Abstract
NASP has been described as a histone H1 chaperone in mammals. However, the molecular mechanisms involved have not yet been characterized. Here, we show that this protein is not only present in mammals but is widely distributed throughout eukaryotes both in its somatic and testicular forms. The secondary structure of the human somatic version consists mainly of clusters of α-helices and exists as a homodimer in solution. The protein binds nonspecifically to core histone H2A-H2B dimers and H3-H4 tetramers but only forms specific complexes with histone H1. The formation of the NASP-H1 complexes is mediated by the N- and C-terminal domains of histone H1 and does not involve the winged helix domain that is characteristic of linker histones. In vitro chromatin reconstitution experiments show that this protein facilitates the incorporation of linker histones onto nucleosome arrays and hence is a bona fide linker histone chaperone.
INTRODUCTION
Histones need to be assembled onto the newly synthesized DNA template during the S phase of the cell cycle (1), and they need to be disassembled and reassembled at other stages to facilitate and permit DNA accessibility during transcription (2), DNA recombination, and repair (3). This dynamic histone exchange is mediated by histone chaperones such as NAP-1** (4), which preferentially binds to H2A-H2B dimers (5), CAF-1/antisilencing factor 1, which binds to H3-H4 tetramers (6–8), and nucleoplasmin (9)/nucleophosmin, which binds to both (10) (see Loyola and Almouzni (11) for a recent comprehensive review on core histone chaperones).
In contrast to the large amount of information on core histone chaperones, very few examples are known of linker histone chaperones, which remain largely uncharacterized. One such protein is NASP, a protein that shares a large degree of homology with the Xenopus histone-binding protein N1/N2 (12,13). NASP binds linker histones that are not bound to DNA in vivo and, hence, may provide a potentially useful reagent for in vitro reconstitution of linker histones onto nucleosome arrays. NASP binds to linker histones, transporting them into the nucleus and exchanging them onto DNA (14,15). This chaperone is present in all mitotic cells, either as an sNASP or a tNASP (16,17). Alternative splicing of the pre-mRNA results in two mature mRNA forms corresponding to the full-length tNASP and the shorter sNASP (16). In humans, sNASP is a 449-amino-acid protein that is essential for cell proliferation, cell cycle regulation, and DNA replication (18,19). During normal cell cycle progression, the level of NASP expressed is under strict control (15,16). Overexpression of NASP leads to a delay in the progression through the G1/S border (15,16). NASP exists in a multichaperone nucleosome-remodeling complex that is composed of not only sNASP but CAF-1 (p150, p60, p48), H3.1, H4, Asfa, Asfb, checkpoint homolog 2, histone acetyltransferase 1, and importin 4 (20,21). This complex coordinates the deposition of H3.1–H4 histones into nucleosomes and allows for the progression of DNA replication through phosphorylation signals at specific checkpoints (22). The implication of this is that completion of S phase in a normal cell cycle requires the presence of NASP (21,23,24). Also, NASP has been found to interact in vivo with Ku70/Ku80 and DNA protein kinase implying that NASP may play a role in DNA repair (14). Intriguingly, it has yet to be shown whether NASP binds DNA directly (22), and NASP sequence possesses motifs indicating possible interaction sites for several essential proteins such as checkpoint homolog 2 and p300 (22).
The distribution of NASP over a broad spectrum of tissues (sNASP) and in testes (tNASP) has been well documented in mammals (16). Here we report that these two species of NASP exhibit a conserved wide distribution in eukaryotes and provide the first biophysical characterization of sNASP, which is shown to be a genuine linker histone chaperone.
MATERIALS AND METHODS
Materials
All of the chemicals and reagents used were of molecular biology grade. HeLa cell RNA was extracted using Qiashredder and RNeasy Mini Kit (Qiagen, Mississauga, ON). cDNA synthesis was conducted using Invitrogen's First Strand cDNA Kit (Invitrogen, Carlsbad, CA). Restriction enzymes were purchased from New England Biolabs (NEB, Pickering, ON). Purification of DNA was done using Qiagen miniprep PCR purification and gel extraction kits. Sequencing was conducted at the DNA Sequencing Facility of Centre for Biomedical Research at the University of Victoria. Rabbit anti-NASP antibody was a gift from Dr. M. O'Rand (Dept. of Cell and Developmental Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC) (16). Goat antirabbit antibodies were purchased from Abcam (Cambridge, MA). Micrococcal nuclease was obtained from Worthington Biochemical (Lakewood, NJ).
PAGE
SDS-PAGE was performed according to Laemmli (25). Native (6.0%)-PAGE (acrylamide:bis-acrylamide 29:1 (w:w)) for nucleosome analysis was carried out in 20 mM sodium acetate, 1mM EDTA, 20 mM Tris-HCl (pH 7.2), E buffer as described elsewhere (26,27).
Construction of expression vectors
The entire coding sequence of human sNASP (GenBank accession number NM_152298) was amplified from HeLa cell cDNA using the following primers and cycle conditions: forward 5′-GCGAATTCGGATCCATGGCCATGGAGTCCACA-3′, reverse 5′-GCCTCGAGGAATTCTTAACATGCAGTGCTTTCAACTG-3′; 3 min at 94°C then a touchdown PCR from 64°C to 52°C annealing temperature for 1 min decreasing 3°C every three cycles then holding at 52°C annealing temperature for 18 cycles. Melting temperatures of 94°C for 1 min and extension temperatures of 72°C for 2 min were used, with a final extension of 10 min at 72°C. PCR products were then ligated into pCR2.1 TOPO vector (Invitrogen) sequenced, then digested with BamHI and XhoI for ligation into pET28a His-tag vector (Novagen, EMD Chemicals, Darmstadt, Germany) for an N-terminal thrombin-cleavable His-tag.
Protein expression and purification
pET28a-sNASP was grown in Escherichia coli Rosetta (DE3) competent cells (Novagen, EMD Chemicals) in TB broth plus 34 μg/ml kanamycin at 37°C to an OD600 of ∼0.6 then induced for 2 h with 0.4 mM isopropyl β-d-1-thiogalactopyranoside. The expressed product had a calculated molecular mass of 52.4 kDa with the His-tag. For purification, cells were suspended in lysis buffer (20 mM Tris-HCl (pH 8.0), 500 mM KCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 15% glycerol (w/v), 1 mM EDTA (pH 8.0), 0.1% NP-40 (w/v), and 1/25 dilution of Complete Protease Inhibitor Tablet (Roche, Basel, Switzerland) (dissolved in 1 mL dH2O), sonicated, and centrifuged. The supernatant was adjusted to 2 mM MgCl2 and filtered through a 0.45-μm membrane. Ni Sepharose High-Performance beads (Invitrogen) were equilibrated with lysis buffer in a column, and the sample was then loaded at ∼5 ml/min. The column was washed with 20 ml of wash buffer (40 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 1/50 Complete protease inhibitors dissolved in 1 ml dH2O (Roche)), 10 ml of wash buffer plus 40 mM imidazole, then the protein was eluted with 20 ml of wash buffer plus 0.5 M imidazole, and 5-ml fractions were collected. sNASP-containing fractions were confirmed by 10% SDS-PAGE, then dialyzed extensively into Buffer A (40 mM Tris-HCl (pH 8.0)). Dialyzed fractions were loaded onto Q-HiTrap 5-ml anion-exchange columns (Amersham, Piscataway, NJ) using the AKTA-fast protein liquid chromatography system (Amersham). A gradient of 20–60% Buffer B (Buffer A plus 1 M NaCl) was used to elute sNASP, which usually came out at ∼40% = 400 mM NaCl. sNASP-containing fractions were confirmed on 10% SDS-PAGE and then dialyzed extensively against 20 mM Tris-HCl (pH 8.0). In several instances, when indicated, the His-tag was removed by thrombin cleavage using a Novagen thrombin kit (Novagen) following the manufacturers' instructions.
