Abstract
The occurrence of late-onset Alzheimer's disease has been related to the lipid homeostasis. We tested whether the membrane lipid environment affects the dynamics and cleavability of a model peptide corresponding to the amino acid sequence 684–726 of the amyloid precursor protein APP reconstituted in liposomes. Solid-state NMR with 2H-Ala713, which is located within the putative transmembrane domain, suggested that the peptide observes less rotational motion in egg phosphatidylcholine (PhC) membranes than in dimyristoyl-phosphatidylcholine (DMPC) bilayers above the main phase transition temperature Tc. The residue 15N-Ala692, which is in the vicinity of the α-cleavage site, i.e., Lys687, showed less motion after reconstitution in distearoyl-phosphatidylcholine liposomes <Tc than in PhC, DMPC, or sphingomyelin vesicles. In all tested liposomal systems the α-cleavage site was accessible for hydrolysis by trypsin. However, the catalytic rate constant was higher in the PhC and DMPC than in the sphingomyelin and distearoyl-phosphatidylcholine systems. In conclusion, the dynamics of APP(684–726) on the transmembrane level as well as the motion of the α-cleavage site and its hydrolysis by a model enzyme are dependent on the bilayer characteristics. This could be relevant for the processing of APP in vivo.
INTRODUCTION
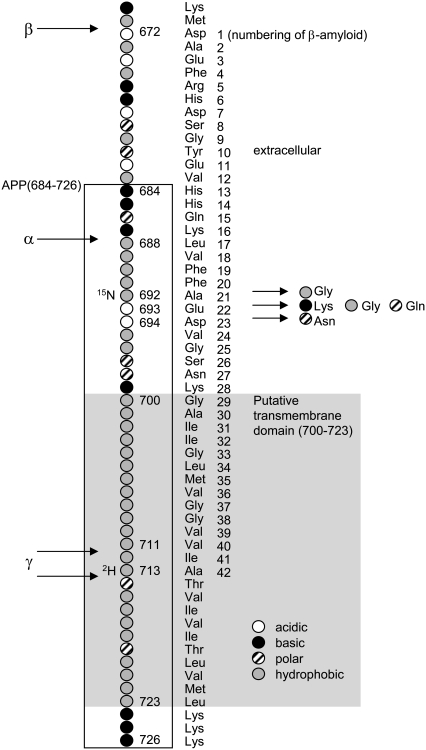
Alzheimer's disease (AD) affects one-fourth to one-third of the population aged 85 years or older in the Western world (1). A hallmark of the disease is the deposition of β-amyloid in the central nervous system. β-Amyloid peptides are generated by the membrane proteases β- and γ-secretase from the amyloid precursor protein (APP), a single span transmembrane protein of ∼770 amino acids. The positions of the cleavage sites with respect to the putative transmembrane domain of APP are shown in Fig. 1. The production of β-amyloid is prevented by the membrane protease α-secretase as it cleaves APP between the β- and γ-cleavage sites. An imbalance between the activities of the secretases and/or between β-amyloid generation and its clearance from the brain, leads to the development of the disease. Early-onset AD, which usually presents in younger individuals, is clearly linked to variations in the genes involved in the processing of APP or in the APP gene itself (1).
FIGURE 1.
APP sequence containing the putative transmembrane domain and the α-, β-, and γ-cleavage sites. The amino acids are labeled according to their physicochemical characteristics, i.e., acidic, basic, polar, and hydrophobic. The gray background indicates the putative transmembrane domain. For NMR, APP(684–726) (box) was labeled with 2H and 15N as indicated. Variants leading to Alzheimer's disease are shown for amino acids 692, 693, and 694. Amino acid counting according to APP770, UniProtKB/Swiss-Prot entry P05067. The amino acid numbering of β-amyloid (1–42) is indicated on the right of the amino acid labels.
The strongest risk factor for sporadic late-onset AD is age. The intense search for genetic factors that increase the risk for late-onset AD continues to bring polymorphisms that are associated with the development of the disease to light (2,3). The one which is best known and probably most frequently involved in sporadic AD (4–6) is the APOE*4 allele of the APOE gene, coding for the apolipoprotein ApoE, which is involved in cholesterol homeostasis. The allele has an increased occurrence among individuals with AD. Beside the genetic risk factors there are dietary, environmental and life style factors that were related to the disease (1). Several reports put forward a link between dietary lipids and the incidence of AD (7–10).
There is strong evidence from clinical, retrospective, animal, and cell culture studies that the processing of APP is related to the cholesterol homeostasis in the brain (8,11–18). According to one of the current models (11) high cholesterol levels in the cell membrane promote the generation of β-amyloid. However, some studies also show an increase in β-amyloid production as a consequence of a moderate depletion of cholesterol in neurons (19,20).
Beside cholesterol, other lipids were linked to the development of AD. Oksman et al. (21) found an advantageous effect, i.e., low β-amyloid levels in APP/PS1 transgenic mice, of docosahexaenoic acid (DHA)-enriched diets but observed increased levels of β-amyloid after a diet rich in saturated fatty acids and cholesterol. DHA had a similar effect on APPsw transgenic mice (22). In general, the degree of saturation of fatty acids and the position of the first double bond in essential fatty acids seem to be critical factors determining the effect of dietary fats on the risk of AD. Unsaturated fats and n-3 double bonds seem to confer protection and an overabundance of saturated fats or n-6 double bonds to increase the risk of developing AD (10). Another lipid that deserves attention is sphingomyelin (Sph): a significant increase in its level has been reported in brains of AD patients during the course of the disease (23).
Lipids and their oxidation products could affect the APP processing via different routes, including signaling functions, regulation of transcription, and translation, or directly via specific or unspecific interactions with the secretases, with APP, or with β-amyloid. For instance, the activity of the β-secretase BACE in hydrolyzing a soluble APP analog in a liposomal assay was directly influenced by the lipid environment of the enzyme (24). Lahdo et al. (25,26) showed that the lipid composition of liposomes and lipid monolayers determines their interactions with APP.
In this work, we focus on the α-cleavage site of APP (Leu687/Lys688, Fig. 1). The nonamyloidogenic processing is carried out by members of the ADAM family, i.e., disintegrin and metalloproteases (27). The protein conformation of APP in the vicinity of the α-cleavage site significantly influences the processing of APP. The introduction of an α-helix-disturbing amino acid in position 692 (Ala → Gly, Ala → Pro) and 690 (Phe → Pro; Fig. 1) increased the ratio of β-amyloid to p3, the latter is the product of α- and γ-cleavage of APP (28). The variant APP Ala692Gly leads to familial AD and cerebral hemorrhages (29).
We address the question whether the lipid composition of the membrane environment of APP could influence the dynamics, accessibility and cleavability of APP at the α-cleavage site. We incorporated the peptide APP(684–726), including the putative transmembrane domain and the α-cleavage site, into liposomes of different phosphocholine lipids. Fig. 1 shows part of APP including the tested peptide. The liposomal system allows us to study the direct influence of the membrane characteristics on APP, excluding indirect effects that could occur in cellular systems.
Our studies were carried out with membranes consisting of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), egg phosphatidylcholine (PhC), and porcine brain Sph, respectively. As shown in Table 1, these lipids differ mainly in their acyl chain length and unsaturation, main phase transition temperature Tc, and their backbone if comparing Sph to the phosphatidylcholines.
TABLE 1.
Acyl chain composition and main transition temperature Tc of the investigated lipids
| Lipid | Acyl chains* | Tc† | Hydrophobic thickness (nm) |
|---|---|---|---|
| DMPC | 14:0 /14:0 | 23°C | 2.30 (Lα),‡ 2.62 (Lα),¶ 2.9 (Lα)** |
| POPC | 16:0 /18:1(C9-C10) | −4°C | 2.58 (Lα)§ |
| PhC | 16:0 (34%), 18:1 (32%), 18:2 (18%) | <0°C | 2.71 (Lα)¶, 2.9 (Lα)** |
| DSPC | 18:0 /18:0 | 54.5°C | 2.95 (Lα),†‖ 3.06 (Lα),** 3.60 (Lβ)‖ |
| Sph | 18:0 (49%), 24:1 (20%) | 35°C | In the range of DSPC as judged from the acyl chain lengths†† |
DMPC, DSPC, POPC, acyl chains at positions 1/2; PhC, Sph, major acyl chains with % occurrence in brackets.
Tc, main transition temperature from the gel (Lβ) to the liquid-crystalline (Lα) phase according to Marsh (72) and Meyer et al. (73).
Lewis and Engelman (74).
Nezil and Bloom (77).
Nagle and Tristram-Nagle (75).
Rinia et al. (78).
Cornell and Separovic (76).
No data available.
The dynamics of the liposome-incorporated APP(684–726) peptide was probed by solid-state NMR with 2H-Ala at position 713 and 15N-Ala at position 692, using magic angle spinning (MAS). These positions are in the center of the putative transmembrane domain and close to the α-cleavage site, respectively (Fig. 1). In parallel, the accessibility and cleavability of the α-cleavage site in the different membrane environments were examined in a kinetic assay with trypsin, which hydrolyses APP at the same position as α-secretase.
We found that the membrane composition has a significant influence on the motion of Ala692, which is in the vicinity of the α-cleavage site, and that the unsaturation of the acyl chains affects the dynamics of the transmembrane domain. The model peptide was hydrolyzed by trypsin in all test systems and Michaelis-Menten analysis showed that differences occurred from the catalytic step, possibly from the release of the enzyme from the membrane surface.
