Abstract
Identification of host factors that interact with pathogens is crucial to an understanding of infectious disease, but direct screening for host mutations to aid in this task is not feasible in mammals. The nematode Caenorhabditis elegans is a genetically tractable alternative for investigating the pathogenic bacterium Pseudomonas aeruginosa. A P. aeruginosa toxin, produced at high cell density under control of the quorum-sensing regulators LasR and RhlR, rapidly and lethally paralyzes C. elegans. Loss-of-function mutations in C. elegans egl-9, a gene required for normal egg laying, confer strong resistance to the paralysis. Thus, activation of EGL-9 or of a pathway that includes it may lead to the paralysis. The molecular identity of egl-9 was determined by transformation rescue and DNA sequencing. A mammalian homologue of EGL-9 is expressed in tissues in which exposure to P. aeruginosa could have clinical effects.
The use of genetic strategies to identify host factors with which pathogens interact has been limited by the fact that traditional disease model hosts, live mammals or their cultured cells, are not amenable to facile genetic analysis. Invertebrate models, such as the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster, offer sophisticated genetic methods but have rarely been used to study infectious disease. Complete sequencing of the genome of C. elegans indicated that at least 36% of the 19,000 predicted proteins have matches in humans (1). Thus, despite the enormous evolutionary distance between humans and nematodes, C. elegans may be a valid model for studies of numerous disease processes.
Pseudomonas aeruginosa is an opportunistic pathogen that infects patients compromised by illness, injury, or inborn genetic defect (2, 3). Hospital-acquired P. aeruginosa pneumonias and septicemias are frequently lethal, and the bacteria infect the lungs of the majority of patients with cystic fibrosis. P. aeruginosa secretes numerous protein virulence factors, including ADP ribosylating enzymes, proteases, and phospholipases, as well as small molecules that include phenazines, rhamnolipid, and cyanide. Production of some of these factors is controlled by one or both of two quorum-sensing systems, LasI/R and RhlI/R, which tie gene expression to high cell density (4).
Recently, Ausubel and colleagues (5–7) described a model for P. aeruginosa pathogenesis that uses C. elegans. Depending on the experimental conditions, strain PA14 kills the nematodes over a period of days (slow killing) or hours (fast killing). The fast killing is mediated by the production of phenazine compounds, which may act through the generation of active oxygen species.
Using a different strain of P. aeruginosa (8), we have identified a third mode by which this species can kill C. elegans. The lethal effect is associated with a rapid neuromuscular paralysis and is caused by the action of one or more diffusible factors whose production requires the Las and Rhl quorum-sensing systems. C. elegans strains carrying loss-of-function mutations in egl-9 are strongly resistant to the paralytic killing. EGL-9 protein, which is expressed in both muscle and neuronal tissues, may thus be a target of the toxic activity.
Materials and Methods
Organisms, Culture Conditions, and Molecular Biological Techniques.
Sources or references for most bacterial strains are shown in Table 1. For lasR complementation assays, PAO-R1 was transformed either with plasmid pPCS1 expressing wild-type lasR (15) or with the vector pUCP18 (16); strain CDP46 is PAO1 transformed with pUCP18. For rhlR complementation, spontaneous rifampicin-resistant derivatives of PAO1 and the rhlR mutant PDO111 (17) were generated to facilitate conjugal plasmid delivery. The strains were then transformed with pMB28, expressing wild-type rhlR (17), or with the vector pLAFR3 (18). For aprA assays, we obtained from A. Lazdunksi (Centre National de la Recherche Scientifique, Marseille, France) an unpublished strain in which the aprA gene was disrupted by insertion of a pUC-family plasmid. The mutation was transduced by using phage F116L (19) into PAO1 to create strain CDP2 and into PAO-B1 to create CDP7. Pseudomonas fluorescens strain SE59, originally isolated by S. Emmons (Albert Einstein College of Medicine, New York), was a gift of R. Hosono (Kanazawa University, Kanazawa, Japan).
Table 1.