Histone purification
Native histone H1, H2A-H2B dimers, and H3-H4 tetramers were purified from HeLa cells, and chicken erythrocyte histone H1 and H5 were obtained from chicken blood. Chromatin obtained from these tissues (28,29) was loaded onto a hydroxyapatite column, and histones were eluted with a NaCl gradient in 10 mM phosphate buffer (pH 6.7) as described elsewhere (30). The trypsin-resistant core (comprising the winged helix domain) of HeLa H1 histones was obtained by digestion with trypsin in the presence of 2 M NaCl plus 20 mM Tris-HCl (pH 7.5) and purified by CMC-25 Sephadex as described by Jutglar et al. (31).
Tissue lysate preparation
Tissue samples were pulverized in liquid nitrogen with a mortar and pestle. The resulting powder was suspended in lysis buffer (20 mM Tris-HCl (pH 7.5) plus 1% SDS) and boiled for 1 min. Samples were then centrifuged for 5 min at 16,000 × g. The pellet was discarded, and the supernatant was used directly for electrophoresis.
Western blotting
Tissue lysates were mixed with 2× loading buffer (125 mM Tris-HCl (pH 6.8), 4% (w/v) SDS, 20% (w/v) glycerol, 1.72 M β-mercaptoethanol, bromophenol blue) and boiled for 5 min. Samples were then run on 10% SDS-PAGE at 100 V. After electrophoresis, gels were transferred to polyvinylidene fluoride membranes at 100 V for 2 h in transfer buffer (192 mM glycine, 25 mM Tris-HCl, 20% (v/v) methanol). Blots were blocked for 30 min at room temperature or overnight at 4°C in blocking buffer (5% (w/v) skim milk in phosphate-buffered saline plus 0.1% (v/v) Tween-20). Rabbit antibodies raised against recombinant mouse NASP (16) were diluted 1:1000 in blocking buffer. Secondary goat antirabbit antibodies (Abcam) were diluted 1:5000 in blocking buffer. Both primary and secondary incubations were done for 1 h at room temperature with three 5-min washes with phosphate-buffered saline plus 0.1% Tween-20 in between and after incubations. Secondary antibodies were detected with enhanced chemiluminescence (GE Healthcare, Baie d'Urfe, QC) and exposure to x-ray film.
Trypsin digestion
Recombinant sNASP in 20 mM Tris-HCl (pH 7.5) buffer was digested with trypsin (EC 3.4.21.4) (type III) (Sigma-Aldrich, St. Louis, MO) for different amounts of time at room temperature using an E:S ratio of 1:1000 (mol:mol). The digests were analyzed by SDS-PAGE. Alternatively they were fractionated by HPLC using a C18 Vydac column (GRACE-Vydac, Hesperia, CA) (32), and the fractions were analyzed by MALDI-time of flight (UVic-Genome BC Proteomics Centre, University of Victoria, Victoria, BC). In some instances the trypsin digests were blotted from the SDS-PAGE onto polyvinylidene fluoride membranes, and the membranes were stained with brilliant blue coomassie stain for a few minutes and air dried. The bands of interest were excised and subjected to N-terminal Edman degradation sequencing (Advanced Protein Technology Centre, Hospital for Sick Children, Toronto, ON).
CD
CD analysis was carried out on a JASCO J720 (Jasco, Easton, MD) as described previously (33). The spectra were analyzed using a Contin LL (34) algorithm.
Extinction coefficients
An extinction coefficient of 20,190 M−1 cm−1 at 276 nm was used for NASP as estimated from its amino acid sequence (35). The extinction coefficient of histone H5 was 48,000 cm−1 M−1 at 230 nm (36). For DNA, an extinction coefficient of 20 cm2 mg−1 at 260 nm was used (37).
Analytical ultracentrifuge
These experiments were performed on an XL-I (Beckman-Coulter, Fullerton, CA) analytical ultracentrifuge. Sedimentation velocity experiments were carried out at 20°C and 44,000 rpm in an An-55 aluminum rotor using double-sector aluminum-filled Epon centerpieces. Sedimentation equilibrium experiments were carried out at 4°C in an An-60 titanium rotor using double-sector aluminum-filled Epon centerpieces. The scans were analyzed by the method of van Holde and Weischet (38) using UltraScan version 9.3 (39) sedimentation data analysis software (Borries Demeler, Missoula, MT). The partial specific volume of NASP (0.720 cm3/g), and the frictional ratio f/f0 were determined using the same software. Sedimentation velocity analysis was also used to determine the molecular mass using a Monte Carlo analysis (40,41).
Secondary and tertiary structure prediction
Secondary structure prediction was performed using the hierarchical neural network protein sequence analysis (42). Tertiary structure prediction was carried out using the SWISS-MODEL server (43) and the Phyre server (44).
Chromatin isolation
Chicken erythrocyte chromatin was prepared as described elsewhere (28). Long chromatin fragments were fractionated using a 5–20% sucrose gradient (45). A heterogeneous fraction with a 23 nucleosome average number was thus obtained. Approximately half of this fraction was stripped of linker histones using CM C-25 Sephadex (GE Healthcare) as described previously (46). The linker histone-containing and -depleted counterparts were analyzed by sedimentation velocity dialysis against different NaCl concentrations in 10 mM Tris-HCl (pH 7.5), 0.2 mM EDTA buffer.
Histone H1 reconstitution
Direct mixing
Linker histone-depleted nucleosome arrays in 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA were directly mixed with chicken erythrocyte histone H5 in the same buffer to a ratio of ∼2 mol H5:1 mol nucleosome. The mixture was incubated for 30 min at room temperature, and aliquots were next dialyzed against different NaCl concentrations in 10 mM Tris-HCl (pH 7.5), 0.2 mM EDTA buffers and analyzed by sedimentation velocity.
NASP-assisted
Histone H5 and NASP were mixed at a ratio of ∼1:1.2 mol:mol in 50 mM NaCl, 10 mM Tris-HCl (pH 7.5) buffer and incubated for 5 min at room temperature. The H5-NASP mixture was then added to the linker histone-depleted nucleosome arrays at a ratio of 2 mol H5:1 mol nucleosome and treated and analyzed as for direct mixing. In some instances (see the results), after incubation, the samples were brought to 4 mM MgCl2 and centrifuged at maximum speed in an Eppendorf (Hamburg, Germany) microcentrifuge at 4°C, and the pellet containing the H1-reconstituted chromatin was resuspended in 10 mM Tris-HCl, 0.2 mM EDTA and used for further analysis (47).
Micrococcal nuclease digestion
Histone H1-reconstituted complexes at different NaCl concentrations in 10 mM Tris-HCl (pH 7.5) buffer with 0.05 mM CaCl2 were digested with micrococcal nuclease at 37°C for different amounts of time. Thirty units of micrococcal nuclease per milligram of DNA was used in these digestions. After digestion, the samples were mixed with an equal volume of a solution containing 30% sucrose, 0.8% SDS, and a mixture of bromphenol blue and cyanol green, incubated at 37°C and loaded directly onto a 4.5% native-PAGE to visualize the DNA composition of the products of digestion.