METHODS
Chemicals
Fmoc-amino acids, Fmoc-Lys(Boc)-Wang resin (100–200 mesh, No. 04-12-2057), and 5-carboxyfluorescein (5-FAM) (No. 01-63-0112) were purchased from Merck Novabiochem (Darmstadt, Germany). Fmoc-PAL-PEG-resin (0.19 mmol/g, No. GEN913383) was purchased from Applied Biosystems (Foster City, CA). Deuterium depleted water, Fmoc-alanine-3,3,3-D3 and Fmoc-alanine-15N derivatives were purchased from Cambridge Isotope Laboratories (Andover, MA). Trifluoroacetic acid (TFA), 2,2,2-trifluoroethanol (TFE), and hexafluoro-2-propanol (HFIP) were from Fluka (Buchs, Switzerland). The phospholipids DMPC, DSPC, POPC, and Sph were from Avanti Polar Lipids (Alabaster, AL). PhC grade 1 was supplied by Lipid Products (Nutfield, UK). All other chemicals were of analytical grade.
Synthesis of APP(684–726) and APP(684–687)
APP(684–726), H-His-His-Gln-Lys-Leu-Val-Phe-Phe-Ala-Glu-Asp-Val-Gly-Ser-Asn-Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met-Val-Gly-Gly-Val-Val-Ile-Ala-Thr-Val-Ile-Val-Ile-Thr-Leu-Val-Met-Leu-Lys-Lys-Lys-NH2, and APP(684–687), H-His-His-Gln-Lys-OH, were synthesized on an Applied Biosystems Peptide Synthesizer Model 431. The peptides were synthesized on a 0.1 mmol scale using standard Fastmoc protocols (30). PAL-PEG and Wang resin were used for APP(684–726) and APP(684–687) yielding peptides with C-terminal amides and acids respectively. Cleavage from the resin, and removal of protecting groups, was accomplished by treatment with TFA/water/ethane dithiol (EDT)/triisopropylsilane (TIS) (volume ratios 94.5:2.5:2.5:1) for 4 h at room temperature. The reaction mixture was filtered and most of the TFA was evaporated in an N2 stream. The peptide was then precipitated by adding cold diethyl ether, collected by centrifugation and lyophilized from an aqueous TFE 10% (v/v) solution. The peptide purity was ∼70% as confirmed by reversed phase HPLC (C4, Vydac 214TP54) and the peptide mass was confirmed by MALDI-TOF and ESI mass spectroscopy. The peptide was not further purified as it accumulated on the HPLC column, resulting in low yields under all tested conditions. For NMR studies 15N-Ala and CD3-Ala were introduced at positions 692 and 713, respectively.
Labeling of APP(684–726) and APP(684–687) with 5-carboxyfluorescein
The coupling of 5-FAM with APP(684–726) and APP(684–687) was carried out according to Fischer et al. (31). Resin bound peptide was equilibrated with a 20% (v/v) piperidine/dimethylformamide (DMF) solution before the addition of a 10 mL DMF solution containing 5-FAM (2.5 eq), 2-(1H-9-azabenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) (2.4 eq), and diisopropylethylamine (DIPEA) (5 eq). The reaction was conducted in the dark for 16 h. The peptide-resins were washed several times with 20% piperidine/DMF to remove the excess 5-FAM. The product was cleaved as described for the untagged peptide and the purity and molecular weight confirmed by reverse phase HPLC and MALDI-TOF and ESI mass spectroscopy.
Preparation of APP(684–726)-liposomes
The peptides APP(684–726), 2H/15N-APP(684–726), and the fluorescent analog FAM-APP(684–726), respectively, were reconstituted into vesicles of different lipid composition (Table 1). For the reconstitution of the peptides in liposomes a protocol for the incorporation of hydrophobic peptides into micelles (32) was adapted. Between 3 and 14 mg peptide was dissolved at a concentration of 1 mg/mL in TFE and incubated for 2 h at room temperature in the dark. The solution was then reduced to the volume of 1 mL in an N2 stream and added to the respective lipid solution in TFE (DMPC, DSPC, and PhC) or HFIP (Sph) to reach a lipid/peptide molar ratio of 100:1. An excess of water (∼50 mL) was added to reconstitute the peptides into stable lipid bilayers. Subsequently all solvent was removed by lyophilization, and the dry powder was rehydrated with 4 mL 10 mM Tris, 1 mM EDTA, 130 mM NaCl, pH 7.4 buffer (Tris/EDTA/NaCl) to form multilamellar vesicles (MLV), which were submitted to 10 cycles of freeze and thaw (>Tc). For NMR measurements, all solutions were prepared with 2H-depleted water.
Density gradient centrifugation was used to confirm peptide incorporation and sample homogeneity. The MLV were therefore transferred with an equal volume of 60% (w/v) sucrose in Tris/EDTA/NaCl into a 12.5 mL centrifuge tube and overlaid with 2 mL each of 20% (w/v), 10% (w/v), 5% (w/v) sucrose and 1.5 mL buffer. After centrifugation at 200,000 × g for 19 h at 4°C in an SW41Ti rotor (Beckman Instruments, Fullerton, CA), MLVs containing the peptide were visible as a clear band in the upper half of the tube. The density of the liposomal fraction was calculated from the measured refractive index (refractometer from Bellingham and Stanley, Turnbridge Wells, UK) according to Birnie and Rickwood (33). The peptide-free control MLVs were located on the top of the sucrose gradient, whereas MLVs containing peptide were concentrated at higher densities, dependent on the type of lipid. DMPC, DSPC, and Sph peptide MLV migrated to a density between 1.03 and 1.05 g/cm3. Peptide-containing PhC vesicles were visible in the upper part between 1.01 and 1.02 g/cm3.
The bands were collected and, after dilution in 10 mL buffer, pelleted three times at 100,000 × g for 1 h at 4°C. Pelleted MLVs containing the labeled peptide were used for NMR studies. For the circular dichroism (CD) measurements and the tryptic cleavage assay, the pellets were resuspended in buffer followed by 10 cycles of freeze and thaw (>Tc). Large unilamellar vesicles (LUV) were prepared by subsequent extrusions through 0.8, 0.4, 0.2, and 0.1 μm pore size polycarbonate membranes (Nucleopore, Whatman, Brentford, UK) using the 10 mL extruder from Lipex Biomembranes (Vancouver, Canada). The final liposomes were characterized by dynamic light scattering with a Zetasizer 3000 HSA (Malvern Instruments, Malvern, UK). The hydrodynamic average mean diameters of all preparations were between 130 and 170 nm (intensity distribution) with a polydispersity index between 0.2 and 0.5, corresponding to a relative standard deviation between 40 and 70% assuming a monomodal size distribution. Quantification of total peptide was carried out with the DC protein assay kit from Bio-Rad (Hercules, CA), which is based on the method of Lowry (34). Lipid concentrations were determined by TLC (35). The final peptide/lipid molar ratios of peptide LUVs were between 1:100 and 1:200.
Solid-state NMR measurements
All NMR spectra were obtained on a Varian Infinity+ 500 spectrometer. 31P-NMR spectra were acquired with a 4 mm double resonance Varian T3 MAS probe using a Hahn-echo pulse sequence (36). 31P pulse lengths were typically 8.0 μs with an inter pulse delay of 100 μs. During acquisition continuous wave decoupling was used at moderate amplitude of 45 kHz to prevent sample heating. A recycle delay of 2.5 sec was used and spectra are the result of 1024 acquisitions. Before Fourier transform the data was left shifted to the top of the echo and processed with 150 Hz linebroadening.
Deuterium and 15N measurements were carried out using a 6 mm double resonance Varian T3 probe. Deuterium spectra were acquired with a 90° pulse of 5 μs and a recycle delay of 1 s. The spectra are a result of 128,000 acquisitions. Before Fourier transform the data were left shifted to the top of the first rotor echo to remove first order phase distortions and a linebroadening of 150 Hz was applied. Cross-polarization 15N spectra were acquired where the protons were excited with a 5 μs pulse and the polarization was subsequently transferred to 15N using an adiabatic cross-polarization step with a spin-lock field centered at 35 kHz (contact time 1 ms). During acquisition 50 kHz TPPM (two pulse phase modulation) decoupling was applied with a 10° phase shift. The recycle delay was 2 s and the spectra are the sum of 64,000 acquisitions. A linebroadening of 150 Hz was applied before Fourier transform.
Analysis of the NMR spectra
The spectra have been processed with Spinsight. The spinning sideband pattern from the MAS experiments was analyzed according to the formalism of Herzfeld and Berger (37). The intensities were calculated in C++ using the GAMMA spin simulation package (38). Powder averaging was carried out according to the method of Cheng et al. (39) using 1154 powder orientations per 2H spectrum and 300 powder orientations in the case of 15N spectra. The fitting of the sideband intensities was carried out with the program MINUIT (MINUIT, CERN Program Library Entry D506). The errors given are those generated by MINUIT and represent 1 SD.
Circular dichroism measurements and analysis
All CD measurements were acquired with a JASCO J720 spectropolarimeter attached to a NESLAB 111 bath circulator. CD spectra were obtained between 260 and 195 nm, with a 0.2 mm path-length cell, a 20 nm/min scanning speed, a response time of 4 s, and a bandwidth of 1 nm. The spectra reported are the averages of four scans each, corrected by subtracting CD spectra of the solvent or the liposomes without peptide. To estimate the secondary structure contents of the peptide, the relevant CD spectra were analyzed with the CDPro software (40). An extended reference set of 56 proteins was used, which included spectra of 13 membrane proteins along with 43 soluble proteins. The analysis was carried out with three deconvolution algorithms, CONTIN/LL, CDSSTR, and SELCON3, in the wavelength range between 260 and 195 nm.
