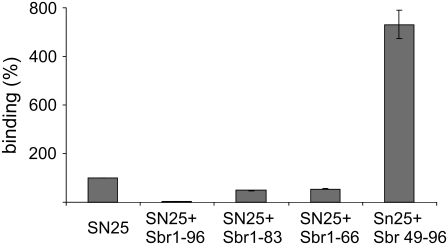
In the final series of experiments, we investigated whether docking, as measured in our assay, requires formation of a trans-complex that nucleates at the N-terminal end of the SNARE motif, as predicted by the zipper hypothesis. It was recently shown that in both solution and liposome fusion, synaptobrevin binding is greatly accelerated if a preassembled acceptor complex of syntaxin and SNAP-25 is present that exhibits a free N-terminal binding site for synaptobrevin (15). To prevent binding of a second syntaxin, a SNARE complex was formed that contained an N-terminally truncated synaptobrevin, allowing full-length synaptobrevin to bind with fast kinetics and subsequently displace the fragment (15). In view of these results, we investigated whether the trans-complex that mediates liposome docking follows the same assembly mechanism. We preformed SNARE complexes on coverslips with different C- and N-terminal synaptobrevin truncations and investigated liposome docking (Fig. 6). As expected, the preformed complex containing full-length synaptobrevin (second column) completely prevented liposome docking (first column). The C-terminal fragment sbr49-96 (fifth column), which has been shown to increase the rates of assembly and liposome fusion by orders of magnitude (15), increased liposome binding almost 10-fold. In contrast, C-terminally truncated synaptobrevin fragments (sbr1-83 and sbr1-66, second and third columns) led to reduced binding, which nevertheless was still higher than that observed when the binding site was blocked with full-length synaptobrevin, again corresponding to the findings made in assembly and fusion assays (15). Taken together, these results indicate that liposome docking is mediated by a trans-SNARE complex that nucleates at the N-terminal end, and thus further support the zipper hypothesis of SNARE function.
FIGURE 6.
Docking of synaptobrevin liposomes to immobilized preassembled complexes containing syntaxin (syx180-262Cys = H3-Cys), SNAP-25, and fragments of synaptobrevin that are either C- or N-terminally truncated as indicated (n = 2, bars indicate range of values).
DISCUSSION
In the study presented here we have shown that SNARE assembly results in the binding of vesicles if one of the SNARE partners is immobilized on a solid support. Using the neuronal SNAREs as an example, our data show that docking requires the formation of a trans-complex that is initiated at the N-terminal end, in accordance with the zipper model of SNARE function. Furthermore, our data show that docking, similarly to in vitro assembly and fusion, displays a certain degree of promiscuity, although some of the tested noncognate SNAREs are less efficient in competing or substituting for the cognate neuronal SNAREs.
As discussed in the Introduction, the extent to which SNARE proteins contribute to vesicle attachment before fusion is still a matter of controversy. In neurons and neuroendocrine cells, the number of vesicles present in active zones is increased rather than decreased upon SNARE cleavage by clostridial neurotoxins (see Neale et al. (43) and references therein), suggesting that at least in these specialized cell types, vesicle docking is mediated primarily by other, still unknown factors. On the other hand, it is possible that in neurons SNARE assembly may contribute to the residence time of a vesicle in an active zone. Evidence for dynamic exchange of docked vesicles in the absence of exocytosis was recently obtained for the neuromuscular junction (44). In nonregulated fusion events, there is no evidence for accumulation of docked vesicles, which suggests that SNARE assembly, once established, normally proceeds toward full fusion.