RESULTS
NASP is widely distributed in eukaryotes
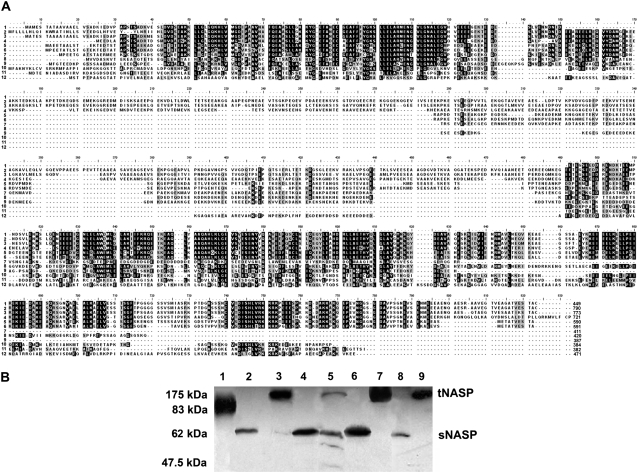
NASP is widely distributed in mammals (48), where it has been shown to exist in a smaller sNASP isoform, ∼400–500 amino acids, and a larger tNASP isoform, which is ∼300 amino acids longer. Although the critical cellular functions assigned to this chaperone and the identification of its activity in Caenorhabditis elegans (48) suggest that the protein should be evenly distributed throughout eukaryotes, a comprehensive analysis of its distribution across different taxonomic groups has been lacking. The analysis shown in Fig. 1 indicates that sequences with a high extent of sequence similarity to mammalian t/sNASP can be identified in both vertebrate and invertebrate organisms including fungi (Fig. 1 A). Furthermore, proteins with similar molecular mass to both tNASP and sNASP can be identified in chordates and more specifically in species representative of each one of the main vertebrate classes from fish to mammals (Fig. 1 B). In all instances the apparent molecular masses in SDS-PAGE of the somatic and testicular forms are larger than the canonical values from the sequences, a fact that reflects the highly negatively charged nature of these proteins.
FIGURE 1.
NASP is widely distributed throughout eukaryotes. (A) Alignment of NASP amino acid sequences of several representative invertebrate and vertebrate organisms. The sequences and Genebank accession numbers are: 1), NM_172164 Homo sapiens (human) isoform 1; 2), NM_152298 Homo sapiens (human) isoform 3; 3), AF349432 Mus musculus (mouse); 4), XM-001235059〈removed hyperlink field〉 Gallus gallus (rooster); 5), NM_001088068 histone-binding protein N1/N2 Xenopus laevis (African clawed frog); 6), CAA28419 Xenopus laevis (African clawed frog); 7), BC068344 Danio rerio (zebrafish); 8), Ensembl Gene Report ENSCINT00000019618 Ciona intestinalis (sea squirt tunicate); 9), XP_791433 Strongylocentrotus purpuratus (sea urchin); 10), XP_970862 Trilobium castaneum (red flour beetle); 11), NM_063979 Caenorhabditis elegans (nematode) isoform 1; 12), XP_961229 Neurospora crassa (red bread mold). (B) Zoowestern analysis of NASP. (lane 1) Ciona intestinalis (mantle); (lane 2) Squalus acanthias (spiny dogfish, heart); (lane 3) S. acanthias (spiny dogfish, testes); (lane 4) X. laevis (frog, liver); (lane 5) Alligator mississippiensis (alligator, liver); (lane 6) G. gallus (chicken, liver); (lane 7) G. gallus (chicken, testes); (lane 8) M. musculus (mouse, liver); (lane 9) M. musculus (mouse, testes).
Importantly, in vertebrates, all the sequences identified as putative NASP bear homology to Xenopus laevis N1/N2 histone-binding protein (Fig. 1 A, line 5) (16,49). N1/N2 and nucleoplasmin are two histone chaperones (50) that are present in the oocytes and eggs of Xenopus, where they are found preferentially associated with core histones H3-H4 and H2A-H2B, respectively (51,52) (see recent reviews (10,50)). In comparison to nucleoplasmin, N1/N2 has been less extensively characterized, and the molecular details of its histone chaperone activity and its involvement in chromatin assembly/remodeling still require elucidation.
Human sNASP consists mainly of α-helical structure and exists as a dimer in solution
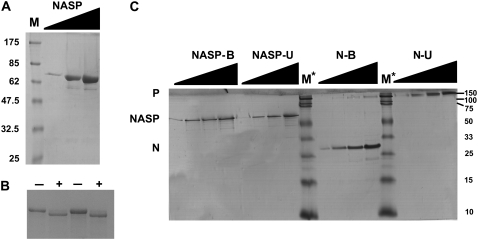
To characterize the structure of human sNASP, we expressed and purified the recombinant version of this protein. Fig. 2, A–B, shows an electrophoretic analysis of the protein thus obtained. In contrast to nucleoplasmin, which forms pentameric complexes that are highly stable in solution in the presence of SDS when not heated (53), recombinant human sNASP remains as a monomer in the presence of SDS with an apparent molecular mass of 60–70 kDa. As in the case of the native form (Fig. 1 B), the apparent molecular mass of the recombinant sNASP is higher than the 52.4 kDa estimated from the sequence of the recombinant version.
FIGURE 2.
Electrophoretic characterization of the recombinant human sNSAP used in this article. (A) SDS (15%)-PAGE of increasing loading amounts (triangles) of purified recombinant sNASP. M is (NEB, broad-range prestained) protein marker. (B) SDS (10%)-PAGE of increasing amounts of human recombinant sNASP before (−) and after (+) treatment with thrombin to remove the N-terminal His-tag. (C) SDS (10%)-PAGE of increasing amounts (triangles) of recombinant sNASP in comparison to recombinant X. laevis nucleoplasmin (N) (85). The proteins were dissolved in SDS sample buffer and loaded directly onto the gel without heat treatment (unboiled, N-U, NASP-U) or previously heated at 100°C for 5 min before loading onto the gel (boiled, N-B, NASP-B). M* is (Bio-Rad Precision Plus Protein Standard) protein marker.
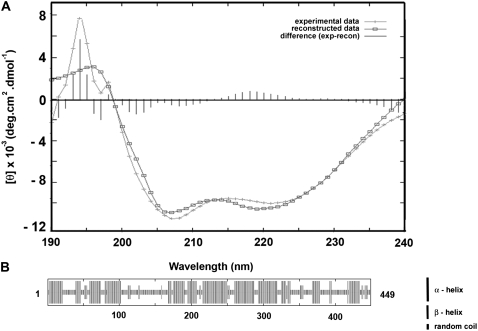
Next we determined the secondary structure of the protein using CD. Fig. 3 A shows the CD spectrum obtained in this way. Fitting of the spectrum using the Contin algorithm (34) revealed that the protein consists of 54% helix and 46% random coil. These values are in very good agreement with the secondary structure of 51% helix, 43% random coil, and 6% extended strand estimated using the Hierarchical Neural Network prediction analysis (42) and with the observations reported earlier using a similar kind of predictive analysis (49).
FIGURE 3.
Determination of the secondary structure of sNASP. (A) CD spectrum. The spectrum was recorded at 20°C in 100 mM NaCl, 40 mM Tris-HCl (pH 8.0) buffer. (B) Secondary structure prediction using the hierarchical neural network protein sequence analysis (42).
