Abstract
The pervasive role of circadian clocks in regulating physiology and behavior is widely recognized. Their adaptive value is their ability to be entrained by environmental cues such that the internal circadian phase is a reliable predictor of solar time. In mammals, both light and nonphotic behavioral cues can entrain the principal oscillator of the hypothalamic suprachiasmatic nuclei (SCN). However, although light can advance or delay the clock during circadian night, behavioral events trigger phase advances during the subjective day, when the clock is insensitive to light. The recent identification of Period (Per) genes in mammals, homologues of dperiod, which encodes a core element of the circadian clockwork in Drosophila, now provides the opportunity to explain circadian timing and entrainment at a molecular level. In mice, expression of mPer1 and mPer2 in the SCN is rhythmic and acutely up-regulated by light. Moreover, the temporal relations between mRNA and protein cycles are consistent with a clock based on a transcriptional/translational feedback loop. Here we describe circadian oscillations of Per1 and Per2 in the SCN of the Syrian hamster, showing that PER1 protein and mRNA cycles again behave in a manner consistent with a negative-feedback oscillator. Furthermore, we demonstrate that nonphotic resetting has the opposite effect to light: acutely down-regulating these genes. Their sensitivity to nonphotic resetting cues supports their proposed role as core elements of the circadian oscillator. Moreover, this study provides an explanation at the molecular level for the contrasting but convergent effects of photic and nonphotic cues on the clock.
Circadian regulation of behavior, metabolism, and physiology is a conserved feature across eukaryotic and some prokaryotic taxa (1–3). In mammals, the principal circadian clock is located within the suprachiasmatic nuclei (SCN) of the hypothalamus (4). The circadian signal generated intrinsically by the SCN is sensitive to light–dark cycles conveyed by direct retinal afferents; thereby, internal time is synchronized (entrained) to solar time. The cell-autonomous nature of the SCN oscillator is demonstrated by the sustained free-running circadian rhythms of electrical firing in individual neurons held in dissociated SCN cultures (5). However, until recently, the molecular elements of the mammalian clockwork were unknown.
In Drosophila, the circadian clock can be explained by a negative transcriptional/translational feedback loop in which the protein products of the clock genes, dperiod and dtimeless, enter the nucleus to suppress the expression of their cognate genes (3). Three mammalian homologues to the fly period gene have now been identified, and a number of lines of evidence indicate that they are critical elements of the core circadian oscillator of the SCN. First, mPer1, mPer2, and mPer3 exhibit spontaneous circadian cycling in the SCN; the first two of these genes are acutely sensitive to light (6–10). Furthermore, at least in the case of mPer1, mRNA and protein levels rise and fall in series with a lag of about 6 hr, consistent with an autoregulatory feedback oscillator (11); light-induced resetting in vivo and glutamate-induced resetting in vitro can be blocked by local infusion of the SCN with antisense oligonucleotides to mPer1 (12). Moreover, genetic mutation of mPer2 ablates free-running circadian rhythmicity in mice (13). Finally, the encoded proteins negatively regulate CLOCK/BMAL-dependent transcription (14, 15). This negative action of mPER on the CLOCK/BMAL-dependent drive to mPer1 and possibly other mPer genes closes the circadian loop, establishing a self-sustaining oscillation in which CLOCK/BMAL-dependent transcriptional activation of mPer genes is periodically suppressed by temporarily increased abundance of their encoded proteins. The rhythmic expression of mPER proteins is also thought to constitute the output of the oscillator, leading to the circadian patterns of expression of clock-controlled genes, such as that encoding the SCN neuropeptide arginine vasopressin (15).
Among the important variables controlled by the clock is the state of rest or activity of the animal. However, it is also known that entirely nonphotic events, such as activity or the associated arousal can feedback and influence the clock (16, 17). Resetting by behavior is a rapid event; the SCN clock adopts a new phase within 1 to 2 hr of presentation of the stimulus (18, 19). Such resetting is mediated by a pathway very different from the retinal glutamatergic system, a pathway dependent primarily on neuropeptide-Y innervation of the SCN from the thalamus, and possibly also serotonergic innervation from the mid-brain (4, 16, 17). Given this sensitivity of the SCN clock to nonphotic resetting, it has been predicted that if mPer genes do encode state variables of the core oscillator, i.e., elements that define, rather than simply reflect, circadian phase, they should be acutely sensitive to nonphotic cues (20). Moreover, that such cues advance the clock during the subjective day when spontaneous expression of these genes is high and protein levels are rising leads to a further prediction: that advances are achieved by rapid suppression of the genes and/or their protein products, thereby accelerating the spontaneous cycle to a (new) phase encoded by lower mRNA or proteins.