Tryptic cleavage assay
Fluorescent FAM-APP(684–726) was reconstituted in LUV of different lipid composition as described above and subjected to proteolysis by trypsin. Sequencing grade modified trypsin (Promega, Madison WI; Part No. 9PIV511, mw 23,400 (41)) was resuspended in Tris/EDTA/NaCl to a concentration of 0.1 μg/mL and incubated for 15 min at 37°C. Of this enzyme preparation, 11 μL were added to 1090 μL FAM-APP(684–726)/liposome suspensions with peptide concentrations between 5 and 80 μM. The final enzyme to peptide molar ratio was ≤1:10,000 to ensure Michaelis-Menten conditions. Kinetic experiments were carried out over 5 h at 37°C. The reaction was stopped at intervals of 1 h by the addition of 20 μL of acidic buffer (10 mM Tris, 1 mM EDTA, 130 mM NaCl, 1% (v/v) TFA) to 200 μL sample. The peptide digests were analyzed by analytical HPLC with a Vydac 214TP54 column (C4, 250 × 4.6 mm i.d., 5 μm particle size, 300 Å pore size), which was equilibrated with 0.1% TFA-acetonitrile (9:1). The solvent flow rate was 1 mL min−1, and the temperature was 40°C. The samples (190 μL) were injected on the column and eluted under the initial conditions for 15 min before acetonitrile was increased in a linear gradient to 1:9 (v/v) within 15 min. The separation was monitored with a fluorescence detector at λex 445 nm and λem 521 nm. The peak corresponding to the peptide fragment FAM-APP(684–687) was identified by co-injection of the synthesized standard and by ESI mass spectrometry. The amounts of the digested product FAM-APP(684–687) were calculated from the integrated fluorescent intensities of a standard curve.
Michaelis-Menten analysis
The kinetics of trypsin cleavage were analyzed according to Michaelis-Menten kinetics (Eqs. 1 and 2), which relate the reaction velocity v with the Michaelis-Menten constant KM and the maximal velocity Vmax (Eq. 2 (42)).
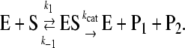 |
(1) |
In Eq. 1, E denotes the enzyme, S the substrate, ES the enzyme-substrate complex and P1 and P2 the products. The rate constants k1 and k−1 determine the affinity between the substrate and the active site of the enzyme and kcat is the rate constant of the catalytic step of the reaction, i.e., the hydrolysis itself and the release of the products and the enzyme.
 |
(2) |
[FAM-APP(684–726)] in Eq. 2 denotes the molar concentration of FAM-labeled substrate. Experimental data were analyzed according to Eq. 2 by nonlinear regression with SigmaPlot v.9 from SPSS.
The rate constant kcat (Eq. 1) was calculated from Vmax according to Eq. 3:
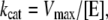 |
(3) |
where [E] is the molar enzyme concentration, i.e., 5 × 10−11 M.
RESULTS
Characterization of APP(684–726) MLVs by 31P-NMR spectroscopy
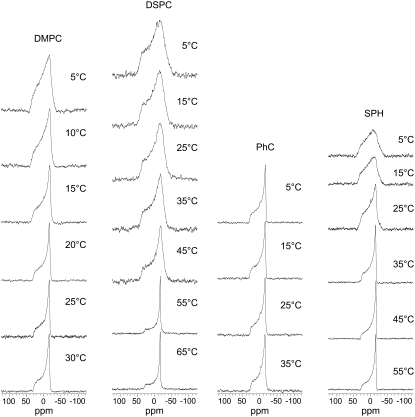
The APP(684–726) MLV preparations were characterized by 31P-NMR to ascertain that the addition of the APP(684–726) had not disrupted the lipid bilayer morphology or lead to significant changes in the phase behavior. At a lipid to protein molar ratio of 100:1 and above the lipid Tc, all the 31P spectra show an axially symmetric lineshape due to a chemical shielding anisotropy (CSA) characterized by the anisotropy parameter δ of approximately −30 ppm and the asymmetry η = 0 (Fig. 2). This is characteristic of the lipids in their liquid crystalline (Lα) phase (43). In the case of DMPC, DSPC, and Sph, lowering the temperature below Tc resulted in a broadening of the features in the spectra, characteristic for the mobility of the lipids in the gel (Lβ) phase (44). The spectra of the PhC and POPC (not shown) vesicles were similar and showed a well-defined, temperature-independent 31P line shape in the examined temperature range reflecting the Lα phase of the membrane (Tc < 0°C). These observations indicate that at the lipid to protein molar ratio of 100:1, the APP(684–726) does not disrupt the bilayer morphology or change significantly the phase transition temperatures.
FIGURE 2.
Temperature-dependence of the static 31P NMR spectra of different lipid MLV with incorporated APP(684–726). The transition temperatures Tc are listed in Table 1. The frequency is given in terms of the chemical shift relative to 85% H3PO4 at 25°C (0 ppm). Spectra of POPC systems were similar to those of PhC systems and are not shown.
Characterization of reconstituted APP(684–726) by circular dichroism
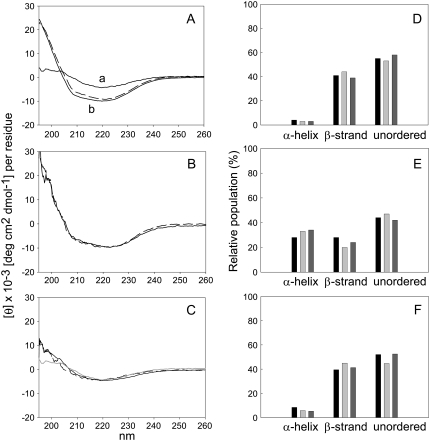
The conformation of solubilized APP(684–726) and of APP(684–726) reconstituted in LUVs of different lipid compositions was investigated by CD. CD spectra are shown in Fig. 3 in the range between 195 and 260 nm that reflects the secondary structural features of peptides. The multicomponent analysis of the spectrum of APP(684–726) dissolved in buffer containing 1% (v/v) acetonitrile indicated that the peptide adopted predominantly β-strands and unordered structures (Fig. 3, A and D).
FIGURE 3.
CD analysis of APP(684–726). CD spectra of APP(684–726) in buffer containing 1% (v/v) acetonitrile at 37°C (A, a) and reconstituted in DMPC LUVs (A, b), and egg PhC LUVs (B), measured at 35°C (solid lines) and 5°C (dashed lines). (C) CD spectra in DSPC LUVs at 60°C (solid line) and 5°C (dashed line). The CD spectrum of the peptide in buffer containing 1% acetonitrile at 37°C (as in A, a) is included for comparison (gray line). Shown is the molar ellipticity per residue corrected for the respective solvent or for the peptide-free liposomes. (D–F) Multicomponent analysis of the CD spectra of APP(684–726) in 1% acetonitrile (D) and after reconstitution in DMPC (E) and DSPC (F) bilayers at 37°C and 60°C, respectively. The CD spectra in A and C were deconvoluted with the algorithms CONTINLL (black bars), SELCON 3 (light gray bars), and CDSSTR (dark gray bars). Bars labeled “unordered” include the estimated percentage of unordered structures and turns. Shown are data from one representative out of three independent experiments each.
Similar spectra and estimated structure characteristics were found for APP(684–726) reconstituted in DSPC liposomes, independent of the phase of the lipids (Fig. 3, C and F). The CD spectra of APP(684–726) reconstituted in DMPC and PhC LUVs, respectively (Fig. 3, A, B, and E), showed typical features of partial α-helical structure, i.e., minima at 222 nm and at 208 nm (45). The analysis of these spectra showed ∼30% α-helix, ∼25% β-, and ∼45% unordered structures and turns. No significant differences were found between the spectra at 5°C and 35°C. Finally, the CD spectra of APP(684–726) incorporated into Sph vesicles were not interpreted as Sph interfered with the CD signal at wavelengths <220 nm. This effect was described before (46). The spectra resulting from the subtraction of the base line (Sph liposomes without peptides) were not reproducible in the low nm range.
Influence of the lipid environment on the dynamics of the transmembrane domain of 2H3-Ala713-APP(684–726)
To investigate the dynamics of the peptide in the proposed transmembrane domain, the 2H NMR spectra of 2H3-Ala713-APP(684–726) were acquired under low-speed MAS (νr < ΔνQ, where νr is the spinning frequency and ΔνQ is the quadrupolar splitting) that allows the characterization of the quadrupolar interaction through an analysis of the spinning sidebands intensities. The scaling of the quadrupolar interaction can provide information about the dynamics of the peptide on the microsecond timescale.