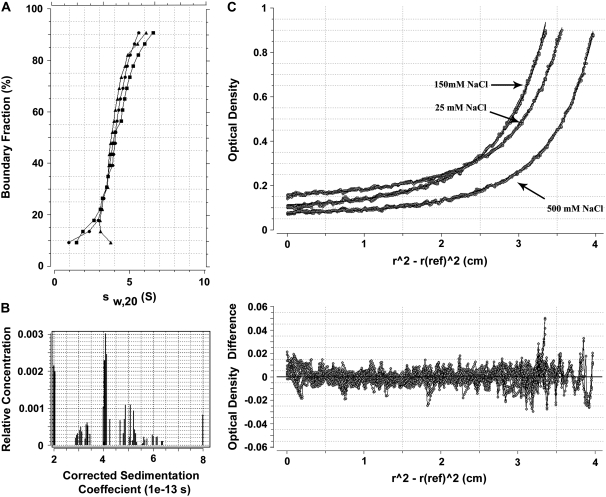
The tertiary and quaternary structures of the protein were analyzed in the analytical ultracentrifuge using both sedimentation velocity and sedimentation equilibrium experiments. The results of such analyses are summarized in Fig. 4. Fig. 4 A shows the integral distribution of the sedimentation coefficient of human sNASP under different ionic strengths, in the range from 25 to 500 mM NaCl, showing that the sedimentation behavior is little affected by the salt concentration in this range. Fig. 4 B shows the result of the Monte Carlo analysis (41) of the sedimentation velocity in 25 mM NaCl. This type of analysis not only allows an accurate analysis of the sedimentation coefficient but also provides a solution to the Lamm equation and allows the determination of the molecular masses of the different macromolecules present in the sample. It was determined that the major species present had an average sedimentation coefficient of s20,w = 4.5 ± 0.2 S corresponding to a molecular mass of 100,000 Daltons. The same sedimentation coefficient was obtained when the N-terminal His-tag of the protein was removed (Fig. 4 A, inset). A similar value of 99,000 Daltons was obtained from the best fits to the sedimentation equilibrium profiles of the sample under the same ionic conditions (Fig. 4 C). Thus, under the different ionic strengths used here, the protein remained as a dimer.
FIGURE 4.
Analytical ultracentrifuge analysis of recombinant human sNASP. (A) Integral distribution of the sedimentation coefficient of sNASP at different ionic strengths: 25 mM NaCl (black triangles); 150 mM NaCl (black circles); 500 mM NaCl, 20 mM Tris-HCl (pH 7.5) buffer (black squares). The inset shows the integral distribution of the sedimentation coefficient of a sNASP sample, before (white squares) and after (black circles) treatment with thrombin to remove the N-terminal His-tag, analyzed in 150 mM NaCl, 20 mM Tris-HCl (pH 7.5) buffer. The sedimentation velocity runs were performed at 44,000 rpm and at 20°C. (B) Monte Carlo analysis of the sedimentation velocity experiment shown in A at 25 mM NaCl. (C) Sedimentation equilibrium analyses of sNASP at different salt concentrations. The top plot shows the absorbance at 230 nm as a function of the square of the radial distance of the sample at any position within the cell (r) minus the square of the radial position at a reference (r(ref)). The continuous line in this plot was obtained by fitting the experimental data (circles) to a single ideal species with a Mr of 99,000 Da. The bottom plot shows χ2 residuals as a function of (r2 − r(ref)2) for the best fit (solid line).The data shown were obtained at 17,000 rpm and at 4°C.
When the value of the sedimentation coefficient s20,w = 4.5 was used in conjunction with the molecular mass of the dimer, using as an average preferential hydration parameter of the protein the value of 0.22 g water/g protein (54), a frictional coefficient ratio f/f0 = 1.68 was determined as described elsewhere (55). This corresponds to a molecule with a prolate ellipsoid shape of axial ratio a/b = 12. The highly asymmetric value thus obtained suggests that the two monomers of sNASP are held together in a head-to-tail type arrangement with the dimer adopting a highly extended conformation.
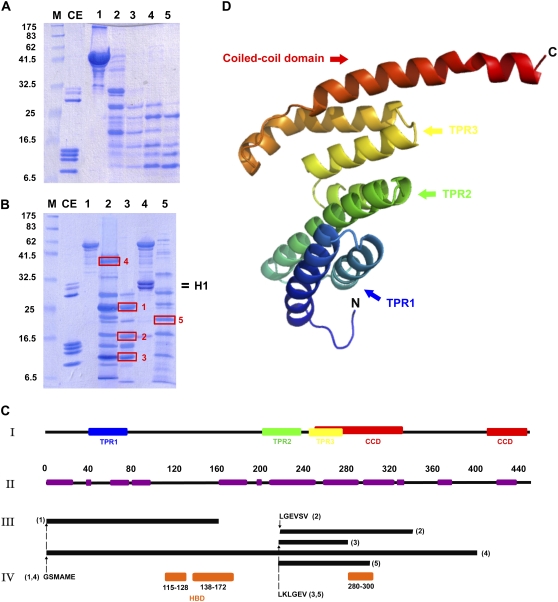
Further insight into the folding of the human sNASP molecule was obtained by trypsin digestion. The results of such analysis are depicted in Fig. 5, A–C. The pattern of digestion obtained (Fig. 5 A) with this protease was very similar regardless of whether the digestion was carried out in the presence of 10 mM NaCl (Fig. 5, A and B, lane 3) or at 2 M NaCl (Fig. 5 B, lane 2 and results not shown). The main tryptic-resistant peptides were identified by a combination of HPLC/SDS-PAGE/MALDI and N-terminal sequencing approach (see Materials and Methods). To help visualize the location of the trypsin-resistant domains determined in this way, a three-dimensional prediction analysis of the NASP monomer was carried out using the SWISS-MODEL server (43) (Fig. 5, C, I, and D). As can be seen in Fig. 5, C and D, the main trypsin-resistant peptides (1, 2, and 3 in Fig. 5 C, III) are defined by the three predicted tetratricopeptide repeat domains and the coiled-coil domain. The previously defined histone-binding domains are contained within trypsin-resistant peptide 1. Interestingly, binding of histone H1 results in the preferential protection of peptide 5, helping us identify a novel histone-binding site that encompasses the region between amino acids 280 and 300 of human sNASP (Fig. 5 C, IV).
FIGURE 5.
Tertiary structure of sNASP. (A) SDS-PAGE analysis of the time course of digestion of NASP by trypsin (E:S 1:1000) in 100 mM NaCl, 20 mM Tris-HCl (pH 7.5). (lane 1) Undigested sample; (lanes 2–5) peptides generated after 5, 10, 20, and 40 min of digestion, respectively. (B) SDS-PAGE analysis of NASP (lane 1) and a NASP-HeLa histone H1 (1 mol:1 mol) complex (lane 4) before (lanes 1 and 4) and after digestion with trypsin (lanes 2, 3, and 5). Digestion was for 20 min under the same conditions as in A (lanes 3 and 5) or in 2 M NaCl, 20 mM Tris-HCl (pH 7.5) (lane 2). CE, chicken erythrocyte histone marker; M, protein marker. The boxes highlight the peptides subjected to N-terminal sequencing analysis. (C) (I) Secondary structure organization derived from the tertiary structure prediction of sNASP using the Phyre program (44) in comparison to (II) the α-helix structure predicted from the hierarchical neural network analysis (see Fig. 3 B). Also shown are (III) the distribution of the trypsin-resistant peptides identified by N-terminal Edman degradation sequencing and MALDI-time of flight as well as (IV) the sites of histone H1 binding (orange boxes, 115–128, 138–172) described by Batova and O'Rand (70) or identified here (orange box, 280–300) (4). The numbers in brackets in III correspond to the peptide numbers identified in B. TPR, tetratricopeptide repeat domain; CCD, coiled-coil domain. (D) Tertiary structure predicted from the primary structure of sNASP using the Phyre program (44).
sNASP forms defined complexes with histone H1 but not with H2A-H2B or H3-H4
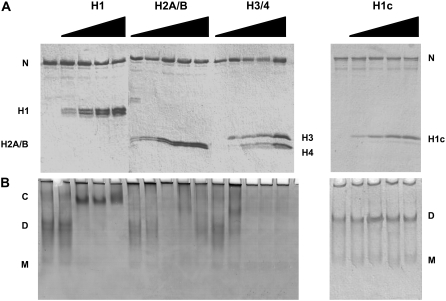
Fig. 6 shows the characterization of the interaction of human sNASP with core and linker histones using native gel electrophoresis. Fig. 6 A shows an SDS-PAGE electrophoretic analysis of the samples used for this analysis. As seen in Fig. 6 B, NASP forms discrete complexes at a ratio of 2 mol of H1/mol NASP dimer. In contrast, H2A-H2B dimers or H3-H4 tetramers bind to NASP forming large aggregates that do not enter the native gels. Thus, it seems that NASP exhibits binding specificity to linker histones, and although it is able to bind to core histones, it does so in a less specific way. The nonspecific binding is not surprising considering the large negative charge of NASP and the positively charged nature of core histones.
