Abstract
The S1 gene nucleotide sequences of 10 type 3 (T3) reovirus strains were determined and compared with the T3 prototype Dearing strain in order to study sequence diversity in strains of a single reovirus serotype and to learn more about structure-function relationships of the two S1 translation products, sigma 1 and sigma 1s. Analysis of phylogenetic trees constructed from variation in the sigma 1-encoding S1 nucleotide sequences indicated that there is no pattern of S1 gene relatedness in these strains based on host species, geographic site, or date of isolation. This suggests that reovirus strains are transmitted rapidly between host species and that T3 strains with markedly different S1 sequences circulate simultaneously. Comparison of the deduced sigma 1 amino acid sequences of the 11 T3 strains was notable for the identification of conserved and variable regions of sequence that correlate with the proposed domain organization of sigma 1 (M.L. Nibert, T.S. Dermody, and B. N. Fields, J. Virol. 64:2976-2989, 1990). Repeat patterns of apolar residues thought to be important for sigma 1 structure were conserved in all strains examined. The deduced sigma 1s amino acid sequences of the strains were more heterogeneous than the sigma 1 sequences; however, a cluster of basic residues near the amino terminus of sigma 1s was conserved. This analysis has allowed us to investigate molecular epidemiology of T3 reovirus strains and to identify conserved and variable sequence motifs in the S1 translation products, sigma 1 or sigma 1s.
Full text
PDF


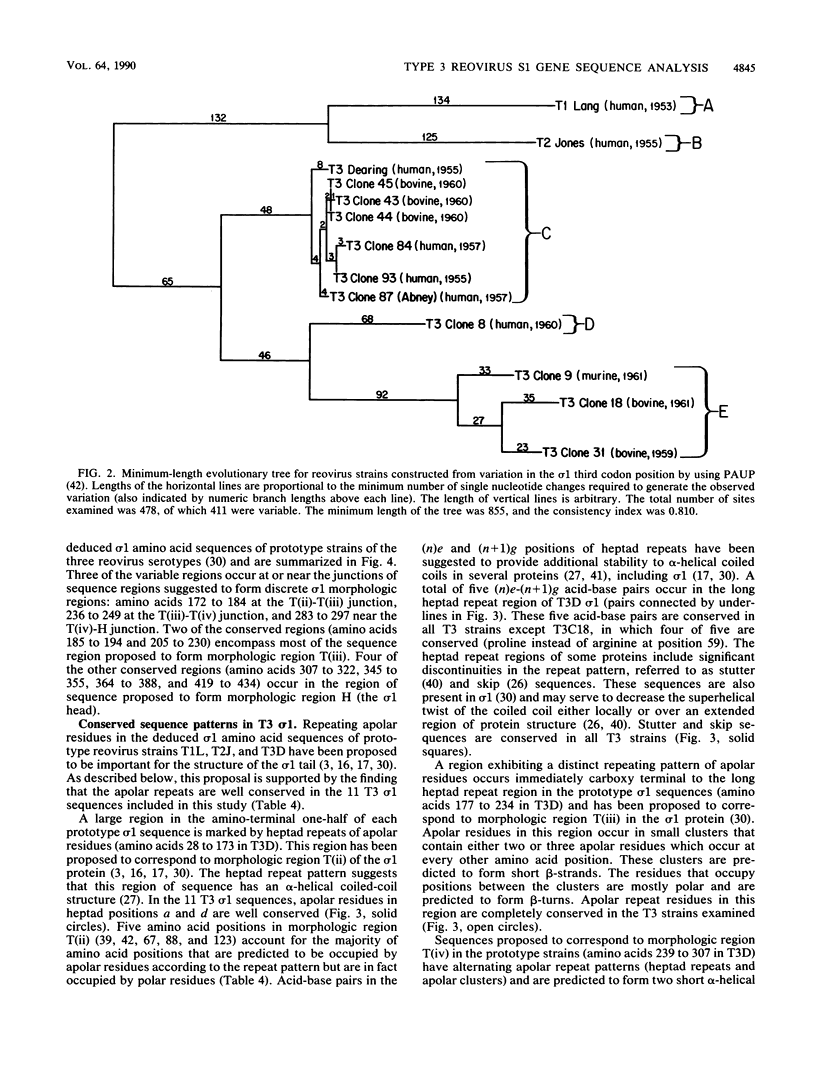




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmüller A., Fitch W. M., Scholtissek C. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol. 1989 Aug;70(Pt 8):2111–2119. doi: 10.1099/0022-1317-70-8-2111. [DOI] [PubMed] [Google Scholar]
- Banerjea A. C., Brechling K. A., Ray C. A., Erikson H., Pickup D. J., Joklik W. K. High-level synthesis of biologically active reovirus protein sigma 1 in a mammalian expression vector system. Virology. 1988 Dec;167(2):601–612. [PubMed] [Google Scholar]
- Bassel-Duby R., Jayasuriya A., Chatterjee D., Sonenberg N., Maizel J. V., Jr, Fields B. N. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. 1985 May 30-Jun 5Nature. 315(6018):421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R., Spriggs D. R., Tyler K. L., Fields B. N. Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J Virol. 1986 Oct;60(1):64–67. doi: 10.1128/jvi.60.1.64-67.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Fitch W. M., Palese P. Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology. 1986 Aug;153(1):12–21. doi: 10.1016/0042-6822(86)90003-6. [DOI] [PubMed] [Google Scholar]
- Burstin S. J., Spriggs D. R., Fields B. N. Evidence for functional domains on the reovirus type 3 hemagglutinin. Virology. 1982 Feb;117(1):146–155. doi: 10.1016/0042-6822(82)90514-1. [DOI] [PubMed] [Google Scholar]