Our data provide a new experimental approach toward the study of trans-SNARE complexes. Although formation of trans-complexes in conjunction with SNARE-mediated docking of liposomes (21) or to supported bilayers (28) has been described previously, to our knowledge this is the first in vitro system that clearly demonstrates the requirement of all three neuronal SNAREs for SNARE-mediated attachment of vesicles, as expected for a SNARE-dependent step leading to fusion. In our assay, the vesicles are frozen in a docked state because they cannot proceed to fusion as a result of the absence of the second membrane. Whereas the structure of these trans-complexes remains to be established, some conclusions can be drawn. First, synaptobrevin is expected to be bent and probably strained, thus resembling the status expected from a docked vesicle engaged in trans-SNARE interactions. In contrast, syntaxin, while being linked to the gold surface at the C-terminal end with a sulfur-mediated dative bond of a strength resembling covalent bonds (33), may also be adsorbed via noncovalent forces. This view is supported by the fact that vesicle docking is also observed when there is no C-terminal cysteine. Thus, syntaxin and the trans-complex may be bound to the gold surface over part or even the entire length of the SNARE motifs. Consequently, we cannot exclude the possibility that despite oriented attachment via the terminal cysteine, the SNARE motif is conformationally constrained due to extended surface contact, although it is evidently still capable of engaging in SNARE complexes. Since the concentration of syntaxin on the coverslip surface is probably high, both 1:1 and 2:1 complexes form after the addition of SNAP-25, with synaptobrevin binding nucleating at the N-terminal end of these acceptor complexes (14). Second, complete zippering is not needed for vesicle docking, although we do not know at present to which of the interacting layers the assembly proceeds during vesicle docking. C-terminal truncation of SNAP-25 by nine amino acids still results in vesicle docking, although the two C-terminal layers cannot form anymore. Further shortening of SNAP-25 then results in the loss of binding, suggesting that zippering must proceed beyond the “0” layer for stable binding. These conclusions are still preliminary, and our assay may be useful in mapping the required regions more precisely using site-directed mutagenesis.
Third, docking of liposomes strictly requires all three SNARE molecules: syntaxin, synaptobrevin, and SNAP-25. No docking was observed when syntaxin instead of synaptobrevin was present in the liposome membrane. Although it is difficult to completely exclude the possibility that SNAP-25 bound to both vesicular and surface-bound syntaxin in our experiments, it appears that the binary 2:1 complex between syntaxin and SNAP-25, although rather stable in vitro (37), does not provide sufficient strength for docking. Since the stability of the binary complex is lower than the stability of the ternary complex as measured by melting temperature and SDS sensitivity, this observation suggests that docking, as monitored by our assay, is (at least initially) reversible and depends on the strength of the interaction between the bridging molecules. Furthermore, omission of SNAP-25 from the assay, competition with soluble synaptobrevin or syntaxin, or cleavage of synaptobrevin on liposomes with toxins reduces liposome binding to background levels. Since our assay has a low time resolution, and no monitoring of single vesicles was performed, we cannot exclude the possibility that some short-lived SNARE-dependent interactions occurred. This may explain the differences between our observations and those of others who measured SNARE-dependent vesicle interactions with planar membranes (21,27,28). The fact that no docking is observed in the absence of SNAP-25 agrees with the finding that without SNAP-25, syntaxin and synaptobrevin do not interact with each other in solution (32).
Our findings agree with the notion that a partially zippered trans-complex forms a metastable intermediate during fusion, which may represent an energy minimum in the reaction path toward fusion. The fact that SNAP-23 cannot substitute for SNAP-25 in docking, although complexes formed in vitro with neuronal SNAREs exhibit similar thermal stability (D. Fasshauer, personal communication, 2003), suggests that the interactions within the trans-complex are considerably weaker, exposing differences between SNAREs that are not obvious in the thermodynamic properties of fully assembled complexes. However, the findings agree well with the observation that in contrast to SNAP-25, SNAP-23 is not able to fully rescue exocytosis in chromaffin cells derived from SNAP-25 deficient mice. In fact, the lack of an exocytotic burst is explained by a high depriming rate of SNAP-23 complexes that may be due to a higher dissociation constant of the complex in trans configuration (29).
Acknowledgments
The authors are indebted to Alexander Stein and Dr. Silvio Rizzoli (Göttingen) for providing the recombinant toxin light chains and R-SNAREs, respectively; to Ursel Ries for expert technical assistance; and to Avishay Pelham and Gudrun Heim for their help with the AFM measurements.
This work was supported by grants from the National Institutes of Health (to R.J.) and the VW Foundation.
Olga Vites's present address is Molecular Neuropathobiology Laboratory, London Research Institute, Lincoln's Inn Fields Laboratories, London, UK.