FIGURE 6.
Binding of histones to sNASP. HeLa cell core histones H2A-H2B, H3-H4, and linker histones H1 and their trypsin-resistant core H1c were mixed with sNASP in the presence of 100 mM NaCl, 10 mM Tris-HCl (pH 7.5) at increasing histone:sNASP molar ratios (0 (lanes 1, 6, 11, and 16), 0.5 (lanes 2, 7, 12, and 17), 1.0 (lanes 3, 8, 13, and 18), 1.5 (lanes 4, 9, 14, and 19), and 2.0 (lanes 5, 10, 15, and 20)) and (A) analyzed by either SDS-(15%) PAGE or (B) native (6%) PAGE. C, histone H1-NASP complex; D, sNASP dimer; M, sNASP monomer; N, sNASP.
Linker histones differ from core histones in their lysine-rich composition and in that their folded protein domains are different. Core histones consist of a histone fold (56), whereas linker histones contain a winged helix motif (57). The specific binding of histone H1 to NASP raises the possibility that the winged helix domain of H1 may involved. To test this possibility, we studied the interaction of the trypsin-resistant core of HeLa cell histone H1 (which encompasses the winged helix domain). As can be seen in Fig. 6 B, this histone domain exhibits no binding to NASP, indicating that the charge effects and probably the distribution of such charge in the histone H1 molecule are responsible for the binding specificity.
sNASP facilitates the assembly of histone H1 onto nucleosome arrays
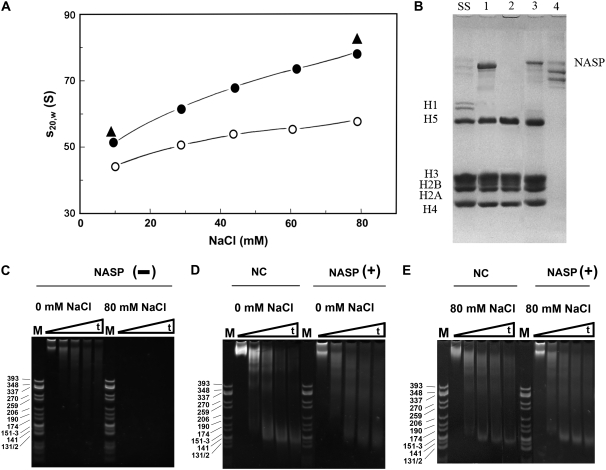
To check whether sNASP facilitates the proper deposition of linker histones onto linker histone-depleted nucleosome arrays, a native chromatin fraction consisting of a weighted average nucleosome number of 23 was obtained by sucrose gradient fractionation of chicken erythrocyte chromatin (45). The NaCl dependence of the sedimentation coefficient of the fraction before and after careful depletion of linker histones (H1 and H5) was examined (Fig. 7 A). The increase in the sedimentation coefficient observed in the range of 0–80 mM NaCl measures the extent of folding of the chromatin fiber (58–60). Linker histones play a critical role in the folding of the chromatin fiber, and in their absence, the nucleosome array complexes exhibit a significantly lower ability to fold (open circles in Fig. 7 A). These results demonstrate the ability of NASP to add linker histones to nucleosome arrays to produce a chromatin fiber that folds in the same way as the native counterpart.
FIGURE 7.
NASP facilitates the deposition of linker histones onto nucleosome arrays. (A) Change in the sedimentation coefficient (s20,W) of chicken erythrocyte chromatin fraction as a function of the sodium chloride concentration. The average number of nucleosomes of the chromatin fraction used was Nw = 23. Data obtained before (solid circles) and after (open circles) removal of linker histones. Values obtained with the linker histone-depleted fraction on addition of histone H5 in the presence of NASP (solid triangles). (B) SDS-PAGE analysis of starting chromatin fraction (lane SS). Linker histone-depleted fraction on addition of 2 mol H5/mol nucleosome in the presence (lane 1) or absence of sNASP (lane 2). Products recovered from MgCl2 (5 mM) precipitation (lanes 3 and 4) of the samples (lanes 1 and 2, respectively). (C–E) Native-(4.5%) PAGE analysis of the DNA from the micrococcal nuclease digestion products of the starting native chromatin fraction (NC) and the linker histone-depleted counterpart on reconstitution with H5 in the presence (NASP (+)) or the absence (NASP (−)) of sNASP at 0 and 80 mM NaCl in 10 mM Tris-HCl, 0.1 mM EDTA (pH 7.5). The triangles indicate increasing time of micrococcal nuclease digestion: 0 (lanes 1 and 6), 5 (lanes 2 and 7), 20 (lanes 3 and 8), 40 (lanes 4 and 9), and 60 (lanes 5 and 10) min.
When the linker-depleted fraction was directly mixed with histone H5 at a molar ratio of 2 mol of H5/mol nucleosome in the absence of sNASP, a sedimentation coefficient of 48 S was obtained in 10 mM Tris-HCl (pH 7.5). This is almost identical to that of the native nonlinker depleted counterpart (result not shown). However, dialysis of this sample against 80 mM NaCl resulted in its complete precipitation, indicative of an anomalous deposition. However, when the same experiment was carried out in the presence of sNASP, the values of the sedimentation coefficients at 0 mM and at 80 mM NaCl in the presence of 10 mM Tris-HCl (pH 7.5) produced values that were slightly higher than those of the native fraction but were still very similar and followed the same increasing trend (Fig. 7 A, solid triangles). The slightly higher values are most likely the result of the native chicken chromatin fraction containing ∼1.3–1.5 mol of linker histone (H1 and H5) per nucleosome (61) compared to the reconstituted complexes that can take slightly higher amounts (47) (see below).
The native chromatin fiber and its fractions are completely precipitated at MgCl2 concentrations above 1.5 mM (60,62,63). In the absence of linker histones, or when they are not properly positioned on the fiber, chromatin exhibits an enhanced MgCl2 solubility. Therefore, such divalent ion-associated behavior can be used to monitor the proper assembly of the chromatin fiber, and it has been used to prepare highly homogeneous reconstituted oligonucleosomal fiber fragments (47,64). As can be seen in Fig. 7 B, sNASP-assisted linker histone deposition (Fig. 7 B, lanes 1 and 3) produced complexes that were precipitated in 5 mM MgCl2 and exhibited a stoichiometry of ∼1.7 mol histone H5/mol nucleosome (lane 3). In contrast, direct mixing (Fig. 7 B, lanes 2 and 4) produced complexes with an altered solubility that resulted in highly insoluble histone aggregates (lane 4). Such complexes were highly resistant to micrococcal nuclease digestion at low salt (Fig. 7 C).
The fidelity of the NASP-assisted reconstitution of chromatin can be assessed by micrococcal nuclease digestion. Fig. 7, D and E, shows the digestion of complexes reconstituted in this way. Although there are slight differences in the nucleosome repeat pattern in Fig. 7, D and E, between the native and NASP+ lanes, the pattern of nucleosome digestion is very close to that of the corresponding native counterpart at both 0 and 80 mM NaCl.