Paralysis assays of P. aeruginosa strains
| Strain | Source or reference | Chromosomal genotype | Plasmid | Mean percentage paralyzed | SD | n |
|---|---|---|---|---|---|---|
| Wild-type strains | ||||||
| PAO1 | 8 | WT | None | 95 | 14 | 5,348 |
| 388 | 9 | WT | None | 99 | 0.9 | 508 |
| PA14 | 10 | WT | None | 1.6 | 2.3 | 495 |
| Virulence factor mutants | ||||||
| PAOΔtox | M. Vasil | toxA∷Gm | None | 95 | 1.6 | 468 |
| 388ΔexoS | 11 | ΔexoS∷Tc | None | 100 | 0.3 | 534 |
| PAO-A1 | 12 | lasA∷Tc | None | 99 | 1.2 | 741 |
| PAO-B1 | 12 | lasB∷Ω | None | 100 | 0.0 | 730 |
| PAO-B1A1 | 12 | lasB∷Ω; lasA∷Tc | None | 100 | 0.0 | 624 |
| CDP2 | This study | aprA∷pUC | None | 100 | 0.0 | 430 |
| CDP7 | This study | lasB∷Ω; aprA∷pUC | None | 100 | 0.0 | 430 |
| ΔSRN | 13 | ΔplcSR∷Tc; ΔplcN∷Gm | None | 54 | 50 | 674 |
| Quorum-sensing mutants | ||||||
| PAO-R1 | 14 | ΔlasR∷Tc | None | 0.8 | 1.8 | 1,502 |
| CDP11 | This study | ΔlasR∷Tc | Vector | 0.0 | 0.0 | 247 |
| CDP10 | This study | ΔlasR∷Tc | lasR+ | 67 | 44 | 469 |
| CDP46 | This study | WT | Vector | 72 | 38 | 514 |
| CDP32 | This study | rhlR∷Tn501-11 | Vector | 0.0 | 0.0 | 347 |
| CDP31 | This study | rhlR∷Tn501-11 | rhlR+ | 18 | 3.4 | 322 |
| CDP30 | This study | WT | Vector | 86 | 24 | 307 |
All mutant strains are derived from PAO1 except 388ΔexoS∷Tc, which is derived from strain 388. Each strain was assayed a minimum of three times for paralysis of adult wild-type nematodes. Plasmids are described in Materials and Methods. Gm, gentamycin resistance; Tc, tetracycline resistance; Ω, streptomycin resistance; WT, wild type. M. Vasil is from University of Colorado, Denver, CO.
Bacteria were grown on brain heart infusion agar (BHI, Difco) or L agar (20). Plasmids were maintained in P. aeruginosa in media supplemented with carbenicillin (100 μg/ml; pUCP18 and derivatives) or tetracycline (100 μg/ml; pLAFR3 and derivatives). Triparental mating with the helper plasmid pRK2013 (21) was used to deliver plasmids for rhlR complementation assays; counterselection employed media containing rifampicin (100 μg/ml). Standard molecular biological methods were used (20).
The C. elegans wild-type Bristol strain N2 and its mutant derivatives were used; culture methods, M9 buffer, and genetic techniques have been described (22, 23). Strains were kept at 20°C, except temperature-sensitive mutants, which were grown permissively at 15°C and restrictively at 25°C.
Standard Paralysis Assay.
P. aeruginosa was grown overnight in BHI broth and then diluted 100-fold into fresh broth. BHI agar plates (100-mm diameter) were spread with 400 μl each of the dilution and then incubated at 37°C for 24 h to form lawns of bacteria. Nematodes were washed off stock plates and suspended in a minimal volume of M9 buffer (pH 6.5). Droplets containing nematodes were placed onto the P. aeruginosa lawns, and the plates were then incubated at room temperature (21–23°C); the droplets dried within 30 min and deposited the nematodes on the lawn. In general, the animals were examined after 4 h at approximately ×25 magnification; they were scored as paralyzed if they did not move spontaneously and did not respond detectably to mechanical stimulation. Because pharyngeal pumping is obscured when nematodes are suspended in liquid, inhibition of pumping was observed by transferring animals directly from stock plates to PAO1 lawns with a wire pick.
Filter Paralysis Assay.