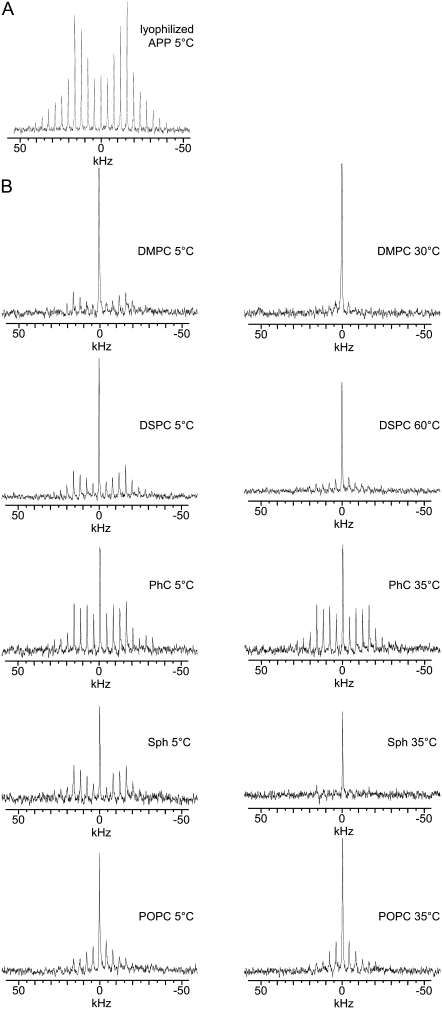
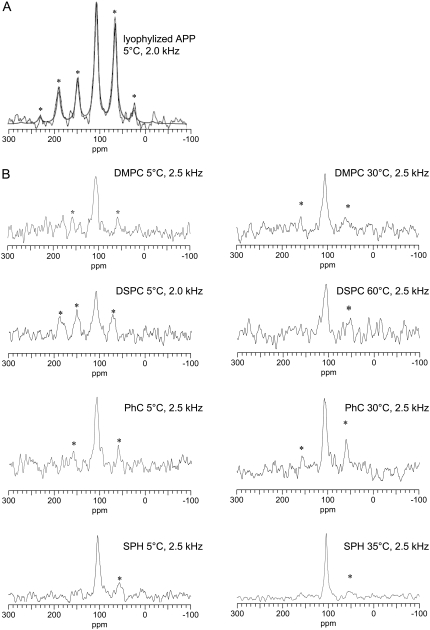
The 2H MAS spectrum for the 2H3-Ala713 of lyophilized APP(684–726) is shown in Fig. 4 A. The spectrum shows a large family of sidebands, with the maximum intensity at ±20 kHz (fifth sideband). Analysis of the spectrum shows ΔνQ = 39.9 ± 0.2 kHz with an asymmetry parameter η = 0.00 ± 0.06, characteristic for a rotating methyl group.
FIGURE 4.
2H NMR MAS spectra of 2H-Ala713-labeled APP(684–726) with νr = 4 kHz. Spectra of lyophilized peptide in the absence of lipids (A) and of the peptide reconstituted in MLV of the indicated lipids at the indicated temperatures (B). The central lines in the spectra of reconstituted peptide are assigned to residual DHO. Spectra were referenced to DHO.
The 2H MAS spectra of APP(684–726) reconstituted into phospholipid vesicles acquired at 5°C appeared as a family of spinning sidebands and a narrow resonance attributed to DHO (Fig. 4). The spectra of APP(684–726) incorporated in DMPC, DSPC and Sph bilayers in the Lβ phase were similar to the spectrum of the lyophilized peptide, i.e., ΔνQ ∼ 40 kHz and η ≈ 0 (data not shown). Deuterium spectra of APP(684–726) in PhC and POPC bilayers at 5°C also showed significant sideband intensities. However, the quadrupolar coupling constant is somewhat smaller as compared to DMPC, DSPC, and Sph indicating the presence of additional motion above those seen in the other lipids analyzed.
Raising the temperature such that the DMPC, DSPC, and Sph enter their Lα phase, leads to significant changes in the spectra, with the sideband family collapsing in a way that the bulk of the intensity is in the central resonance and the intensity of the sidebands decreases rapidly with increasing sideband order. This is indicative of the presence of significant motions on the microsecond timescale that average the quadrupolar interaction. Increasing the temperature of the PhC and POPC systems from 5 to 35°C had limited effects on the spectra. These lipids are in their Lα phase at both temperatures.
The relatively high DHO signal was assigned to the natural abundance of deuterium in the buffer components and in the residual water on the glassware used for the preparation of the liposomes.
Motional analysis of 15N-Ala692 of APP(684–726) in different lipid environments
To investigate the effect of lipid bilayer composition on the dynamics in APP(684–726) close to the α-cleavage site, the 15N CSA of the amide group of Ala692 was investigated in the low-speed MAS cross-polarization spectra. Fitting the spectrum of the lyophilized peptide gave an anisotropy of CSA, δ = −97 ± 3 ppm and a η = 0.0 (Fig. 5 A).
FIGURE 5.
15N CP-MAS spectra of lyophilized and liposome-incorporated 15N-Ala692-labeled APP(684–726). (A) Superimposition of an experimental spectrum of lyophilized peptide (scattering lines) with the fitted 15N MAS spectrum (smooth lines, details see text). (B) Spectra of the liposome-reconstituted peptide were recorded at the indicated spinning speeds and temperatures. Asterisks indicate the spinning sidebands. The frequency is given in terms of the chemical shift relative to the 15N signal of glycine at 107 ppm.
The 15N MAS spectra of 15N-Ala692-APP(684–726) reconstituted in lipid vesicles at different temperatures are shown in Fig. 5 B. On reconstitution into lipid bilayers, the 15N-Ala692 located in the extramembrane part of the peptide observed more mobility as compared to the 15N-Ala692 of the lyophilized peptide (Fig. 5 A). Due to the low signal/noise ratio of the spectra no precise analysis of the corresponding CSA parameters was possible. However, the simulation of the 15N tensor for the peptide reconstituted into DMPC at 30°C, for instance, indicated a reduced δ, with −99 ppm < δ < −79 ppm (assuming η = 0).
The 15N spectra of APP in PhC, DMPC, and Sph at 5°C did not show any significant differences and were similar to the respective spectra at higher temperatures. The 15N spectra of APP(684–726) in DSPC showed a phase-dependent scaling of the CSA, which suggests a higher mobility of the peptide ectodomain in the Lα phase (at 60°C) than in the Lβ phase (5°C), taking into account the similarity in its conformation in the two phases as seen in the CD spectra.
Influence of the lipid environment on the cleavage by trypsin of reconstituted FAM-APP(684–726)
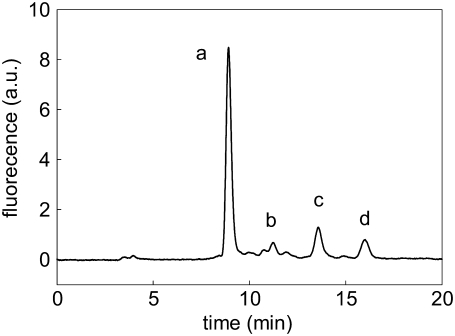
Trypsin cleaves APP(684–726) at the α-cleavage site between Lys687 and Leu688 and is used here as a probe for the accessibility and cleavability of the α-cleavage site of the peptide reconstituted in different lipid bilayers. The cleavage of solubilized FAM-APP(684–726) and of LUV-reconstituted FAM-labeled peptide was investigated under Michaelis-Menten conditions. Fig. 6 shows the isocratic HPLC elution profile of a representative liposomal peptide preparation after incubation for 5 h with trypsin. FAM-APP(684–687) represents the main soluble product of the enzymatic digestion as confirmed by mass spectrometry. The side peaks were not further analyzed, however, none of them corresponded to 5-carboxyfluorescein. The peaks shown in Fig. 6 were negligible in control experiments without enzyme and immediately after enzyme addition. After the addition of the enzyme, the peak areas increased linearly up to 5 h of incubation and were dependent on the initial FAM-APP(684–726) concentration.
FIGURE 6.
HPLC separation of the products from the tryptic digestion of FAM-APP(684–726). Sph liposomes with incorporated peptide were analyzed after 5 h of incubation with trypsin. Peaks a, b, c and d were eluted within 20 min and represent the fluorescent products of the digestion. a.u., arbitrary units.
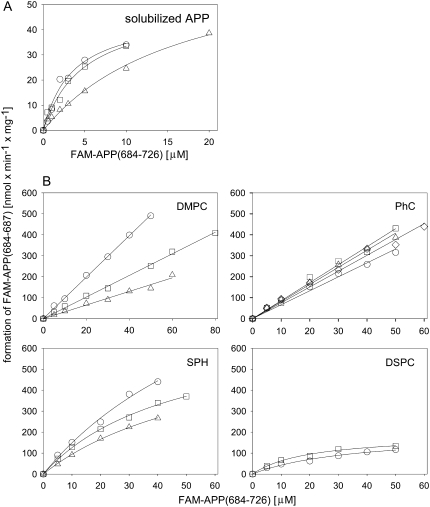
The cleavage velocities v were calculated from the FAM-APP(684–687) concentrations at the various time points and were plotted against the initial FAM-APP(684–726) concentrations. Fig. 7 A shows the Michaelis-Menten analysis of the cleavage of solubilized peptide. The Michaelis-Menten constant KM was ∼4 μM when the peptide was solubilized in buffer containing 1% acetonitrile and ∼10 μM at 5% acetonitrile, respectively. The corresponding Vmax values were ∼50 and ∼70 nmol/min per mg enzyme, corresponding to kcat values of ∼0.02 and ∼0.03 s−1, respectively (Table 2).
FIGURE 7.
Michaelis-Menten kinetic analysis of the hydrolysis of solubilized and liposome-incorporated FAM-APP(684–726) to FAM-APP(684–687) by trypsin. The peptide was solubilized in Tris buffer, pH 7.4 (A), containing 1% (○ and □) or 5% (▵) acetonitrile or was reconstituted in liposomes as indicated (B) and subsequently incubated at different peptide concentrations with trypsin at 37°C. The formation of the cleavage product FAM-APP(684–687) was followed by HPLC and plotted as a function of the initial FAM-APP(684–726) concentration. Shown are two to four independent experiments each (○, □, ▵, and ⋄).
TABLE 2.