DISCUSSION
Knowledge about the histone deposition and chromatin assembly mediated by histone chaperones in an in vivo setting within the cell is still very incomplete. Most of what is known comes from information on core histone chaperones that bind to either core histones H3-H4, H2A-H2B, or to both. Linker histones (histone H1 family) play a critical role in proper chromatin folding and dynamics (19) and have been shown to be deposited simultaneously with core histones on newly replicated DNA (65). Yet the mechanisms involved in histone H1 deposition and those involved in the interactions with its sole genuine chaperone (NASP) described to date for this histone family (16) remain undetermined.
We surmised that if NASP is a key player in chromatin assembly during DNA replication (22), then the protein should be ubiquitously distributed throughout the eukaryotic domain. The high cross-reactivity of NASP antibody across different vertebrate and chordate species as well as our ability in silico to identify related proteins in organisms ranging from fungi to mammals (Fig. 1) support this notion. The relatively large extent of sequence variability of NASP-like proteins observed in invertebrates (Fig. 1 A) is not surprising because these proteins correspond to chaperones for invertebrate H1 histones, which constitute the fastest evolving group of histones and exhibit a high extent of interspecies sequence variability (66,67).
In contrast to core histone chaperones such as nucleoplasmin, which has an almost exclusively β-strand secondary structure (9), or NAP-1, which consists of a mixture of α-helix and β-strand (4), the structural organization of sNASP is almost exclusively α-helical (Fig. 3). Thus, when compared to other histone chaperones, this represents a distinctive feature of this molecule. However, as with NAP-1 (4,68), the protein exists in solution mainly as a dimer (Fig. 4) with a highly extended conformation. The dimeric nature does not come as a surprise because there is a predicted leucine zipper (between residues 230 and 251) in the protein sequence. An extended conformation of the homodimer would fully expose the three tetratricopeptide protein-protein interaction domains TPR1–3 (14,16) that are present in the molecule (Fig. 5 C), making them available for interaction with other proteins. Indeed, several NASP nonhistone partners have been identified such as heat shock protein 90, DNA-activated protein kinase, and ATP-dependent DNA helicase II (70-kDa subunit) (14). In addition, NASP has been shown to be part of the C. elegans transformer-4-NASP– class I histone deacetylase complex, which participates in sex determination (69), and NASP is also found in the multisubunit complexes CAF-1 and histone cell cycle regulation defective homolog A that participate in histone H3.1 and H3.3 nucleosome assembly, respectively (20). The existence of so many interacting partners attests to the multiple functionality of this protein (22).
Trypsin digestion of sNASP was useful in corroborating some of the prediction analysis regarding the secondary structure organization of sNASP (Fig. 5, A and B). Only in the case of one of the trypsin-resistant peptides (peptide 1, Fig. 5, B and C) did the resistance extend beyond the predicted helical domains (Fig. 5 C, I and II). However, the protection observed in this instance spanning from residue 100 to 160 is most likely the result of the presence of glutamic acid clusters in this region corresponding to the regions 115–128 and 138–172, which had been previously identified as the putative histone H1 binding domains (16,70) and which correspond to similar glutamic acid-rich domains (108–119) and (296–326) of X. laevis N1 (71) (see also Fig. 1 A). More importantly, this type of analysis helped us uncover a new glutamic acid-rich region spanning approximately from residue 280 to 300 (Fig. 5).
Our histone-binding results strongly suggest that although sNASP can bind nonspecifically to core and linker histones, it forms specific complexes only with the latter (Fig. 6 B). Linker histones are the native substrate of NASP in vivo (16,17). A similar situation has been observed with other histone chaperones with regard to histone preference. In vivo, NAP-1 and nucleoplasmin have been shown to be preferentially bound to H2A-H2B (72–74), whereas the Xenopus N1/N2 heterodimer and ACF-1 are found associated with H3-H4 (9,11,51). However, both NAP-1 and nucleoplasmin have also been shown to be able to bind to H3-H4 (9,68,75,76) and to histone H1 (77–79) in vitro.
The ambivalent nature of all these proteins for their histone-binding substrate, especially in experiments carried out in vitro, is not surprising because regardless of their structural organization, all of them have a low pI, which undoubtedly will lead to some binding redundancy for basic proteins. The binding data also indicate that the binding of histone H1 with sNASP appears to involve the N-terminal and C-terminal domains of this histone. This is similar to what has been observed with NAP-1, where the histone tails appear to be critical for binding (75,80). The way histone tails provide binding specificity considering their disordered conformation in solution may involve the charge distribution signature conferred by their underlying primary structure. Also, they can exhibit a significant extent of secondary structure organization when interacting with DNA in the case of both linker histones (81,82) and core histones (83). Therefore, the possibility that such organization could also participate during binding to their chaperones cannot be excluded.
The binding of core histones to NAP-1 and of linker histones to NASP contrasts with that of core histones to nucleoplasmin in which the histone fold appears to be critical for binding recognition (76). This most likely reflects the differences in the diverse structural levels of folding between these chaperones to which we have previously referred. Interestingly, the three sets of domains for H1 binding to sNASP (Fig. 5 C, IV) exhibit an interesting arrangement in which the two N-terminal binding sites are separated from the C-terminal domain by ∼100 amino acids. We would like to speculate here that the two distinct binding domains defined in this way serve for binding to the C-terminal and N-terminal charged regions of linker histones, with their winged fold domain occupying the region in between.
Our chromatin reconstitution experiments using a native chromatin fraction depleted of linker histones show that NASP can efficiently put the linker histones back to produce a chromatin fraction that exhibits a NaCl folding dependence almost identical to that of the native counterpart (Fig. 7). This not only attests to its physiological role in histone H1 deposition in the cellular setting (15) but also provides a nice in vitro tool that may complement the protocols already available for proper reconstitution of linker histones onto nucleosome arrays reconstituted onto sequence-defined DNA templates (47,64,84).
The results presented in this article provide strong support for the functional role(s) proposed for NASP. In vivo this molecule has been shown to transport histone H1 into nuclei (14), where it is exchanged with DNA (15). Such an exchange is likely to play a critical role in the modulation of the proper chromatin fiber folding that is critical to fundamental metabolic processes of the cell such as replication and DNA repair.
Acknowledgments
We are very thankful to Dr. Michael G. O'Rand (University of North Carolina at Chapel Hill) for kindly providing us with NASP antibody. We are also grateful to Dr. Borries Demeler (The University of Texas Health Science Center at San Antonio) for helping us with the Monte Carlo analysis of the sedimentation velocity data. This work was supported by the Canadian Institutes of Health Research (CIHR) grant MOP 57718 (to J.A.).
Ron M. Finn and Kristen Brown contributed equally to this work.
Kristen Brown's present address is Zoology Department, University of British Columbia, 2350 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada.
Abbreviations used: (t/s)NAP-1, (testes/somatic) nucleosome assembly protein 1; CAF-1, chromatin assembly factor 1; CD, circular dichroism; MALDI, matrix-assisted laser desorption/ionization; (t/s)NASP, (testes/somatic) nuclear autoantigenic sperm protein; PAGE, polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; rpm, revolutions per minute; SDS, sodium dodecyl sulfate.
Editor: Stuart M. Lindsay.