Nitrocellulose filters with 0.025-μm pores (Millipore type VSWP) were laid on lawns of PAO1 grown as for the standard assay. Adult nematodes were collected in M9, and 500 μl of suspended animals were placed on the filters with a pipette; during the assay, M9 was added as necessary to keep the nematodes suspended. After an 8-h incubation at room temperature, suspended nematodes were placed on agar plates with a pipette for scoring of paralysis. Just before scoring, a 50-μl aliquot of the liquid was spread on L agar containing chloramphenicol (10 μg/ml) to assay the presence of viable P. aeruginosa (which is naturally chloramphenicol resistant). No colonies were observed, indicating that fewer than approximately 10 P. aeruginosa cells passed through each filter.
Mutagenesis of C. elegans.
Strain N2 was mutagenized with ethyl methanesulfonate (23). F2 progeny were assayed for paralysis essentially as described above, except that P. fluorescens SE59 was used (this strain was the first found to paralyze C. elegans; see below), and the bacteria were grown at 30°C. Survivors were propagated, and the broods were retested for resistance; only one strain per mutagenized parent was preserved. Mutants were shown subsequently to be resistant to P. aeruginosa PAO1. The original isolate of egl-9(sa307) was outcrossed five times to create strain JT307; the original isolate of egl-9(sa330) was outcrossed twice to create strain JT330. Strains carrying egl-9 alleles n586 and n571 were provided by H. R. Horvitz (Massachusetts Institute of Technology, Cambridge, MA).
Transgenic Rescue of egl-9 Mutations.
The mutation lin-15(n765ts), which confers an easily scored multivulva (Muv) phenotype at restrictive temperature (24), was used as the transformation marker. Animals of genotype egl-9(sa330); lin-15(n765ts) were grown at permissive temperature and injected (25) with solutions containing test DNA plus 60 ng/μl of pbLH98, which carries wild-type lin-15 (24). Broods were grown at restrictive temperature; non-Muv transformants were picked; and the resulting transgenic lines were assayed for sensitivity to P. aeruginosa. Gene F22E12.4 was amplified by long PCR (TaqPlus Long, Stratagene) in two overlapping segments from total DNA from a yeast strain bearing the yeast artificial chromosome (YAC) clone Y32F6 (ref. 26; http://genome.wustl.edu/gsc/). One segment was amplified with the primer pair 5′-TAGCTTCGTTCTATCCAATGC and 5′-GGAGGAAGATTCTCATGAGG; the other was amplified with 5′-GACGGAATTTGGACGGAGG and 5′-AGGCAAGTCTTGGGTGAGG. The products were coinjected at 5 ng/μl or 20 ng/μl each.
cDNA Sequencing and Molecular Identification of egl-9 Mutations.
Cycle sequencing with dye terminators was used. cDNA yk130h5 was first sequenced in its entirety. To identify mutations in egl-9 alleles, PCR primers were designed to amplify exons, splice donors, and splice acceptors as predicted by comparing the cDNA to genomic sequence (GenBank accession nos. Z71180 and AL021475); the PCR products were sequenced directly. All point mutations were confirmed by sequencing the complementary strand of DNA. The deletion in sa307 was discovered by sequencing and confirmed by Southern hybridization.
Construction and Analysis of an egl-9∷gfp (Green Fluorescent Protein) Fusion.
YAC Y32F6 DNA was cloned into phage LambdaGEM-12 (Promega). The resultant library was screened by plaque hybridization and PCR, and one phage clone was identified that carried the entire egl-9 sequence. A 13.2-kilobase (kb) BsaHI fragment containing egl-9 as well as 4.6 kb of upstream sequence and 0.7 kb of downstream sequence was subcloned into the 3.6-kb BsaHI fragment of pCM570, a pACYC177 derivative lacking the BamHI site, to generate pCCW1. pCCW1 was then digested with PvuII and ApaI to remove all egl-9 sequences downstream of the second exon and was fused to a 1.9-kb MscI–ApaI fragment of pPD95.69 (A. Fire, S. Xu, J. Aynn, and G. Seydoux, personal communication), which encodes a GFP containing a nuclear localization signal (NLS) and messenger polyadenylation signals. The resulting construct, pCCW5, encodes the first 35 amino acids of EGL-9 fused to NLS-GFP. Transgenic animals were generated as described above with strain MT1642 lin-15(n765ts). pCCW5 and pbLH98 were coinjected at concentrations of 100 ng/μl and 60 ng/μl, respectively, and transgenic animals were isolated at room temperature.
Results
A Diffusible Product of P. aeruginosa Paralyzes C. elegans.