Michaelis-Menten parameters of the cleavage by trypsin of FAM-APP(684–726) at the α-cleavage site
| System | KM [μM] | KM′ [μM]* | Vmax [nmol/min] per mg enzyme | kcat [s−1] | Vmax/KM [mL/min] per mg enzyme | Vmax/KM′ [mL/min] per mg enzyme |
|---|---|---|---|---|---|---|
| Tris buffer containing 1% acetonitrile | 3.51/4.85 | — | 46.7/49.7 | 0.018/0.019 | 13.3/10.2 | 13.3/10.2 (Vmax/KM) |
| DMPC liposomes | >60 | >30 | >1000 | >0.39 | 6.1 ± 3.4 | 12.2 ± 6.8 |
| PhC liposomes | >60 | >30 | >1000 | >0.39 | 7.7 ± 0.8 | 15.5 ± 1.7 |
| DSPC liposomes | 36.5/19.9 | 18.2/10.0 | 198/189 | 0.077/0.074 | 5.4/9.5 | 10.8/18.9 |
| Sph liposomes | 63.9 ± 15.9 | 31.9 ± 8.0 | 918 ± 355 | 0.36 ± 0.14 | 14.3 ± 3.1 | 28.5 ± 6.1 |
Mean values from three independent experiments with SD are shown, except for soluble APP and DSPC with values from two independent experiments.
KM′, KM corrected for the estimated fraction of peptide oriented with the cleavage site outside the liposomes (KM′= KM/2).
In all liposomal systems the peptide was hydrolyzed by the protease. Fig. 7 B shows the Michaelis-Menten plots for the release of FAM-APP(684–687) by the proteolytic cleavage of the peptide reconstituted in DMPC, PhC, Sph, and DSPC bilayers. No enzyme saturation was observed in the studied concentration range up to 60 μM in the case of DMPC and PhC LUVs. Both lipids are in the Lα phase at 37°C and have a lower hydrophobic thickness than brain Sph and DSPC bilayers at the same temperature (Table 1). The slope of the v versus concentration plot corresponds to the enzyme efficiency Vmax/KM of the reaction. The respective values are listed in Table 2.
Typical Michaelis-Menten saturation kinetics were observed for Sph- and DSPC-reconstituted peptide in the same concentration range. The Tc of Sph is close to the incubation temperature, i.e., 35°C, Tc of DSPC is at 54.5°C (Table 1). In contrast to DMPC and PhC these two systems are not or not exclusively in their Lα phase at the assay temperature and the hydrophobic thickness of the bilayers is larger than in DMPC and PhC membranes. The fitted KM and Vmax values and the corresponding Vmax/KM values are shown in Table 2. Although the KM and Vmax values of the examined systems vary strikingly, Vmax/KM values, i.e., the initial slopes in the Michaelis-Menten plots in Fig. 7, are within a factor of three.
DISCUSSION
The APP sequence APP(684–726) was reconstituted in lipid vesicles consisting of different phosphocholine lipids to investigate the influence of the lipid bilayer characteristics on the conformation and dynamics of the peptide and on the accessibility and cleavability of the α-cleavage site. Solid-state NMR with labels in the putative transmembrane domain and close to the α-cleavage site, CD spectroscopy and enzymatic hydrolysis at the α-cleavage site showed that the lipid acyl chain unsaturation and length have an influence on the global secondary structure of the peptide, on the dynamics of the transmembrane domain, on the motion of Ala692 in the vicinity of the α-cleavage site and on the hydrolysis kinetics of the α-cleavage site by trypsin.
According to the CD spectra, our model peptide adopts mainly β-strand and unordered structures in buffer containing 1% acetonitrile and in DSPC bilayers. We did not further analyze whether the test peptide formed aggregates, protofilament structures or fibrils in buffer. APP(683-699) and (683-713) corresponding to β-amyloid (12–28) and (12–42), respectively (47,48), as well as APP(705-713) corresponding to β-amyloid (34–42) (49) were described to form fibrils. These sequences (except residue 683) are contained in our model peptide APP(684–726). When reconstituted in liposomes and analyzed by density centrifugation, all detectable peptide migrated together with the liposomes, indicating complete membrane incorporation in all liposomal systems.
Incorporation into DMPC and PhC bilayers induced a conformational transition reflected by the appearance of typical features of an α-helical structure in the CD measurements. According to the spectra the peptide adopted a stable conformation with α-helical and unordered domains in these two systems. This is in agreement with structure analysis results of APP or APP fragments. The transmembrane domain is assumed to form an α-helix (50) and according to structural prediction algorithms the sequence adjacent to the exofacial membrane surface does not display any standard secondary structure (51). Comparison of the CD spectra in DMPC and PhC showed no significant differences in the structural content. In all three analyzed liposome systems the structure was independent of the lipid phase (below or above Tc).
Transmembrane peptides reconstituted in lipid membranes typically undergo fast rotations about the bilayer normal and/or about the peptide long axis according to the thermotropic properties of the membrane environment (52,53). 2H NMR spectroscopy is well-suited to study the dynamic properties of membrane proteins in phospholipid bilayers (54) with the quadrupolar splitting and line shapes sensitive to motions on the microsecond timescale (55,56). The alanine residue at position 713 of APP(684–726) was deuterated and used as a nonperturbing probe for the dynamics of the peptide within the bilayer. In fact, the methyl side chain is a useful probe of protein dynamics as it is directly attached to the protein backbone, thus reflecting the motions observed by the peptide in this region and not just by the amino acid side chains (57).
The mobility of the transmembrane part of APP(684–726) depended strongly on the phase of the lipids. The 2H spectra of liposome-incorporated peptide at temperatures <Tc (DMPC, DSPC, Sph) were similar to the spectrum of lyophilized dry peptide. The only motion present <Tc resulted from the methyl group rotation. A significant scaling of the quadrupolar interaction was observed at a temperature above or near the Tc of the investigated lipid indicating an increase in the density of motion on the microsecond timescale observed by the APP(684–726) within the bilayer. Such an averaging of quadrupolar interactions to a few kHz as seen for DMPC and Sph would be expected if the peptide assumes an α-helical conformation at the site of Ala713 and it observes faster rotational motion around the helix axis upon the lipids transition from the Lβ to Lα phase (52,58,59). Indeed, this is in agreement with the secondary structural predictions that suggest the formation of an α-helical transmembrane domain between residues 700 and 723 (see above) and the presence of α-helical contributions in the CD spectra of the APP(684–726) in DMPC. It should be noted, however, that this averaging of the quadrupolar interactions could also be due to the introduction of a range of different motions or changes in the conformation of the system.
The scaling of the quadrupolar interaction observed in the 2H spectra of APP(684–726) in DSPC in the Lα phase is weaker than for DMPC. This could be due to a different secondary structure of APP(684–726) in the two systems, as suggested by the CD spectra, and/or to different average motions.
Interestingly, on reconstitution into PhC bilayers, which are in the Lα phase at all tested temperatures, the quadrupol interaction was only moderately reduced with respect to the value for the lyophilized peptide. Similar spectra were observed upon incorporation of APP(684–726) into vesicles composed of POPC. The incomplete averaging of the quadrupol interaction found for the peptide incorporated in bilayers composed of unsaturated phosphatidylcholine could arise through increases in either the radius of the complex or bilayer viscosity both of which would reduce the rotational correlation time of the APP(684–726). Saturation-transfer ESR measurements of integral membrane proteins indicate that differences between the membrane viscosity of saturated and unsaturated lipids are small (differing by no more than a factor of 2) (60). This suggests the differences observed between the 2H spectra of APP(684–726) in saturated and unsaturated bilayers may arise from the formation of complexes with a larger radius within the unsaturated bilayer as compared to the other systems >Tc.
APP contains GlyxxxGly motifs in the putative transmembrane region (x stands for any amino acid). In general, this motif can lead to the dimerization of the respective transmembrane domain (61). Experiments in cell culture showed that ∼50% of the APP is dimerized (62) whereas the sAPPα truncate, which is the soluble product of α-cleavage, does not dimerize in solution (51). Dimerization and monomerization of APP might be involved in signal transduction across the membrane (51). Experiments with Gly → Ala mutants showed that dimerization had no influence on the total generation of β-amyloid in cultured cells. However, the ratios of β-amyloids consisting of 38, 40, and 42 amino acids, respectively, were significantly different in the mutants and the wild-type cells. The reduction in dimerization by Gly → Ala mutations resulted in higher ratios of shorter to longer β-amyloid peptides. No or only minor changes up to 14% increase, depending on the APP mutant, were observed for the generation of sAPPα, i.e., for the extent of α-cleavage (62). Based on these reports, it can be concluded that within the cell some of the APP dimerizes on the level of the transmembrane domain and that this has no influence on the accessibility of the α-cleavage site by the α-secretase but determines the position of γ-cleavage.
Our findings with 2H solid-state NMR and with the tryptic assay are in agreement with the above. The low amplitude of the motion of Ala713 observed in the PhC bilayer could be due to dimerization/oligomerization within the PhC membrane. The reduced motion of Ala713 in the PhC membrane compared to the DMPC membrane had no effect on the hydrolysis kinetics in the trypsin assay as the kinetics were similar for the two systems. If indeed, and this needs further investigations, the lipid composition dictates the monomer/dimer equilibrium of the peptide, the membrane environment of APP could affect the position of γ-processing via the extent of dimerization in the respective membrane environment. If β-cleaved APP and γ-secretase colocalize in a membrane environment that promotes dimerization, the ratio of shorter to 42-amino acid β-amyloid, the one that is most toxic, would be in disfavor for the individual.