To test these predictions, we first examined the spontaneous cycle of expression of mPer1 and mPer2 (as mRNA and protein) in the SCN of Syrian hamsters. We then tested the pattern of expression of these genes in the SCN of hamsters subjected to a much-studied and potent nonphotic resetting cue, namely confinement to a running wheel that generally elicits considerable activity and arousal, driving the clock to a new phase (16, 22). The results demonstrate rhythmic expression of mPer1 and mPer2 in the hamster SCN, with mRNA and protein encoded by mPer1 exhibiting a delayed phase relationship consistent with the negative-feedback model for the mammalian clock. Moreover, we describe an acute down-regulation of these two genes at the level of mRNA, identifying them as a common target for both photic and nonphotic resetting cues, and therefore as likely central elements of the core clock mechanism.
Materials and Methods
All experiments on animals were conducted in accordance with local codes of practice and within the framework of the Animals (Scientific Procedures) Act of 1986 and the Canadian Council on Animal Welfare.
Experiment 1.
To define the basic profile of mPer1 and mPer2 expression in the SCN, adult Syrian hamsters (Charles River) were caged individually with food and water available ad libitum, and entrained to a 14-hr light/10-hr dark schedule, in which lights off is defined as Zeitgeber time (ZT) 12. At selected intervals (every 2 hr), 3 or 4 hamsters (total sample size = 34) were killed by cervical dislocation, and their brains were dissected free from the skull and rapidly frozen on dry ice before storage at −50°C.
Experiment 2.
To test the effect of a nonphotic behavioral resetting cue on mPer expression, adult male Syrian hamsters (Harlan Sprague-Dawley) were housed in cages equipped with a 17.5-cm diameter running wheel (21). After 3 weeks, hamsters were confined to a novel running wheel at ZT 4; lights were turned off at this time for the rest of the experiment (19, 22). Control animals remained in their home cages, but experienced the same lighting change at the same time as the experimental group. Groups matched for the amount of running in the novel wheel were assigned for the measurement of any subsequent phase shift or for sampling of brain tissue at ZT 7, 20, or 24. Control groups of hamsters that had never been in the novel wheels were also killed at these times, as well as at ZT 4. Brains were dissected free from the skull and rapidly frozen on dry ice before storage at below −50°C. There were no significant differences between the groups in running during the 3 hr in the novel wheels. Mean values for all groups were >8,400 revolutions, and no individual studied here made <5,590 revolutions. This level is reliably associated with subsequent phase shifts (16). It was confirmed by the animals that were retained for recording of locomotor activity patterns.
In situ Hybridization and Immunocytochemistry.
Brains were sectioned at 16 μm, and every fourth section was processed for in situ hybridization for mPer1 or mPer2 expression by using 35S-labeled antisense riboprobes directed against mouse mPer sequences (6, 7). Film autoradiographs were generated by exposure to Beta-max Hyperfilm (Amersham) for 5–6 days, whilst emulsion images (K5, Ilford) were exposed for 6 weeks. The relative intensity of the hybridization signal on film images viewed through a Hamamatsu camera was assessed by using National Institutes of Health (NIH) image software (gift of W. Roshband, NIH), run on an Apple Macintosh IIci. Gray-scale density averaged over the whole SCN identified by Nissl stain was expressed as a ratio of gray-scale density of the adjacent medial hypothalamus. Immunocytochemical analyses of mPER1 protein levels in the SCN were done by using an antiserum raised against mouse mPER1 (11), which also generates specific immunostaining on Syrian hamster and Siberian hamster (M.H.H., unpublished data) brain tissue. Cryostat sections adjacent to those used for in situ hybridization were fixed by immersion in 4% paraformaldehyde, rinsed in buffered saline, and then processed for immunoreactivity (ir) as described for free-floating and slide-mounted sections (11, 23). The number of mPER-ir nuclei in individual SCN sections was determined with image software, and expressed as a total for the whole 1-in-4 series of SCN sections.