A pseudomonad that contaminated C. elegans cultures was fortuitously observed to exert a toxic effect on the nematodes (S. Emmons, personal communication). The contaminant, SE59, was subsequently identified as a strain of P. fluorescens (P. A. W. Martin, personal communication). After finding that a similar toxicity is produced by several strains of P. aeruginosa (Table 1 and data not shown), we conducted most of our investigations with P. aeruginosa PAO1, a genetically well characterized strain whose genome sequence is essentially complete (http://www.pseudomonas.com/). A P. aeruginosa strain (PA14) that was found by Ausubel and coworkers (5–7) to kill C. elegans under different conditions did not cause the paralysis (Table 1).
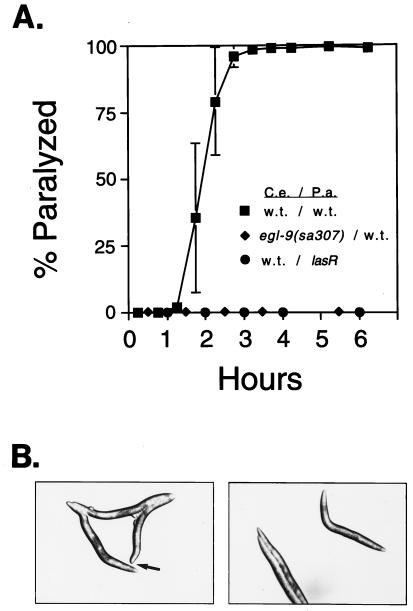
Nematodes grown on lawns of Escherichia coli feed by a rapid contraction of pharyngeal muscles that pump bacteria into the intestine. When the animals are placed on a dense lawn of PAO1, pumping becomes sporadic within seconds and ceases altogether within minutes. The nematodes also cease defecation and egg laying. Locomotion, either spontaneous or induced by touch or other disturbance, becomes progressively sluggish. The reduction in locomotion is sometimes accompanied by spasmodic twitching of the animals. After 4 h on the lawn, almost all animals are completely paralyzed (Fig. 1A). The fully paralyzed animals seem to be dead, because transfer to E. coli fails to restore movement. In many paralyzed nematodes, the terminal body posture is kinked, and in some, the nose becomes blunt (Fig. 1B). These features indicate aberrant hypercontraction of body-wall muscles.
Figure 1.
(A) Time course of paralysis of C. elegans by P. aeruginosa. Error bars show SDs of three independent trials. w.t., wild type. (B) Postures of paralyzed nematodes. Arrow indicates a hypercontracted nose.
The speed of the lethal effect suggested that death is caused by a diffusible bacterial activity rather than by an infection. We tested whether contact between nematodes and bacteria is required for paralysis by overlaying lawns of PAO1 with filters preventing bacterial passage and then suspending C. elegans in buffer on the filters (see Materials and Methods). In two trials, an aggregate of 99.6% of the nematodes was killed (n = 261), indicating that a diffusible activity is sufficient for paralysis.
Paralysis Requires the Quorum Sensors LasR and RhlR.
We first examined whether known P. aeruginosa virulence genes play a role in nematode pathogenesis. Paralysis was produced, usually at wild-type levels, by mutants defective in the production of exotoxin A (encoded by the toxA gene), exoenzyme S (exoS), staphylolytic protease (lasA), elastase (lasB), alkaline protease (aprA), hemolytic phospholipase C (plcS), and nonhemolytic phospholipase C (plcN; Table 1). Thus, none of these secreted proteins seems to be required for the paralytic killing.
We next examined mutants defective for the LasR or RhlR quorum-sensing proteins and found that both regulators are required for toxin production (Table 1 and Fig. 1A). Mutation in either gene abolished paralysis, and each defect was at least partially complemented by a plasmid carrying the corresponding wild-type gene. Among the known protein targets of LasR regulation are elastase (14), staphylolytic protease (27), and alkaline protease (28), but as shown above, none of these is required for paralysis. The roles of small molecules regulated by RhlR, including cyanide, pyocyanin, and rhamnolipid (17, 29, 30), require further study.
Isolation and Molecular Identification of Nematode Mutants Resistant to Paralysis.