The mobility of the amino acid Ala692 of APP(684–726), which is between the α-cleavage site and the putative transmembrane domain, showed a temperature-dependence when reconstituted in DSPC vesicles. Thus, at the temperature <Tc, the molecular motion of the 15N site was restricted as shown by the large CSA compared to that observed in the Lα phase. No such influence of the bilayer phase was observed with the DMPC system. The difference between the two systems is the bilayer thickness. The hydrophobic thickness of DSPC in the Lβ phase is 3.6 nm (Table 1) whereas it is significantly less for DMPC and PhC or DSPC in the Lα phase. It seems that, without making any assumption on the structure of APP(684–726) reconstituted into DSPC, the DSPC bilayer in the Lβ phase is thick enough to restrict the motion of Ala692. The hydrophobic thickness of rafts was estimated to ∼3.6 nm (63). Depending on the lipid surrounding, variations in the motion and localization of the α-cleavage site relative to the membrane surface can therefore be expected in the cell. This may have an influence on the interaction of APP with the α-secretase.
Trypsin was used to probe the accessibility of the amino acid sequence forming the α-cleavage site of APP. The peptide concentration was varied by altering the liposome concentration. The surface density of the peptide was therefore constant at all peptide concentrations. Table 3 shows the characteristics of the liposomal systems in the kinetic assay. The peptide as well as the number of liposomes was always in excess of the enzyme. The surface density of the peptide was such that the average distance between two peptides was larger than the diameter of the enzyme. As trypsin does not bind to the surface of PhC liposomes (64), we assume that the enzyme acts in a “hopping mode” (65), i.e., it leaves the vesicle surface after one cycle of hydrolysis before it proceeds with the next hydrolysis cycle with a peptide from the same or another vesicle. Based on these conditions and on the assumption that the ratios of outside to inside oriented α-cleavage sites are similar in all systems, i.e., 1:1, we applied Michaelis-Menten analysis as for ideal solutions.
TABLE 3.
Concentrations and molar ratios of enzyme, peptide, lipids, and liposomes in the cleavage experiments
| Enzyme concentration | 5 × 10−11 M |
| Peptide concentration | 5 × 10−6 M to 6 × 10−5 M |
| Lipid concentration | 5 × 10−4 M to 6 × 10−3 M |
| Lipids per liposome | 2 × 105 (assuming a sphere with 150 nm diameter)* |
| Average number of peptides per liposome | 2000 (at a lipid/peptide ratio of 100) |
| Average distance between two peptides | 6 nm† |
| Diameter of the enzyme | 4 nm‡ |
| Vesicle concentration | 3 × 10−9 M to 3 × 10−8 M¶ |
| Enzyme/peptide (M/M) | 1/105 to 1/106 |
| Enzyme/vesicle (M/M) | 1/60 to 1/600 |
Calculated with an average surface area of 0.7 nm2 per lipid (79).
Calculated from the average liposome surface area and the average number of peptides per liposome.
Diaz and Balkus (80).
Calculated from the average outer vesicle surface, the average lipid surface area, and the lipid concentration.
In all investigated systems the model peptide was hydrolyzed by trypsin between Lys687 and Leu688, i.e., at the α-cleavage site. However, we observed significant differences in the kinetics of the tested systems. The parameters KM′ (the observed KM multiplied with 0.5, assuming 50% of the peptide was oriented with the α-cleavage site on the outer membrane surface of the liposome) and Vmax were lowest in solution, intermediate in the systems with Sph and DSPC membranes and highest with DMPC and PhC.
Finehout et al. (66) reported a KM and kcat for the hydrolysis of FITC-casein by trypsin at 37°C and pH 7.5 of 2.09 ± 0.14 μM and 0.40 ± 0.01 s−1, respectively. In comparison, the kcat of the cleavage of FAM-APP(684–726) in our study was lower in 1% acetonitrile and in DSPC liposomes, i.e., ∼0.02 s−1 and 0.08 s−1, respectively, but was in the same range in the Sph liposome system and higher in the DMPC and PhC liposome systems. The Michaelis-Menten constant KM was similar in solution but was significantly higher in all liposomal systems.
Comparing the kinetic parameters in the tested systems and assuming Michaelis-Menten conditions, the changes in KM and Vmax at a similar ratio Vmax/KM is typical for alterations in the rate of the catalytic step of the reaction, i.e., changes in the rate constant kcat (42). Indeed, mathematical simulations of the experimental data were only possible by altering kcat but not by varying k1 and/or k−1 (data not shown). Based on mechanistic considerations the rate constant kcat depends on the actual hydrolysis rate and on the release of the shortened membrane-associated peptide APP(688-726) and the soluble FAM-APP(684–687), including their replacement by a water molecule in the active site.
The low kcat values in 1% acetonitrile and in DSPC liposomes may be related to the peptide conformation that was different from that in DMPC and PhC liposomes as judged from the CD measurements. In this case, the membrane characteristics would via their influence on the peptide conformation affect the kinetics of trypsin cleavage.
Alternatively, differences in the micro environment and particularly in the pH at the position of the enzyme-peptide complex depending on its distance from the membrane surface could affect the hydrolysis rate. The KM for the hydrolysis of FITC-labeled calcein by the modified trypsin was pH-dependent in a range between pH 7.0 and 8.6 (66). However, kcat only varied between 0.32 ± 0.01 and 0.44 ± 0.01 s−1 in the same pH range. Any slight differences in pH at the positions where hydrolysis takes place in our systems are therefore not expected to cause the observed variations in Vmax and kcat.
Regarding the release of the products from the active site, we do not expect a direct influence of the bilayer, however, the residence time of the enzyme at the membrane surface could be dependent on the characteristics of the bilayer. The catalytic rate (kcat) was faster in the systems with the thinner bilayers (DMPC and PhC) than in those with the thicker bilayers (DSPC and Sph). One possibility is that the enzyme interacts with the membrane surface, depending on the distance of the cleavage site from the surface. When interacting with the peptide incorporated in the thicker membranes, the enzyme may sense an intimate contact with the surface, forming hydrogen bonds and ionic interactions. It may need more energy and time to detach from the cleaved peptide and the membrane surface than the enzyme cleaving a peptide reconstituted in a thinner membrane. In other words, the hopping rate is slower with the thicker than with the thinner membranes, leading to the observed differences in kcat.
Based on our data we can not conclude whether the membrane characteristics affect the hydrolysis kinetics via their influence on the peptide conformation or via their influence on the distance of the α-cleavage site from the membrane surface. Trypsin was used in our study as a probe for the accessibility and cleavability of the α-cleavage site. We do not expect that the kinetics of hydrolysis catalyzed by α-secretase would show the same characteristics as we found with trypsin. However, based on our results from the 15N-NMR analysis and from the tryptic cleavage assay, we conclude that the membrane thickness influences the motion and disposition of the α-cleavage site. This could have an influence on its recognition and processing by α-secretase.
On the cellular level the generation of β-amyloid was found to occur in cholesterol-rich, detergent-resistant membrane domains, also called rafts, and was reduced by the treatment of the cells with statins, which inhibit the biosynthesis of cholesterol (67–69). In addition, cholesterol depletion by β-methyl cyclodextrin of cells and inhibition of cholesterol biosynthesis with statins, increased the products of α-cleavage (15). The activity of α-secretase was related to the membrane fluidity. Membrane fluidization by cholesterol depletion enhanced the α-secretase activity whereas cholesterol replacement by sterols lacking membrane-rigidifying effects did not reduce the α-secretase activity (15). Considering our findings, it will be interesting to see whether beside the membrane fluidity also the membrane thickness has an influence on the α-cleavage in cultured cells.
It has been hypothesized that the nonamyloidogenic processing of APP, i.e., α-cleavage, occurs outside rafts (70). In HEK cells, ADAM 10 was mainly localized in detergent soluble domains and only a small fraction of ADAM 10 proform was found in rafts (15). Also ADAM 17, another enzyme with suggested α-secretase activity, was localized in nonraft fractions (71). According to (19) APP is protected from β- and γ-cleavage by the separation of the respective enzymes into rafts whereas APP resides in detergent-soluble membrane domains. Based on our study it may be that not only the physical separation of APP and α-secretase from the β- and γ-secretase plays a role for the processing of APP but also the membrane environment of APP in that it influences its properties as a substrate for α- and γ-secretase, respectively.
In conclusion, the rotational motion of the transmembrane domain of the model peptide APP(684–726) is reduced in phosphatidylcholine bilayers with unsaturated acyl chains as compared to bilayers consisting of saturated lipids (>Tc) at a similar or larger membrane thickness. This may indicate that peptide dimerization or oligomerization is dependent on the properties of the membrane hydrophobic core. The membrane thickness on the other hand affected the dynamics of the α-cleavage site as well as the kinetics of its tryptic cleavage. Extrapolated to the in vivo situation, the nonamyloidogenic processing of APP may be affected by the lipid environment of the protein.
Acknowledgments
We thank Barbara Bianchi, Guido Grassi, and Maja Günthert for technical support and Maja for carefully reading the manuscript.
Marco Marenchino's present address is Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425.
Philip T. F. Williamson's present address is School of Biological Sciences, University of Southampton, Bassett Crescent East, Southampton, SO16 7PX, United Kingdom.
Editor: Arthur G. Palmer III.