- Cashdollar L. W., Chmelo R. A., Wiener J. R., Joklik W. K. Sequences of the S1 genes of the three serotypes of reovirus. Proc Natl Acad Sci U S A. 1985 Jan;82(1):24–28. doi: 10.1073/pnas.82.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody T. S., Nibert M. L., Bassel-Duby R., Fields B. N. A sigma 1 region important for hemagglutination by serotype 3 reovirus strains. J Virol. 1990 Oct;64(10):5173–5176. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Nakajima K., Alfino P., Pedersen F. S., Haseltine W. A., Hannoun C., Palese P. Biochemical evidence that "new" influenza virus strains in nature may arise by recombination (reassortment). Proc Natl Acad Sci U S A. 1978 Jul;75(7):3341–3345. doi: 10.1073/pnas.75.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R., Horne D., Cashdollar L. W., Joklik W. K., Lee P. W. Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology. 1990 Feb;174(2):399–409. doi: 10.1016/0042-6822(90)90093-7. [DOI] [PubMed] [Google Scholar]
- Ernst H., Shatkin A. J. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc Natl Acad Sci U S A. 1985 Jan;82(1):48–52. doi: 10.1073/pnas.82.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner D. V., Jurka J. Multiple aligned sequence editor (MASE). Trends Biochem Sci. 1988 Aug;13(8):321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- Fraser R. D., Furlong D. B., Trus B. L., Nibert M. L., Fields B. N., Steven A. C. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol. 1990 Jun;64(6):2990–3000. doi: 10.1128/jvi.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong D. B., Nibert M. L., Fields B. N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988 Jan;62(1):246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch J. R., Fields B. N. Genetic diversity in natural populations of mammalian reoviruses: tryptic peptide analysis of outer capsid polypeptides of murine, bovine, and human type 1 and 3 reovirus strains. J Virol. 1984 Mar;49(3):641–651. doi: 10.1128/jvi.49.3.641-651.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy D. B., Rosen L., Fields B. N. Polymorphism of the migration of double-stranded RNA genome segments of reovirus isolates from humans, cattle, and mice. J Virol. 1979 Jul;31(1):104–111. doi: 10.1128/jvi.31.1.104-111.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy D. B., Rubin D. H., Fields B. N. Molecular basis of reovirus neurovirulence: role of the M2 gene in avirulence. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1298–1302. doi: 10.1073/pnas.79.4.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs B. L., Samuel C. E. Biosynthesis of reovirus-specified polypeptides: the reovirus s1 mRNA encodes two primary translation products. Virology. 1985 May;143(1):63–74. doi: 10.1016/0042-6822(85)90097-2. [DOI] [PubMed] [Google Scholar]
- Jayasuriya A. K., Nibert M. L., Fields B. N. Complete nucleotide sequence of the M2 gene segment of reovirus type 3 dearing and analysis of its protein product mu 1. Virology. 1988 Apr;163(2):591–602. doi: 10.1016/0042-6822(88)90300-5. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss S., Webster R. G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989 Nov;63(11):4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Studies on the origin of pandemic influenza. 3. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973 Feb;51(2):383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- Lee P. W., Hayes E. C., Joklik W. K. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981 Jan 15;108(1):156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J Mol Biol. 1983 Mar 15;164(4):605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975 Oct 25;98(2):293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Mah D. C., Lee P. W. Molecular cloning and sequencing of the reovirus (serotype 3) S1 gene which encodes the viral cell attachment protein sigma 1. Nucleic Acids Res. 1984 Nov 26;12(22):8699–8710. doi: 10.1093/nar/12.22.8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Pon R. T., Lee P. W. Analysis of functional domains on reovirus cell attachment protein sigma 1 using cloned S1 gene deletion mutants. Virology. 1987 Sep;160(1):162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- Nibert M. L., Dermody T. S., Fields B. N. Structure of the reovirus cell-attachment protein: a model for the domain organization of sigma 1. J Virol. 1990 Jun;64(6):2976–2989. doi: 10.1128/jvi.64.6.2976-2989.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera T., Toniolo A., Ray U. R., Jenson A. B., Knazek R. A., Notkins A. L. Virus-induced diabetes mellitus. XX. Polyendocrinopathy and autoimmunity. J Exp Med. 1981 Jun 1;153(6):1457–1473. doi: 10.1084/jem.153.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R. W., Lee P. W. Glycophorin is the reovirus receptor on human erythrocytes. Virology. 1987 Jul;159(1):94–101. doi: 10.1016/0042-6822(87)90351-5. [DOI] [PubMed] [Google Scholar]
- ROSEN L., ABINANTI F. R., HOVIS J. F. Further observations on the natural infection of cattle with reoviruses. Am J Hyg. 1963 Jan;77:38–48. doi: 10.1093/oxfordjournals.aje.a120294. [DOI] [PubMed] [Google Scholar]
- ROSEN L., HOVIS J. F., MASTROTA F. M., BELL J. A., HUEBNER R. J. Observations on a newly recognized virus (Abney) of the reovirus family. Am J Hyg. 1960 Mar;71:258–265. doi: 10.1093/oxfordjournals.aje.a120109. [DOI] [PubMed] [Google Scholar]
- Ramig R. F., Cross R. K., Fields B. N. Genome RNAs and polypeptides of reovirus serotypes 1, 2, and 3. J Virol. 1977 Jun;22(3):726–733. doi: 10.1128/jvi.22.3.726-733.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Pelletier J., Bassel-Duby R., Jayasuriya A., Fields B. N., Sonenberg N. Identification of a new polypeptide coded by reovirus gene S1. J Virol. 1985 Jun;54(3):720–725. doi: 10.1128/jvi.54.3.720-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs D. R., Bronson R. T., Fields B. N. Hemagglutinin variants of reovirus type 3 have altered central nervous system tropism. Science. 1983 Apr 29;220(4596):505–507. doi: 10.1126/science.6301010. [DOI] [PubMed] [Google Scholar]
- Spriggs D. R., Fields B. N. Attenuated reovirus type 3 strains generated by selection of haemagglutinin antigenic variants. Nature. 1982 May 6;297(5861):68–70. doi: 10.1038/297068a0. [DOI] [PubMed] [Google Scholar]
- Steinert P. M., Steven A. C., Roop D. R. The molecular biology of intermediate filaments. Cell. 1985 Sep;42(2):411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- Steven A. C., Trus B. L., Maizel J. V., Unser M., Parry D. A., Wall J. S., Hainfeld J. F., Studier F. W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988 Mar 20;200(2):351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- Tyler K. L., McPhee D. A., Fields B. N. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science. 1986 Aug 15;233(4765):770–774. doi: 10.1126/science.3016895. [DOI] [PubMed] [Google Scholar]