We screened 8,000 mutagenized C. elegans genomes for resistance to paralysis and isolated two resistant strains carrying recessive mutations (alleles sa307 and sa330) affecting a single complementation group (Table 2 and Fig. 1A; see Materials and Methods). Further mapping and complementation studies indicated that each is an allele of egl-9, which was previously identified on the basis of an egg-laying-defective (Egl) phenotype (ref. 31 and data not shown). The two alleles of egl-9 isolated previously, n586 and n571, also confer resistance to P. aeruginosa paralysis (Table 2). However, of the recently identified alleles, only sa307 confers a visible Egl phenotype. Animals carrying the three alleles that confer an Egl phenotype are also slightly scrawny and tend to clump into clusters on lawns of E. coli food bacteria (32).
Table 2.
Resistance to paralysis of C. elegans egl-9 mutants
| Strain | Genotype | Mean percentage paralyzed | SD | n |
|---|---|---|---|---|
| JT307 | egl-9(sa307) | 0.0 | 0.0 | 336 |
| JT330 | egl-9(sa330) | 0.0 | 0.0 | 476 |
| MT1201 | egl-9(n571) | 2.1 | 3.6 | 193 |
| MT1216 | egl-9(n586) | 0.2 | 0.4 | 646 |
| N2 | WT | 100 | 0.3 | 589 |
Synchronously growing adult nematodes were assayed in three independent trials. WT, wild type.
We established the molecular identity of egl-9 by transformation rescue (see Materials and Methods). DNA from the YAC clone Y32F6 rescued sensitivity to paralysis of sa330 mutants, but no rescue was obtained with cosmid clones overlapping the YAC (data not shown). A gene not represented in the cosmid physical map of C. elegans, F22E12.4 (GenBank accession no. Z71180), was amplified in overlapping segments by PCR. This DNA restored sa330 animals to P. aeruginosa sensitivity in three independent transgenic lines (Table 3), suggesting that the overlapping DNAs recombined in vivo (25) to produce a functional wild-type egl-9 transgene.
Table 3.
Rescue of egl-9 by gene F22E12.4
| Strain | Genotype | Percentage paralyzed | n |
|---|---|---|---|
| N2 | WT | 95 | 851 |
| MT1642 | lin-15(n765ts) | 93 | 468 |
| JT330 | egl-9(sa330) | 2.8 | 432 |
| JT9592 | egl-9(sa330); lin-15(n765ts) | 1.7 | 468 |
| JT10023 | egl-9(sa330); lin-15(n765ts); saEx433 | 100 | 24 |
| JT10024 | egl-9(sa330); lin-15(n765ts); saEx434 | 89 | 89 |
| JT10025 | egl-9(sa330); lin-15(n765ts); saEx435 | 100 | 117 |
Aggregate paralysis in two to five trials is shown. saEx433, saEX434, and saEx435 are independently generated extrachromosomal arrays containing lin-15(+) and PCR-amplified gene F22E12.4. WT, wild type.
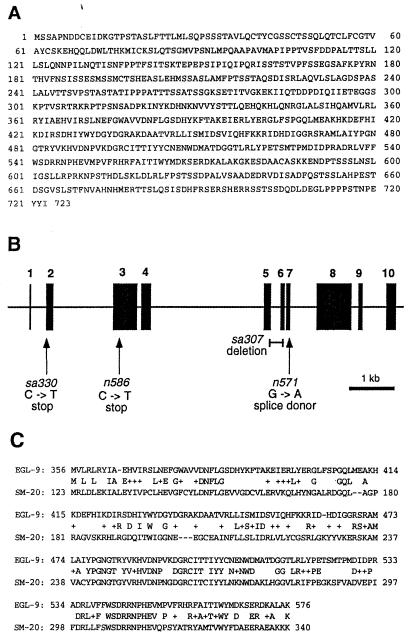
The sequence of F22E12.4 corresponds to cDNA clone yk130h5 (GenBank accession no. D68399), which we sequenced to predict a primary amino acid sequence for EGL-9 (Fig. 2A). Genomic sequencing of egl-9 mutants confirmed the existence of mutations in F22E12.4 in all four alleles (Fig. 2B). sa307 is a 243-bp deletion (nucleotides 2,775 through 3,017; GenBank accession no. AL021475). n586 is a C-to-T mutation (nucleotide 16,172; GenBank accession no. Z71180) that creates a nonsense codon at amino acid 131 of the predicted protein sequence. n571 is a G-to-A mutation (nucleotide 3,187; GenBank accession no. AL021475) that alters a splice donor site. sa330 is a C-to-T mutation (nucleotide 14,566; GenBank accession no. Z71180) that creates a nonsense codon at amino acid 38.