Editor: Lukas K. Tamm.
References
- 1.Bonifacino, J. S., and B. S. Glick. 2004. The mechanisms of vesicle budding and fusion. Cell. 116:153–166. [DOI] [PubMed] [Google Scholar]
- 2.Jahn, R., and R. H. Scheller. 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. [DOI] [PubMed] [Google Scholar]
- 3.Rizo, J., X. Chen, and D. Arac. 2006. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 16:339–350. [DOI] [PubMed] [Google Scholar]
- 4.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 395:347–353. [DOI] [PubMed] [Google Scholar]
- 5.Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. R. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9:107–111. [DOI] [PubMed] [Google Scholar]
- 6.Zwilling, D., A. Cypionka, W. H. Pohl, D. Fasshauer, P. J. Walla, M. C. Wahl, and R. Jahn. 2007. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 26:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strop, P., S. E. Kaiser, M. Vrljic, and A. T. Brunger. 2008. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water filled cavities. J. Biol. Chem. 283:1113–1119. [DOI] [PubMed] [Google Scholar]
- 8.Fasshauer, D., R. B. Sutton, A. T. Brunger, and R. Jahn. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 95:15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock, J. B., H. T. Matern, A. A. Peden, and R. H. Scheller. 2001. A genomic perspective on membrane compartment organization. Nature. 409:839–841. [DOI] [PubMed] [Google Scholar]
- 10.Hong, W. 2005. SNAREs and traffic. Biochim. Biophys. Acta. 1744:120–144. [DOI] [PubMed] [Google Scholar]
- 11.Kloepper, T. H., C. N. Kienle, and D. Fasshauer. 2007. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell. 18:3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahn, R., and P. I. Hanson. 1998. Membrane fusion. SNAREs line up in new environment. Nature. 393:14–15. [DOI] [PubMed] [Google Scholar]
- 13.Sørensen, J. B. 2004. Formation, stabilisation and fusion of the readily releasable pool of secretory vesicles. Pflugers Arch. 448:347–362. [DOI] [PubMed] [Google Scholar]
- 14.Fasshauer, D., and M. Margittai. 2004. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem. 279:7613–7621. [DOI] [PubMed] [Google Scholar]
- 15.Pobbati, A. V., A. Stein, and D. Fasshauer. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 313:673–676. [DOI] [PubMed] [Google Scholar]
- 16.Weber, T., B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Söllner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. [DOI] [PubMed] [Google Scholar]
- 17.Parlati, F., O. Varlamov, K. Paz, J. A. McNew, D. Hurtado, T. H. Söllner, and J. E. Rothman. 2002. Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl. Acad. Sci. USA. 99:5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacia, K., C. G. Schuette, N. Kahya, R. Jahn, and P. Schwille. 2004. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J. Biol. Chem. 279:37951–37955. [DOI] [PubMed] [Google Scholar]
- 19.Tucker, W. C., T. Weber, and E. R. Chapman. 2004. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 304:435–438. [DOI] [PubMed] [Google Scholar]
- 20.Chen, X., D. Arac, T. M. Wang, C. J. Gilpin, J. Zimmerberg, and J. Rizo. 2005. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 90:2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennison, S. M., M. E. Bowen, A. T. Brunger, and B. Lentz. 2006. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys. J. 90:1661–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua, S. Y., D. A. Raciborska, W. S. Trimble, and M. P. Charlton. 1998. Different VAMP/synaptobrevin complexes for spontaneous and evoked transmitter release at the crayfish neuromuscular junction. J. Neurophysiol. 80:3233–3246. [DOI] [PubMed] [Google Scholar]
- 23.Xu, T., B. Rammner, M. Margittai, A. R. Artalejo, E. Neher, and R. Jahn. 1999. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 99:713–722. [DOI] [PubMed] [Google Scholar]