References
- 1.Annunziato, A. T. 2005. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 280:12065–12068. [DOI] [PubMed] [Google Scholar]
- 2.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 20:2009–2017. [DOI] [PubMed] [Google Scholar]
- 3.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 402:555–560. [DOI] [PubMed] [Google Scholar]
- 4.Park, Y. J., and K. Luger. 2006. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA. 103:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park, Y. J., J. V. Chodaparambil, Y. Bao, S. J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A–H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817–1825. [DOI] [PubMed] [Google Scholar]
- 6.Ridgway, P., and G. Almouzni. 2000. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci. 113:2647–2658. [DOI] [PubMed] [Google Scholar]
- 7.English, C. M., M. W. Adkins, J. J. Carson, M. E. Churchill, and J. K. Tyler. 2006. Structural basis for the histone chaperone activity of Asf1. Cell. 127:495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natsume, R., M. Eitoku, Y. Akai, N. Sano, M. Horikoshi, and T. Senda. 2007. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 446:338–341. [DOI] [PubMed] [Google Scholar]
- 9.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell. 8:841–853. [DOI] [PubMed] [Google Scholar]
- 10.Frehlick, L. J., J. M. Eirin-Lopez, and J. Ausio. 2007. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 29:49–59. [DOI] [PubMed] [Google Scholar]
- 11.Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 1677:3–11. [DOI] [PubMed] [Google Scholar]
- 12.O'Rand, M. G., R. T. Richardson, L. J. Zimmerman, and E. E. Widgren. 1992. Sequence and localization of human NASP: conservation of a Xenopus histone-binding protein. Dev. Biol. 154:37–44. [DOI] [PubMed] [Google Scholar]
- 13.Kleinschmidt, J. A., C. Dingwall, G. Maier, and W. W. Franke. 1986. Molecular characterization of a karyophilic, histone-binding protein: cDNA cloning, amino acid sequence and expression of nuclear protein N1/N2 of Xenopus laevis. EMBO J. 5:3547–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alekseev, O. M., R. T. Richardson, M. R. Pope, and M. G. O'Rand. 2005. Mass spectrometry identification of NASP binding partners in HeLa cells. Proteins. 61:1–5. [DOI] [PubMed] [Google Scholar]
- 15.Alekseev, O. M., D. C. Bencic, R. T. Richardson, E. E. Widgren, and M. G. O'Rand. 2003. Overexpression of the linker histone-binding protein tNASP affects progression through the cell cycle. J. Biol. Chem. 278:8846–8852. [DOI] [PubMed] [Google Scholar]
- 16.Richardson, R. T., I. N. Batova, E. E. Widgren, L. X. Zheng, M. Whitfield, W. F. Marzluff, and M. G. O'Rand. 2000. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J. Biol. Chem. 275:30378–30386. [DOI] [PubMed] [Google Scholar]
- 17.Richardson, R. T., D. C. Bencic, and M. G. O'Rand. 2001. Comparison of mouse and human NASP genes and expression in human transformed and tumor cell lines. Gene. 274:67–75. [DOI] [PubMed] [Google Scholar]
- 18.Fan, Y., T. Nikitina, J. Zhao, T. J. Fleury, R. Bhattacharyya, E. E. Bouhassira, A. Stein, C. L. Woodcock, and A. I. Skoultchi. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 123:1199–1212. [DOI] [PubMed] [Google Scholar]
- 19.Bustin, M., F. Catez, and J. H. Lim. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell. 17:617–620. [DOI] [PubMed] [Google Scholar]
- 20.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 116:51–61. [DOI] [PubMed] [Google Scholar]
- 21.Groth, A., D. Ray-Gallet, J. P. Quivy, J. Lukas, J. Bartek, and G. Almouzni. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 17:301–311. [DOI] [PubMed] [Google Scholar]
- 22.Richardson, R. T., O. M. Alekseev, G. Grossman, E. E. Widgren, R. Thresher, E. J. Wagner, K. D. Sullivan, W. F. Marzluff, and M. G. O'Rand. 2006. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 281:21526–21534. [DOI] [PubMed] [Google Scholar]
- 23.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 11:341–351. [DOI] [PubMed] [Google Scholar]
- 24.Hoek, M., and B. Stillman. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA. 100:12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- 26.Li, A., A. H. Maffey, W. D. Abbott, N. Conde e Silva, A. Prunell, J. Siino, D. Churikov, A. O. Zalensky, and J. Ausio. 2005. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry. 44:2529–2535. [DOI] [PubMed] [Google Scholar]
- 27.Yager, T. D., and K. E. van_Holde. 1984. Dynamics and equilibria of nucleosomes at elevated ionic strength. J. Biol. Chem. 259:4212–4222. [PubMed] [Google Scholar]
- 28.Ausió, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451–463. [DOI] [PubMed] [Google Scholar]
- 29.Ausió, J., and K. E. van Holde. 1986. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 25:1421–1428. [DOI] [PubMed] [Google Scholar]
- 30.Thambirajah, A. A., D. Dryhurst, T. Ishibashi, A. Li, A. H. Maffey, and J. Ausio. 2006. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 281:20036–20044. [DOI] [PubMed] [Google Scholar]
- 31.Jutglar, L., J. I. Borrell, and J. Ausió. 1991. Primary, secondary, and tertiary structure of the core of a histone H1-like protein from the sperm of Mytilus. J. Biol. Chem. 266:8184–8191. [PubMed] [Google Scholar]
- 32.Saperas, N., M. Chiva, M. T. Casas, J. L. Campos, J. M. Eirin-Lopez, L. J. Frehlick, C. Prieto, J. A. Subirana, and J. Ausio. 2006. A unique vertebrate histone H1-related protamine-like protein results in an unusual sperm chromatin organization. FEBS J. 273:4548–4561. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., S. C. Moore, M. Laszckzak, and J. Ausió. 2000. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 275:35013–35020. [DOI] [PubMed] [Google Scholar]
- 34.Provencher, S. W., and J. Glockner. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 20:33–37. [DOI] [PubMed] [Google Scholar]
- 35.Gill, S. C., and P. H. von_Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326. [DOI] [PubMed] [Google Scholar]
- 36.Carter, G. J., and K. van Holde. 1998. Self-association of linker histone H5 and of its globular domain: evidence for specific self-contacts. Biochemistry. 37:12477–12488. [DOI] [PubMed] [Google Scholar]
- 37.Ausio, J., D. Seger, and H. Eisenberg. 1984. Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. J. Mol. Biol. 176:77–104. [DOI] [PubMed] [Google Scholar]
- 38.van Holde, K. E., and W. O. Weischet. 1978. Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers. 17:1387–1403. [Google Scholar]
- 39.Demeler, B. 2005. UltrScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. Modern Analytical Ultracentrifugation: Techniques and Methods. D. J. Scott, S. E. Hardfing, and A. J. Rowe, editors. Royal Society of Chemistry, London, UK. 210–229.
- 40.Cao, W., and B. Demeler. 2005. Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution of the Lamm equation. Biophys. J. 89:1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demeler, B., and E. Brookes. 2007. Monte Carlo analysis of sedimentation experiments. Prog. Colloid Polym. Sci. In press.