Figure 2.
(A) Amino acid sequence of EGL-9 as predicted by cDNA yk130h5. (B) Structure of egl-9 gene. Dark boxes indicate exons; arrows show locations of point mutations; the bar shows genomic DNA deleted in allele sa307. (C) Alignment of homologous regions of EGL-9 and rat SM-20 (GenBank accession no. U06713).
The protein predicted by the cDNA contains 723 amino acids, with an unmodified molecular mass of 80 kDa and an abundance of serine and threonine residues. Database searches with the blastp algorithm (33) did not identify similarities to proteins of known function. However, a 221-amino acid portion of EGL-9 is 42% identical to the protein corresponding to rat cDNA SM-20 (Fig. 2C), which was isolated from cultured smooth muscle cells in a differential-expression screen (34). Although the function of SM-20 is unknown, the protein is widely expressed in rat muscle tissue (including skeletal muscle and lung vascular and bronchiolar smooth muscle) and is also found in bronchial epithelium (35). EGL-9 also contains a zinc finger-like motif near its amino terminus (D. D'Argenio, personal communication); it has no apparent signal sequence or membrane-spanning sequences. In searches of expressed sequence tag databases, numerous uncharacterized human clones were found that are similar to EGL-9, indicating that EGL-9 has human homologues.
Expression of an egl-9∷gfp Fusion.
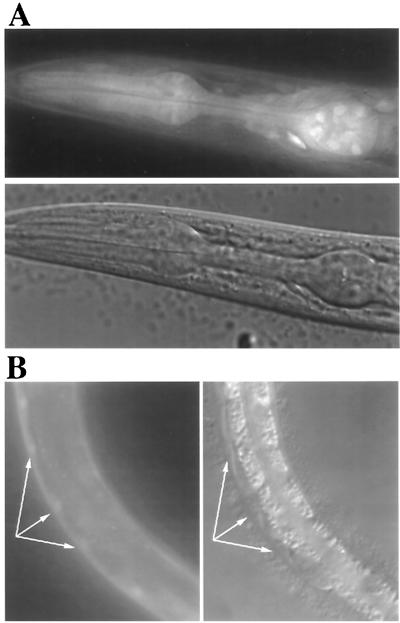
We examined the expression pattern of egl-9 with a gfp reporter gene fusion (see Materials and Methods). A DNA fragment containing the first and second exons of egl-9, along with 4.6 kb of upstream sequence, was fused to a gfp construct encoding a nuclear localization signal. Transgenic animals were generated, and two independent lines were examined for GFP expression. In both L1 and adult hermaphrodites, we observed a complex expression pattern that included significant GFP fluorescence in pharyngeal muscle (Fig. 3A) and body wall muscle (Fig. 3B), as well as in vulval hypodermis, the gonadal distal tip cells, and several sensory neurons and interneurons of the head and tail (data not shown). The gfp analyzed here is fused to the second exon of egl-9, and we cannot exclude the possibility that egl-9 expression is more widespread than we have detected because, for example, of the production of alternative mRNAs.
Figure 3.
(A) Digital epifluorescence and Nomarski images of egl-9∷gfp expression in the pharyngeal muscles. Nuclear localization is incomplete, resulting in fluorescence of entire cells. The epifluorescence image has been deconvoluted with deltavision 2.1 software to eliminate light scattering from other focal planes. Fluorescence from several anterior ganglion neurons can also been seen. (B) Epifluorescence and Nomarski 35-mm images of egl-9∷gfp expression in body wall muscle. Arrows indicate muscle cell nuclei.
Discussion
This report describes a pathogenesis model in which C. elegans is rapidly killed by the bacterium P. aeruginosa. The killing is preceded by a rapid inhibition of pharyngeal pumping, sluggish locomotion, and hypercontraction of body wall muscles, indicating that the toxic activity targets a neuromuscular function.