- 24.Collins, K. M., and W. T. Wickner. 2007. Trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc. Natl. Acad. Sci. USA. 104:8755–8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuette, C. G., K. Hatsuzawa, M. Margittai, A. Stein, D. Riedel, P. Küster, M. König, C. Seidel, and R. Jahn. 2004. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. USA. 101:2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fix, M., T. J. Melia, J. K. Jaiswal, J. Z. Rappoport, D. You, T. H. Söllner, J. E. Rothman, and S. M. Simon. 2004. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. USA. 101:7311–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, T., W. C. Tucker, A. Bhalla, E. R. Chapman, and J. C. Weisshaar. 2005. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys. J. 89:2473–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowen, M. E., K. Weninger, A. T. Brunger, and S. Chu. 2004. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J. 87:3569–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sørensen, J. B., G. Nagy, F. Varoqueaux, R. B. Nehring, N. Brose, M. C. Wilson, and E. Neher. 2003. Differential control of the releasable vesicle pools by SNAP-25 splice variants and SNAP-23. Cell. 114:75–86. [DOI] [PubMed] [Google Scholar]
- 30.Fasshauer, D., W. Antonin, M. Margittai, S. Pabst, and R. Jahn. 1999. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem. 274:15440–15446. [DOI] [PubMed] [Google Scholar]
- 31.Higushi, R. 1990. Recombinant PCR. In PCR Protocols: A Guide to Methods and Applications. M. A. Innis, D. H. Gelfand, J. J. Sinisky, and T. J. White, editors. Academic Press, New York. 177–183.
- 32.Fasshauer, D., W. K. Eliason, A. T. Brunger, and R. Jahn. 1998. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 37:10354–10362. [DOI] [PubMed] [Google Scholar]
- 33.Hermanson, G. T. 1996. Bioconjugate Techniques. Academic Press, New York.
- 34.Dulubova, I., S. Sugita, S. Hill, M. Hosaka, I. Fernandez, T. C. Südhof, and J. Rizo. 1999. A confromational switch in syntaxin during exocytosis: role of Munc18. EMBO J. 18:4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margittai, M., D. Fasshauer, S. Pabst, R. Jahn, and R. Langen. 2001. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem. 276:13169–13177. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, W., M. A. Poirier, M. K. Bennett, and Y. K. Shin. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol. 8:308–311. [DOI] [PubMed] [Google Scholar]
- 37.Fasshauer, D., W. Antonin, V. Subramaniam, and R. Jahn. 2002. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol. 9:144–151. [DOI] [PubMed] [Google Scholar]
- 38.Binz, T., J. Blasi, S. Yamasaki, A. Baumeister, E. Link, T. C. Sudhof, R. Jahn, and H. Niemann. 1994. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J. Biol. Chem. 269:1617–1620. [PubMed] [Google Scholar]
- 39.Marxen, P., F. Bartels, G. Ahnert-Hilger, and H. Bigalke. 1991. Distinct targets for tetanus and botulinum A neurotoxins within the signal transducing pathway in chromaffin cells. Naunyn Schmiedebergs Arch. Pharmacol. 344:387–395. [DOI] [PubMed] [Google Scholar]
- 40.Xu, T., T. Binz, H. Niemann, and E. Neher. 1998. Multiple kinetic components of exocytosis distinguished by neurotoxin sensitivity. Nat. Neurosci. 1:192–200. [DOI] [PubMed] [Google Scholar]
- 41.Banerjee, A., J. A. Kowalchyk, B. R. DasGupta, and T. F. Martin. 1996. SNAP-25 is required for a late postdocking step in Ca2+-dependent exocytosis. J. Biol. Chem. 271:20227–20230. [DOI] [PubMed] [Google Scholar]
- 42.Schiavo, G., M. Matteoli, and C. Montecucco. 2000. Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80:717–766. [DOI] [PubMed] [Google Scholar]
- 43.Neale, E. A., L. M. Bowers, M. Jia, K. E. Bateman, and L. C. Williamson. 1999. Botulinum neurotoxin A blocks synaptic vesicle exocytosis but not endocytosis at the nerve terminal. J. Cell Biol. 147:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizzoli, S. O., and W. J. Betz. 2004. The structural organization of the readily releasable pool of synaptic vesicles. Science. 303:2037–2039. [DOI] [PubMed] [Google Scholar]