References
- 1.Blennow, K., M. J. de Leon, and H. Zetterberg. 2006. Alzheimer's disease. Lancet. 368:387–403. [DOI] [PubMed] [Google Scholar]
- 2.Thomas, P., and M. Fenech. 2007. A review of genome mutation and Alzheimer's disease. Mutagenesis. 22:15–33. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, L., M. B. McQueen, K. Mullin, D. Blacker, and R. E. Tanzi. 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 39:17–23. [DOI] [PubMed] [Google Scholar]
- 4.Raber, J., Y. D. Huang, and J. W. Ashford. 2004. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol. Aging. 25:641–650. [DOI] [PubMed] [Google Scholar]
- 5.Corder, E. H., A. M. Saunders, W. J. Strittmatter, D. E. Schmechel, P. C. Gaskell, G. W. Small, A. D. Roses, J. L. Haines, and M. A. Pericakvance. 1993. Gene dose of apolipoprotein-E type-4 allele and the risk of Alzheimers-disease in late-onset families. Science. 261:921–923. [DOI] [PubMed] [Google Scholar]
- 6.Poirier, J., J. Davignon, D. Bouthillier, S. Kogan, P. Bertrand, and S. Gauthier. 1993. Apolipoprotein-E polymorphism and Alzheimers disease. Lancet. 342:697–699. [DOI] [PubMed] [Google Scholar]
- 7.Petot, G. J., and R. P. Friedland. 2004. Lipids, diet and Alzheimer disease: an extended summary. J. Neurol. Sci. 226:31–33. [DOI] [PubMed] [Google Scholar]
- 8.Rojo, L., M. K. Sjoberg, P. Hernandez, C. Zambrano, and R. B. Maccioni. 2006. Roles of cholesterol and lipids in the etiopathogenesis of Alzheimer's disease. J. Biomed. Biotechnol. 2006:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mielke, M. M., and C. G. Lyketsos. 2006. Lipids and the pathogenesis of Alzheimer's disease: Is there a link? Int. Rev. Psychiatry. 18:173–186. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, J. L. 2003. Dietary lipids in the etiology of Alzheimer's disease—implications for therapy. Drugs Aging. 20:399–418. [DOI] [PubMed] [Google Scholar]
- 11.Puglielli, L., R. E. Tanzi, and D. M. Kovacs. 2003. Alzheimer's disease: the cholesterol connection. Nat. Neurosci. 6:345–351. [DOI] [PubMed] [Google Scholar]
- 12.Sambamurti, K., A. C. Granholm, M. S. Kindy, N. R. Bhat, N. H. Greig, D. K. Lahiri, and J. E. Mintzer. 2004. Cholesterol and Alzheimer's disease: clinical and experimental models suggest interactions of different genetic, dietary and environmental risk factors. Curr. Drug Targets. 5:517–528. [DOI] [PubMed] [Google Scholar]
- 13.Whitfield, J. F. 2006. Can statins put the brakes on Alzheimer's disease? Expert Opin. Investig. Drugs. 15:1479–1485. [DOI] [PubMed] [Google Scholar]
- 14.Famer, D., S. Meaney, M. Mousavi, A. Nordberg, I. Bjorkhem, and M. Crisby. 2007. Regulation of α- and β-secretase activity by oxysterols: cerebrosterol stimulates processing of APP via the α-secretase pathway. Biochem. Biophys. Res. Commun. 359:46–50. [DOI] [PubMed] [Google Scholar]
- 15.Kojro, E., G. Gimpl, S. Lammich, W. Marz, and F. Fahrenholz. 2001. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the α-secretase ADAM 10. Proc. Natl. Acad. Sci. USA. 98:5815–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., D. Cao, D. W. Garber, H. Kim, and K. Fukuchi. 2003. Association of aortic atherosclerosis with cerebral β-amyloidosis and learning deficits in a mouse model of Alzheimer's disease. Am. J. Pathol. 163:2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Refolo, L. M., M. A. Pappolla, B. Malester, J. LaFrancois, T. Bryant-Thomas, R. Wang, G. S. Tint, K. Sambamurti, and K. Duff. 2000. Hypercholesterolemia accelerates the Alzheimer's amyloid pathology in a transgenic mouse model. Neurobiol. Dis. 7:321–331. [DOI] [PubMed] [Google Scholar]
- 18.Sparks, D. L., S. W. Scheff, J. C. Hunsaker, H. C. Liu, T. Landers, and D. R. Gross. 1994. Induction of Alzheimer-like β-amyloid immunoreactivity in the brains of rabbits with dietary cholesterol. Exp. Neurol. 126:88–94. [DOI] [PubMed] [Google Scholar]
- 19.Abad-Rodriguez, J., M. D. Ledesma, K. Craessaerts, S. Perga, M. Medina, A. Delacourte, C. Dingwall, B. De Strooper, and C. G. Dotti. 2004. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J. Cell Biol. 167:953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaether, C., and C. Haass. 2004. A lipid boundary separates APP and secretases and limits amyloid β-peptide generation. J. Cell Biol. 167:809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oksman, M., H. Livonen, E. Hogyes, Z. Amtul, B. Penke, I. Leenders, L. Broersen, D. Lutjohann, T. Hartmann, and H. Tanila. 2006. Impact of different saturated fatty acid, polyunsaturated fatty acid and cholesterol containing diets on β-amyloid accumulation in APP/PS1 transgenic mice. Neurobiol. Dis. 23:563–572. [DOI] [PubMed] [Google Scholar]
- 22.Lim, G. P., F. Calon, T. Morihara, F. S. Yang, B. Teter, O. Ubeda, N. Salem, S. A. Frautschy, and G. M. Cole. 2005. A diet enriched with the omega-3 fatty acid docosahexaenoic acid reduces amyloid burden in an aged Alzheimer mouse model. J. Neurosci. 25:3032–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettegrew, J. W., K. Panchalingam, R. L. Hamilton, and R. J. McClure. 2001. Brain membrane phospholipid alterations in Alzheimer's disease. Neurochem. Res. 26:771–782. [DOI] [PubMed] [Google Scholar]
- 24.Kalvodova, L., N. Kahya, P. Schwille, R. Ehehalt, P. Verkade, D. Drechsel, and K. Simons. 2005. Lipids as modulators of proteolytic activity of BACE—involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J. Biol. Chem. 280:36815–36823. [DOI] [PubMed] [Google Scholar]
- 25.Lahdo, R., S. Coillet-Matillon, J. P. Chauvet, and L. de La Fourniere-Bessueille. 2002. The amyloid precursor protein interacts with neutral lipids—liposomes and monolayer studies. Eur. J. Biochem. 269:2238–2246. [DOI] [PubMed] [Google Scholar]
- 26.Lahdo, R., and L. De la Fourniere-Bessueille. 2004. Insertion of the amyloid precursor protein into lipid monolayers: effects of cholesterol and apolipoprotein E. Biochem. J. 382:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooper, N. M. 2005. Roles of proteolysis and lipid rafts in the processing of the amyloid precursor protein and prion protein. Biochem. Soc. Trans. 33:335–338. [DOI] [PubMed] [Google Scholar]
- 28.Haass, C., A. Y. Hung, D. J. Selkoe, and D. B. Teplow. 1994. Mutations associated with a locus for familial Alzheimers-disease result in alternative processing of amyloid β-protein precursor. J. Biol. Chem. 269:17741–17748. [PubMed] [Google Scholar]
- 29.Roks, G., F. Van Harskamp, I. De Koning, M. Cruts, C. De Jonghe, S. Kumar-Singh, A. Tibben, H. Tanghe, M. F. Niermeijer, A. Hofman, J. C. Van Swieten, C. Van Broeckhoven, and C. M. Van Duijn. 2000. Presentation of amyloidosis in carriers of the codon 692 mutation in the amyloid precursor protein gene (APP692). Brain. 123:2130–2140. [DOI] [PubMed] [Google Scholar]
- 30.Chan, W. C., and P. D. White, editors. 2000. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford University Press, Oxford.
- 31.Fischer, R., O. Mader, G. Jung, and R. Brock. 2003. Extending the applicability of carboxyfluorescein in solid-phase synthesis. Bioconjug. Chem. 14:653–660. [DOI] [PubMed] [Google Scholar]
- 32.Killian, J. A., T. P. Trouard, D. V. Greathouse, V. Chupin, and G. Lindblom. 1994. A general method for the preparation of mixed micelles of hydrophobic peptides and sodium dodecyl-sulfate. FEBS Lett. 348:161–165. [DOI] [PubMed] [Google Scholar]
- 33.Birnie, G. D., and D. Rickwood. 1978. Centrifugal Separations in Molecular and Cell Biology. Butterworths, London.
- 34.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 35.Krämer, S. D., J. A. Hurley, N. J. Abbott, and D. J. Begley. 2002. Lipids in blood-brain barrier models in vitro I: thin-layer chromatography and high-performance liquid chromatography for the analysis of lipid classes and long-chain polyunsaturated fatty acids. In Vitro Cell. Dev. Biol. 38:557–565. [DOI] [PubMed] [Google Scholar]
- 36.Hahn, E. L. 1950. Spin echoes. Phys. Rev. 80:580–594. [Google Scholar]
- 37.Herzfeld, J., and A. E. Berger. 1980. Sideband intensities in NMR-spectra of samples spinning at the magic angle. J. Chem. Phys. 73:6021–6030. [Google Scholar]
- 38.Smith, S. A., T. O. Levante, B. H. Meier, and R. R. Ernst. 1994. Computer-simulations in magnetic-resonance—an object-oriented programming approach. J. Magn. Reson. A. 106:75–105. [Google Scholar]
- 39.Cheng, V. B., H. H. Suzukawa, and M. Wolfsber. 1973. Investigations of a nonrandom numerical-method for multidimensional integration. J. Chem. Phys. 59:3992–3999. [Google Scholar]
- 40.Sreerama, N., and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252–260. [DOI] [PubMed] [Google Scholar]
- 41.Rice, R. H., G. E. Means, and W. D. Brown. 1977. Stabilization of bovine trypsin by reductive methylation. Biochim. Biophys. Acta. 492:316–321. [DOI] [PubMed] [Google Scholar]
- 42.Cornish-Bowden, A. 2004. Fundamentals of Enzyme Kinetics. Portland Press Ltd., London.