Our results indicate that paralysis is caused by a diffusible activity that is under regulation of two quorum-sensing systems, LasR and RhlR. By assaying mutant bacteria, we eliminated as candidates for the toxic activity the proteins exotoxin A, exoenzyme S, elastase, staphylolytic protease, alkaline protease, hemolytic phospholipase C, and nonhemolytic phospholipase C. Further study with different genetic and biochemical approaches is required to identify the paralytic activity.
The major goal of our work was to establish that informative mutations in the nematode host could be obtained and analyzed to help dissect the paralytic phenomenon. Our screen of 8,000 mutagenized genomes was modest by C. elegans standards, but it produced two recessive mutations that confer essentially complete resistance to paralysis. The mutations causing resistance to paralysis were found to affect egl-9, a gene required for normal egg laying (31). The molecular identify of egl-9 was determined, but analysis of the sequence of the gene has not suggested a specific function for the encoded protein. However, the data are consistent with a role for EGL-9 as a nonessential modulator of muscle contraction in wild-type animals. All of the egl-9 alleles contain apparent loss-of-function mutations: one is an internal deletion; one is predicted to cause a splicing defect; and two are nonsense mutations. Three of the mutations result in a moderately decreased frequency of egg laying compared with the frequency in wild-type animals. The fourth mutation (in sa330) creates an early nonsense codon, but animals bearing this allele are non-Egl. The apparent partial function of the mutant gene could be explained in several ways, including the production of alternative transcripts in different tissues.
The genetics of egl-9 indicate that the paralytic toxin is unlikely to act by blocking EGL-9 function, because in that case, loss-of-function mutations would not confer resistance. We therefore hypothesize that the toxin aberrantly activates muscle contraction by acting either directly on EGL-9 or on a pathway that includes it. It is also possible that EGL-9 is required for transport or activation of the toxin. These models are supported by our finding that an egl-9∷gfp fusion is expressed in both pharyngeal and body wall muscle, as well as in neuronal tissues.
In parallel to our investigations, Ausubel and colleagues (5–7) developed a C. elegans model for P. aeruginosa pathogenesis by using PA14, a different strain of the bacterium. Two modes of killing, slow and fast, were described. Neither mode seems to be the same as the egl-9-dependent paralysis we describe. Slow killing by PA14 requires days, not hours, and egl-9 mutants are not resistant to this effect (F. Ausubel, personal communication). Fast killing by PA14, although similar in its kinetics to paralysis by PAO1, also seems to be a different phenomenon, for three reasons. First, paralysis by PAO1 is LasR dependent (Table 1), but fast killing by PA14 is not (7); second, egl-9 mutants are not resistant to fast killing (F. Ausubel, personal communication); and third, PA14 does not produce egl-9-dependent paralysis when assayed under the conditions we describe (Table 1). Thus, it seems that there are at least three separate modes by which P. aeruginosa can kill C. elegans.
Phenazines, a large class of molecules produced by a variety of pseudomonads (36, 37), are implicated in fast killing (5). The role of phenazines in paralysis by PAO1, if any, has not been determined, and it is possible that distinct phenazines are responsible for fast killing by PA14 and paralytic killing by PAO1. Intriguingly, the sputum of cystic fibrosis patients has been shown to contain two phenazines (pyocyanin and 1-hydroxyphenazine) that induce bronchoconstriction in sheep (38). Pyocyanin also inhibits nitric oxide-dependent vasodilation (39, 40). Thus, constriction (or inhibition of relaxation) of muscle by P. aeruginosa products has been shown in mammals, suggesting that the hypercontractile paralysis we describe in C. elegans may be relevant to human infections. Furthermore, a homologue of EGL-9 (SM-20) is expressed in rat vascular and bronchiolar smooth muscle, as well as in bronchial epithelium (35). The mammalian protein is thus expressed in tissues that are in close proximity to P. aeruginosa during pulmonary infections.
Our results indicate the feasibility of using a genetically tractable model organism to screen directly for potential host targets of a bacterial pathogen. The identification of such host factors is a key step in delineating the mechanisms by which bacteria can cause disease.