- Tyler K. L., Virgin H. W., 4th, Bassel-Duby R., Fields B. N. Antibody inhibits defined stages in the pathogenesis of reovirus serotype 3 infection of the central nervous system. J Exp Med. 1989 Sep 1;170(3):887–900. doi: 10.1084/jem.170.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Ault K. A., Fields B. N. Interaction of reovirus with cell surface receptors. I. Murine and human lymphocytes have a receptor for the hemagglutinin of reovirus type 3. J Immunol. 1980 May;124(5):2143–2148. [PubMed] [Google Scholar]
- Weiner H. L., Drayna D., Averill D. R., Jr, Fields B. N. Molecular basis of reovirus virulence: role of the S1 gene. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5744–5748. doi: 10.1073/pnas.74.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Fields B. N. Neutralization of reovirus: the gene responsible for the neutralization antigen. J Exp Med. 1977 Nov 1;146(5):1305–1310. doi: 10.1084/jem.146.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Ramig R. F., Mustoe T. A., Fields B. N. Identification of the gene coding for the hemagglutinin of reovirus. Virology. 1978 May 15;86(2):581–584. doi: 10.1016/0042-6822(78)90099-5. [DOI] [PubMed] [Google Scholar]
- Wenske E. A., Chanock S. J., Krata L., Fields B. N. Genetic reassortment of mammalian reoviruses in mice. J Virol. 1985 Nov;56(2):613–616. doi: 10.1128/jvi.56.2.613-616.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener J. R., Joklik W. K. Comparison of the reovirus serotype 1, 2, and 3 S3 genome segments encoding the nonstructural protein sigma NS. Virology. 1987 Dec;161(2):332–339. doi: 10.1016/0042-6822(87)90125-5. [DOI] [PubMed] [Google Scholar]
- Wiener J. R., Joklik W. K. Evolution of reovirus genes: a comparison of serotype 1, 2, and 3 M2 genome segments, which encode the major structural capsid protein mu 1C. Virology. 1988 Apr;163(2):603–613. doi: 10.1016/0042-6822(88)90301-7. [DOI] [PubMed] [Google Scholar]
- Wiener J. R., Joklik W. K. The sequences of the reovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology. 1989 Mar;169(1):194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- Williams W. V., Guy H. R., Rubin D. H., Robey F., Myers J. N., Kieber-Emmons T., Weiner D. B., Greene M. I. Sequences of the cell-attachment sites of reovirus type 3 and its anti-idiotypic/antireceptor antibody: modeling of their three-dimensional structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6488–6492. doi: 10.1073/pnas.85.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Krystal M., Fitch W. M., Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988 Mar;163(1):112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- Yeung M. C., Gill M. J., Alibhai S. S., Shahrabadi M. S., Lee P. W. Purification and characterization of the reovirus cell attachment protein sigma 1. Virology. 1987 Feb;156(2):377–385. doi: 10.1016/0042-6822(87)90417-x. [DOI] [PubMed] [Google Scholar]
- Yeung M. C., Lim D., Duncan R., Shahrabadi M. S., Cashdollar L. W., Lee P. W. The cell attachment proteins of type 1 and type 3 reovirus are differentially susceptible to trypsin and chymotrypsin. Virology. 1989 May;170(1):62–70. doi: 10.1016/0042-6822(89)90352-8. [DOI] [PubMed] [Google Scholar]
- Young J. F., Palese P. Evolution of human influenza A viruses in nature: recombination contributes to genetic variation of H1N1 strains. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6547–6551. doi: 10.1073/pnas.76.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]