- 43.Seelig, J. 1978. P-31 Nuclear magnetic-resonance and head group structure of phospholipids in membranes. Biochim. Biophys. Acta. 515:105–140. [DOI] [PubMed] [Google Scholar]
- 44.Dixon, G. S., S. G. Black, C. T. Butler, and A. K. Jain. 1982. A differential AC calorimeter for biophysical studies. Anal. Biochem. 121:55–61. [DOI] [PubMed] [Google Scholar]
- 45.Greenfield, N. J. 1996. Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal. Biochem. 235:1–10. [DOI] [PubMed] [Google Scholar]
- 46.Litman, B. J., and Y. Barenholz. 1975. Optical activity of D-erythro-sphingomyelin and its contribution to circular-dichroism of sphingomyelin-containing systems. Biochim. Biophys. Acta. 394:166–172. [DOI] [PubMed] [Google Scholar]
- 47.Li, L. P., T. A. Darden, L. Bartolotti, D. Kominos, and L. G. Pedersen. 1999. An atomic model for the pleated β-sheet structure of A β amyloid protofilaments. Biophys. J. 76:2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabanal, F., J. M. Tusell, L. Sastre, M. R. Quintero, M. Cruz, D. Grillo, M. Pons, F. Albericio, J. Serratosa, and E. Giralt. 2002. Structural, kinetic and cytotoxicity aspects of 12–28 β-amyloid protein fragment: A reappraisal. J. Pept. Sci. 8:578–588. [DOI] [PubMed] [Google Scholar]
- 49.Halverson, K., P. E. Fraser, D. A. Kirschner, and P. T. Lansbury. 1990. Molecular determinants of amyloid deposition in Alzheimers-disease—conformational studies of synthetic β-protein fragments. Biochemistry. 29:2639–2644. [DOI] [PubMed] [Google Scholar]
- 50.Kang, J., H. G. Lemaire, A. Unterbeck, J. M. Salbaum, C. L. Masters, K. H. Grzeschik, G. Multhaup, K. Beyreuther, and B. Mullerhill. 1987. The precursor of Alzheimers-disease amyloid-A4 protein resembles a cell-surface receptor. Nature. 325:733–736. [DOI] [PubMed] [Google Scholar]
- 51.Gralle, M., and S. T. Ferreira. 2007. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog. Neurobiol. 82:11–32. [DOI] [PubMed] [Google Scholar]
- 52.Williamson, P. T. F., G. Zandomeneghi, F. J. Barrantes, A. Watts, and B. H. Meier. 2005. Structural and dynamic studies of the gamma-M4 trans-membrane domain of the nicotinic acetylcholine receptor. Mol. Membr. Biol. 22:485–496. [DOI] [PubMed] [Google Scholar]
- 53.Lewis, B. A., G. S. Harbison, J. Herzfeld, and R. G. Griffin. 1985. NMR structural-analysis of a membrane-protein—bacteriorhodopsin peptide packbone orientation and motion. Biochemistry. 24:4671–4679. [DOI] [PubMed] [Google Scholar]
- 54.Prosser, R. S., and J. H. Davis. 1994. Dynamics of an integral membrane peptide—a deuterium NMR relaxation study of gramicidin. Biophys. J. 66:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beshah, K., E. T. Olejniczak, and R. G. Griffin. 1987. Deuterium NMR-study of methyl-group dynamics in L-alanine. J. Chem. Phys. 86:4730–4736. [Google Scholar]
- 56.Keniry, M. A., A. Kintanar, R. L. Smith, H. S. Gutowsky, and E. Oldfield. 1984. Nuclear magnetic-resonance studies of amino-acids and proteins—deuterium nuclear magnetic-resonance relaxation of deuteriomethyl-labeled amino-acids in crystals and in Halobacterium-Halobium and Escherichia coli cell-membranes. Biochemistry. 23:288–298. [DOI] [PubMed] [Google Scholar]
- 57.Jones, D. H., A. C. Rigby, K. R. Barber, and C. W. M. Grant. 1997. Oligomerization of the EGF receptor transmembrane domain: a H-2 NMR study in lipid bilayers. Biochemistry. 36:12616–12624. [DOI] [PubMed] [Google Scholar]
- 58.Cornell, B. A., F. Separovic, A. J. Baldassi, and R. Smith. 1988. Conformation and orientation of gramicidin-a in oriented phospholipid-bilayers measured by solid-state C-13 NMR. Biophys. J. 53:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, R., F. Separovic, T. J. Milne, A. Whittaker, F. M. Bennett, B. A. Cornell, and A. Makriyannis. 1994. Structure and orientation of the pore-forming peptide, melittin, in lipid bilayers. J. Mol. Biol. 241:456–466. [DOI] [PubMed] [Google Scholar]
- 60.Spooner, P. J. R., R. H. E. Friesen, J. Knol, B. Poolman, and A. Watts. 2000. Rotational mobility and orientational stability of a transport protein in lipid membranes. Biophys. J. 79:756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911–919. [DOI] [PubMed] [Google Scholar]
- 62.Munter, L. M., P. Voigt, A. Harmeier, D. Kaden, K. E. Gottschalk, C. Weise, R. Pipkorn, M. Schaefer, D. Langosch, and G. Multhaup. 2007. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of A β 42. EMBO J. 26:1702–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Duyl, B. Y., D. T. S. Rijkers, B. de Kruijff, and J. A. Killian. 2002. Influence of hydrophobic mismatch and palmitoylation on the association of transmembrane α-helical peptides with detergent-resistant membranes. FEBS Lett. 523:79–84. [DOI] [PubMed] [Google Scholar]
- 64.Liu, D., and L. Huang. 1992. Trypsin-induced lysis of lipid vesicles—effect of surface-charge and lipid-composition. Anal. Biochem. 202:1–5. [DOI] [PubMed] [Google Scholar]
- 65.Gelb, M. H., M. K. Jain, A. M. Hanel, and O. G. Berg. 1995. Interfacial enzymology of glycerolipid hydrolases—lessons from secreted phospholipases A2. Annu. Rev. Biochem. 64:653–688. [DOI] [PubMed] [Google Scholar]
- 66.Finehout, E. J., J. R. Cantor, and K. H. Lee. 2005. Kinetic characterization of sequencing grade modified trypsin. Proteomics. 5:2319–2321. [DOI] [PubMed] [Google Scholar]
- 67.Simons, M., P. Keller, B. De Strooper, K. Beyreuther, C. G. Dotti, and K. Simons. 1998. Cholesterol depletion inhibits the generation of β-amyloid in hippocampal neurons. Proc. Natl. Acad. Sci. USA. 95:6460–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fassbender, K., M. Simons, C. Bergmann, M. Stroick, D. Lutjohann, P. Keller, H. Runz, S. Kuhl, T. Bertsch, K. von Bergmannn, M. Hennerici, K. Beyreuther, and T. Hartmann. 2001. Simvastatin strongly reduces levels of Alzheimer's disease β-amyloid peptides A β 42 and A β 40 in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 98:5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ehehalt, R., P. Keller, C. Haass, C. Thiele, and K. Simons. 2003. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 160:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolozin, B. 2001. A fluid connection: cholesterol and A β. Proc. Natl. Acad. Sci. USA. 98:5371–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Tresckow, B., K. J. Kallen, E. P. von Strandmann, P. Borchmann, H. Lange, A. Engert, and H. P. Hansen. 2004. Depletion of cellular cholesterol and lipid rafts increases shedding of CD30. J. Immunol. 172:4324–4331. [DOI] [PubMed] [Google Scholar]
- 72.Marsh, D. 1990. Handbook of Lipid Bilayers. CRC Press, Boston.
- 73.Meyer, H. W., H. Bunjes, and A. S. Ulrich. 1999. Morphological transitions of brain sphingomyelin are determined by the hydration protocol: ripples re-arrange in plane, and sponge-like networks disintegrate into small vesicles. Chem. Phys. Lipids. 99:111–123. [DOI] [PubMed] [Google Scholar]
- 74.Lewis, B. A., and D. M. Engelman. 1983. Lipid bilayer thickness varies linearly with acyl chain-length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166:211–217. [DOI] [PubMed] [Google Scholar]
- 75.Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cornell, B. A., and F. Separovic. 1983. Membrane thickness and acyl chain-length. Biochim. Biophys. Acta. 733:189–193. [DOI] [PubMed] [Google Scholar]
- 77.Nezil, F. A., and M. Bloom. 1992. Combined influence of cholesterol and synthetic amphiphillic peptides upon bilayer thickness in model membranes. Biophys. J. 61:1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinia, H. A., R. A. Kik, R. A. Demel, M. M. E. Snel, J. A. Killian, J. P. J. M. van der Eerden, and B. de Kruijff. 2000. Visualization of highly ordered striated domains induced by transmembrane peptides in supported phosphatidylcholine bilayers. Biochemistry. 39:5852–5858. [DOI] [PubMed] [Google Scholar]
- 79.Huang, C., and J. T. Mason. 1978. Geometric packing constraints in egg phosphatidylcholine vesicles. Proc. Natl. Acad. Sci. USA. 75:308–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diaz, J. F., and K. J. Balkus, Jr. 1996. Enzyme immobilization in MCM-41 molecular sieve. J. Mol. Catal., B Enzym. 2:115–126. [Google Scholar]