Results
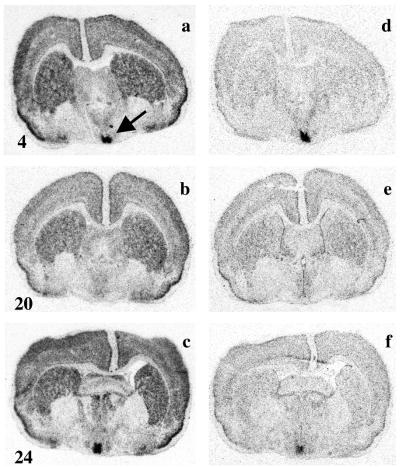
Strong hybridization signals for mPer1 and mPer2 were detected in the piriform cortex (Fig. 1) and hippocampus (data not shown) of the Syrian hamster. The signal for mPer1 was more extensive, with strong hybridization also in the striatum, neocortex (Fig. 1), and thalamus (data not shown). For both genes, the signal was specific: there was no hybridization to sense probe. The intensity of signal in these areas did not change appreciably with time of day (Fig. 1 a and f). However, the highest level of expression for both genes was in the SCN, where there was a strong cycle of expression, with peak levels in the early light phase (ZT 4, Fig. 1 a and d), and a nadir in subjective night (ZT 16–ZT 20, Fig. 1 b and e). Levels of expression started to rise again by the end of circadian night (Fig. 1 c and f). As a result, there was a pronounced daily cycle in the relative intensity of the hybridization signal for both genes in the SCN (Fig. 2a). Two-way ANOVA revealed a highly significant time effect (F = 42.5, P < 0.0001), a gene effect (F = 13.9, P < 0.001), and a highly significant interaction between time and gene (F = 10.6, P < 0.001). The interaction arose from differential phasing of the two rhythms, with mPer1 rising earlier and declining sooner than mPer2. The phase delay of mPer2 relative to mPer1 was approximately 3 to 4 hr.
Figure 1.
Rhythmic expression of mPer1 and mPer2 hybridization signal in hamster SCN. Representative film autoradiographs of the mPer1 (a–c) and mPer2 (d–f) hybridization signals generated from adjacent coronal sections of the Syrian hamster brain, sampled at ZT 4 (a and d), ZT 20 (b and e), or ZT 24 (c and f). Note the widespread, constitutive expression in most sites, but the pronounced daily rhythm of expression in the SCN, marked by arrow in a.
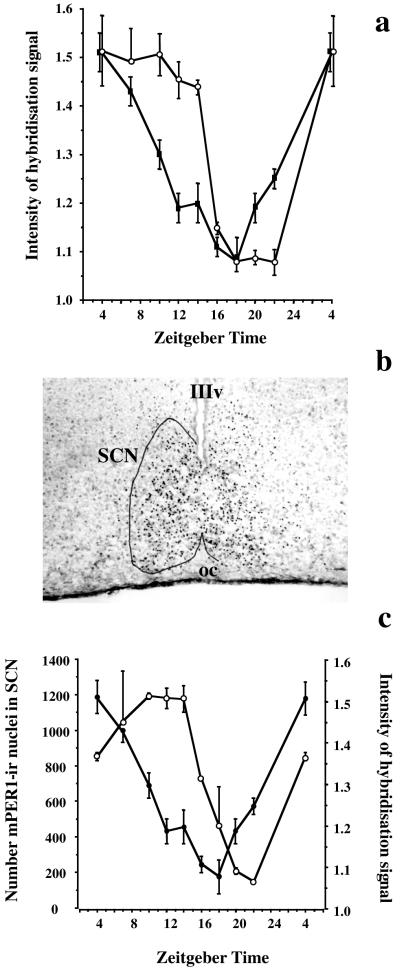
Figure 2.
Daily rhythms of mPer1, mPer2, and mPER1-ir in hamster SCN are differentially phased. (a) Relative intensity of hybridization signal (mean ± SEM) for mPer1 (■) and mPer2 (○) in hamster SCN as a function of Zeitgeber time (ZT). Note the phase delay of mPer2, relative to mPer1. (b) Representative coronal section of hamster SCN, sampled at ZT 7 and processed for mPER1-ir. Note nuclear localization of ir profiles extending across the SCN, the outline of which is drawn on left-hand side. IIIv, Third ventricle. oc, optic chiasm. (c) Daily cycle of abundance (mean ± SD) of mPER1-ir cells in hamster SCN (○), with peak expression in mid to late subjective day (ZT 10–14) and nadir at end of subjective night (ZT 20–22). The mPer1 mRNA hybridization data from Fig. 2a are replotted here (●) for comparison.
mPER1-ir was detected in the SCN of sections adjacent to those used for in situ hybridization (Fig. 2b). The immunoreaction was exclusively nuclear, and was also observed in tissue outside the SCN, including in the piriform cortex, striatum, and thalamus (data not shown). The abundance of mPER1-ir cells in the SCN varied significantly with the circadian phase (ANOVA time effect F = 22.9, P < 0.001). Levels started to rise in the early light phase, peaked between ZT 10 and ZT 14, and then started to fall to a nadir at ZT 22 (Fig. 2c). Comparison with the mRNA cycle revealed a phase delay of the protein cycle of about 6 hr. As protein levels reached a peak at ZT 10, the mRNA signal had already started to decline and it did not increase again until protein levels were at their nadir at ZT 20–22. This phase relationship is consistent with a negative-feedback interaction between mPER1 protein and mRNA expression.