Acknowledgments
We are indebted to S. Emmons for communicating his unpublished discovery of paralysis of C. elegans. We thank F. Ausubel, M.-W. Tan, R. Baran, A. Davies, J. Rand, and the Caenorhabditis elegans Sequencing Consortium for sharing data before publication. We thank S. Lory for encouragement and advice, E. P. Greenberg for suggesting the lasR mutant tests, and many of our colleagues for comments on the manuscript. We are grateful to P. Navas for YAC Y32F6 DNA. For strains and clones, we thank S. Chissoe, A. Coulson, A. Fire, D. Frank, H. R. Horvitz, Y. Kohara, B. Iglewski, A. Lazdunski, S. Lory, D. Ohman, P. Okkema, L. Passador, P. Sternberg, M. Vasil, and the Caenorhabditis Genetics Center. C.M. was funded by the Royalty Research Fund of the University of Washington and a Pilot Project Grant from the Cystic Fibrosis Foundation. C.D. was supported by a National Science Foundation predoctoral fellowship. C.L.C. was supported by a National Research Service Award postdoctoral fellowship from the National Institutes of Health.
Abbreviations
- YAC
yeast artificial chromosome
- kb
kilobase
- GFP
green fluorescent protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF178536).
References
- 1.Caenorhabditis elegans Sequencing Consortium. Science. 1998;282:2012–2018. [Google Scholar]
- 2.Campra M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an Opportunistic Pathogen. New York: Plenum; 1993. [Google Scholar]
- 3.Pollack M. In: Infectious Diseases. 2nd Ed. Gorbach S L, Bartlett J G, Blacklow N R, editors. Philadelphia: Saunders; 1998. pp. 1824–1837. [Google Scholar]
- 4.Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 5.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 6.Tan M-W, Mahajan-Miklos S, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan M-W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holloway B W, Krishnapillai V, Morgan A F. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iglewski B H, Sadoff J, Bjorn M J, Maxwell E S. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 11.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 12.Toder D S, Ferrell S J, Nezezon J L, Rust L, Iglewski B H. Infect Immun. 1994;62:1320–1327. doi: 10.1128/iai.62.4.1320-1327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shortridge V D, Lazdunski A, Vasil M L. Mol Microbiol. 1992;6:863–871. doi: 10.1111/j.1365-2958.1992.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 14.Gambello M J, Iglewski B H. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seed P C, Passador L, Iglewski B H. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweizer H P. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 17.Brint J M, Ohman D E. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 19.Krishnapillai V. Mol Gen Genet. 1971;114:134–143. doi: 10.1007/BF00332784. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Figursky D H, Helinski D R. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood W B, editor. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 24.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello C, Fire A. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. New York: Academic; 1995. pp. 451–482. [Google Scholar]
- 26.Coulson A, Waterston R, Kiff J, Sulston J, Kohara Y. Nature (London) 1988;335:184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- 27.Toder D S, Gambello M J, Iglewski B H. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 28.Gambello M J, Kaye S, Iglewski B H. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsner U A, Koch A K, Fiechter A, Reiser J. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 31.Trent C, Tsung N, Horvitz H R. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas J H, Birnby D A, Vowels J J. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wax S D, Rosenfield C-L, Taubman M B. J Biol Chem. 1994;269:13041–13047. [PubMed] [Google Scholar]
- 35.Wax S D, Tsao L, Lieb M E, Fallon J T, Taubman M B. Lab Invest. 1996;74:797–808. [PubMed] [Google Scholar]
- 36.Leisinger T, Margraff R. Microbiol Rev. 1979;43:422–442. doi: 10.1128/mr.43.3.422-442.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budzikiewicz H. FEMS Microbiol Rev. 1993;10:209–228. doi: 10.1111/j.1574-6968.1993.tb05868.x. [DOI] [PubMed] [Google Scholar]
- 38.Forteza R, Lauredo I T, Burch R, Abraham W M. Am J Respir Crit Care Med. 1994;149:687–693. doi: 10.1164/ajrccm.149.3.8118638. [DOI] [PubMed] [Google Scholar]
- 39.Gryglewski R J, Zembowicz A, Salvemini D, Taylor G W, Vane J R. Br J Pharmacol. 1992;106:838–845. doi: 10.1111/j.1476-5381.1992.tb14422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bozinovski J, Brien J F, Marks G S, Nakatsu K. Can J Physiol Pharmacol. 1994;72:746–752. doi: 10.1139/y94-106. [DOI] [PubMed] [Google Scholar]