- 42.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150. [DOI] [PubMed] [Google Scholar]
- 43.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett-Lovsey, R. M., A. D. Herbert, M. J. Sternberg, and L. A. Kelley. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 70:611–625. [DOI] [PubMed] [Google Scholar]
- 45.Garcia Ramirez, M., F. Dong, and J. Ausió. 1992. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J. Biol. Chem. 267:19587–19595. [PubMed] [Google Scholar]
- 46.Libertini, L. J., and E. W. Small. 1980. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 8:3517–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huynh, V. A., P. J. Robinson, and D. Rhodes. 2005. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 345:957–968. [DOI] [PubMed] [Google Scholar]
- 48.Lee, Y. H., and M. G. O'Rand. 1993. Ultrastructural localization of a nuclear autoantigenic sperm protein in spermatogenic cells and spermatozoa. Anat. Rec. 236:442–448. [DOI] [PubMed] [Google Scholar]
- 49.Welch, J. E., L. J. Zimmerman, D. R. Joseph, and M. G. O'Rand. 1990. Characterization of a sperm-specific nuclear autoantigenic protein. I. Complete sequence and homology with the Xenopus protein, N1/N2. Biol. Reprod. 43:559–568. [DOI] [PubMed] [Google Scholar]
- 50.Prado, A., I. Ramos, L. J. Frehlick, A. Muga, and J. Ausio. 2004. Nucleoplasmin: a nuclear chaperone. Biochem. Cell Biol. 82:437–445. [DOI] [PubMed] [Google Scholar]
- 51.Kleinschmidt, J. A., and W. W. Franke. 1982. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 29:799–809. [DOI] [PubMed] [Google Scholar]
- 52.Laskey, R. A., B. M. Honda, A. D. Mills, and J. T. Finch. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 275:416–420. [DOI] [PubMed] [Google Scholar]
- 53.Prieto, C., N. Saperas, C. Arnan, M. H. Hills, X. Wang, M. Chiva, R. Aligue, J. A. Subirana, and J. Ausió. 2002. Nucleoplasmin interaction with protamines. Involvement of the polyglutamic tract. Biochemistry. 41:7802–7810. [DOI] [PubMed] [Google Scholar]
- 54.Eisenberg, H. 1990. Thermodynamics and the structure of biological macromolecules. Rozhinkes mit mandeln. Eur. J. Biochem. 187:7–22. [DOI] [PubMed] [Google Scholar]
- 55.Ausió, J., D. A. Malencik, and S. R. Anderson. 1992. Analytical sedimentation studies of turkey gizzard myosin light chain kinase and telokin. Biophys. J. 61:1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arents, G., and E. N. Moudrianakis. 1995. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA. 92:11170–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramakrishnan, V., J. T. Finch, V. Graziano, P. L. Lee, and R. M. Sweet. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 362:219–223. [DOI] [PubMed] [Google Scholar]
- 58.Butler, P. J., and J. O. Thomas. 1980. Changes in chromatin folding in solution. J. Mol. Biol. 140:505–529. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, J. O., and P. J. Butler. 1980. Size-dependence of a stable higher-order structure of chromatin. J. Mol. Biol. 144:89–93. [DOI] [PubMed] [Google Scholar]
- 60.Ausio, J., N. Borochov, D. Seger, and H. Eisenberg. 1984. Interaction of chromatin with NaCl and MgCl2. Solubility and binding studies, transition to and characterization of the higher-order structure. J. Mol. Biol. 177:373–398. [DOI] [PubMed] [Google Scholar]
- 61.Bates, D. L., and J. O. Thomas. 1981. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 9:5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borochov, N., J. Ausió, and H. Eisenberg. 1984. Interaction and conformational changes of chromatin with divalent ions. Nucleic Acids Res. 12:3089–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fletcher, T. M., and J. C. Hansen. 1996. The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr. 6:149–188. [DOI] [PubMed] [Google Scholar]
- 64.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 37:14776–14787. [DOI] [PubMed] [Google Scholar]
- 65.Bavykin, S., L. Srebreva, T. Banchev, R. Tsanev, J. Zlatanova, and A. Mirzabekov. 1993. Histone H1 deposition and histone-DNA interactions in replicating chromatin. Proc. Natl. Acad. Sci. USA. 90:3918–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eirín-López, J. M., L. J. Frehlick, and J. Ausió. 2006. Protamines, in the footsteps of linker histone evolution. J. Biol. Chem. 281:1–4. [DOI] [PubMed] [Google Scholar]
- 67.Eirín-López, J. M., A. M. Gonzalez-Tizon, A. Martinez, and J. Mendez. 2004. Birth-and-death evolution with strong purifying selection in the histone H1 multigene family and the origin of orphon H1 genes. Mol. Biol. Evol. 21:1992–2003. [DOI] [PubMed] [Google Scholar]
- 68.Toth, K. F., J. Mazurkiewicz, and K. Rippe. 2005. Association states of nucleosome assembly protein 1 and its complexes with histones. J. Biol. Chem. 280:15690–15699. [DOI] [PubMed] [Google Scholar]
- 69.Large, E. E., and L. D. Mathies. 2007. Chromatin regulation and sex determination in Caenorhabditis elegans. Trends Genet. 23:314–317. [DOI] [PubMed] [Google Scholar]
- 70.Batova, I., and M. G. O'Rand. 1996. Histone-binding domains in a human nuclear autoantigenic sperm protein. Biol. Reprod. 54:1238–1244. [DOI] [PubMed] [Google Scholar]
- 71.Kleinschmidt, J. A., and A. Seiter. 1988. Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 7:1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang, L., S. S. Loranger, C. Mizzen, S. G. Ernst, C. D. Allis, and A. T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 36:469–480. [DOI] [PubMed] [Google Scholar]
- 74.Mosammaparast, N., K. R. Jackson, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2001. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McBryant, S. J., Y. J. Park, S. M. Abernathy, P. J. Laybourn, J. K. Nyborg, and K. Luger. 2003. Preferential binding of the histone (H3–H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 278:44574–44583. [DOI] [PubMed] [Google Scholar]
- 76.Arnan, C., N. Saperas, C. Prieto, M. Chiva, and J. Ausio. 2003. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 278:31319–31324. [DOI] [PubMed] [Google Scholar]
- 77.Kepert, J. F., J. Mazurkiewicz, G. L. Heuvelman, K. F. Toth, and K. Rippe. 2005. NAP1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J. Biol. Chem. 280:34063–34072. [DOI] [PubMed] [Google Scholar]
- 78.Shintomi, K., M. Iwabuchi, H. Saeki, K. Ura, T. Kishimoto, and K. Ohsumi. 2005. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc. Natl. Acad. Sci. USA. 102:8210–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramos, I., A. Prado, R. M. Finn, A. Muga, and J. Ausio. 2005. Nucleoplasmin-mediated unfolding of chromatin involves the displacement of linker-associated chromatin proteins. Biochemistry. 44:8274–8281. [DOI] [PubMed] [Google Scholar]
- 80.McQuibban, G. A., C. N. Commisso-Cappelli, and P. N. Lewis. 1998. Assembly, remodeling, and histone binding capabilities of yeast nucleosome assembly protein 1. J. Biol. Chem. 273:6582–6590. [DOI] [PubMed] [Google Scholar]
- 81.Vila, R., I. Ponte, M. Collado, J. L. Arrondo, M. A. Jimenez, M. Rico, and P. Suau. 2001. DNA-induced alpha-helical structure in the NH2-terminal domain of histone H1. J. Biol. Chem. 276:46429–46435. [DOI] [PubMed] [Google Scholar]
- 82.Vila, R., I. Ponte, M. Collado, J. L. Arrondo, and P. Suau. 2001. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J. Biol. Chem. 276:30898–30903. [DOI] [PubMed] [Google Scholar]
- 83.Baneres, J. L., A. Martin, and J. Parello. 1997. The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J. Mol. Biol. 273:503–508. [DOI] [PubMed] [Google Scholar]
- 84.Howe, L., M. Iskandar, and J. Ausió. 1998. Folding of chromatin in the presence of heterogeneous histone H1 binding to nucleosomes. J. Biol. Chem. 273:11625–11629. [DOI] [PubMed] [Google Scholar]
- 85.Saperas, N., M. Chiva, Aligu, T. Itoh, C. Katagiri, J. A. Subirana, and J. Ausió. 1999. Physicochemical and functional comparison of Xenopus laevis nucleoplasmin obtained from oocytes and from overexpression in bacteria. Arch. Biochem. Biophys. 361:135–141. [DOI] [PubMed] [Google Scholar]