Transfer to darkness at ZT 4 had a small advancing effect on the circadian activity rhythm of home-cage control hamsters (mean ± SEM 0.93 ± 0.17 hr, n = 5, Fig. 3a). However, confinement to a novel wheel for 3 hr, starting 8 hr before lights off, produced much larger phase advances (2.58 ± 0.43 hr, n = 6, Fig. 3b); these were significantly greater than those of control animals (P < 0.01 two-tailed t test).
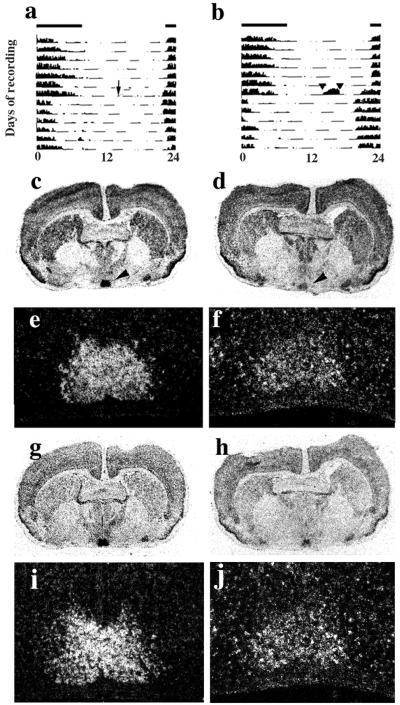
Figure 3.
Nonphotic resetting of the clock induces suppression of mPer1 and mPer2 hybridization signals in hamster SCN. (a) Representative single-plotted actograms of the circadian rhythm of activity of home cage controls transferred to continuous darkness at ZT 4 (arrow). Note small phase advance. (b) Representative actogram of animal confined to novel wheel for 3 hr, commencing at ZT 4 (indicated by arrowheads and enhanced wheel-running) and simultaneously transferred to continuous darkness, leading to a large phase advance of free-running activity rhythm. Bars above actograms indicate light:dark cycle before transfer. (c, e, g, and i) Representative in situ hybridization images (c and g, film; e and i, emulsion) from adjacent coronal sections of hamster brain reveal high levels of expression of mPer1 (c and e) and mPer2 (g and i) in the SCN (arrowheads) of home cage controls sampled at ZT 7. (d, f, h, and j) Confinement to a novel running wheel is associated with suppression of mPer1 (d and f) and mPer2 (h and j) signals at ZT 7.
In situ hybridization again revealed widespread expression of mPer1 in the hamster brain, including the cerebral cortex, striatum, and SCN (Fig. 3 c and d). Moreover, just as in the first experiment, the intensity of hybridization signal for mPer1 in the SCN of hamsters kept in their home cages varied with circadian phase (there were no obvious changes in other brain areas). Levels were high in the subjective day (ZT 4 and 7, Fig. 3 c and e) and low during subjective night (ZT 20), rising again at the start of the following subjective day (ZT 24). In hamsters sampled at ZT 7, immediately after confinement to a novel wheel, the intensity of mPer1 hybridization signal in the SCN was dramatically reduced (Fig. 3 d and f), compared with home-cage controls (Fig. 3 c and e). The signal intensity elsewhere in the brain did not show any obvious change. Statistical analysis of the relative intensity of mPer1 hybridization signal revealed a highly significant time effect (two-way ANOVA time effect F = 33.4, P < 0.01), but no overall difference between treatment groups [F = 0.9, not significant (n.s.)]. However, there was a highly significant interaction between time and treatment (ANOVA interaction F = 8.7, P < 0.01), arising from a significant suppression of hybridization signal at ZT 7 after running in the novel wheel (Fig. 4a). The nonphotic cue had no significant detectable effect on the mPer1 signal between ZT 20 and ZT 24, as the cycle progressed from the nadir to the phase of peak expression.
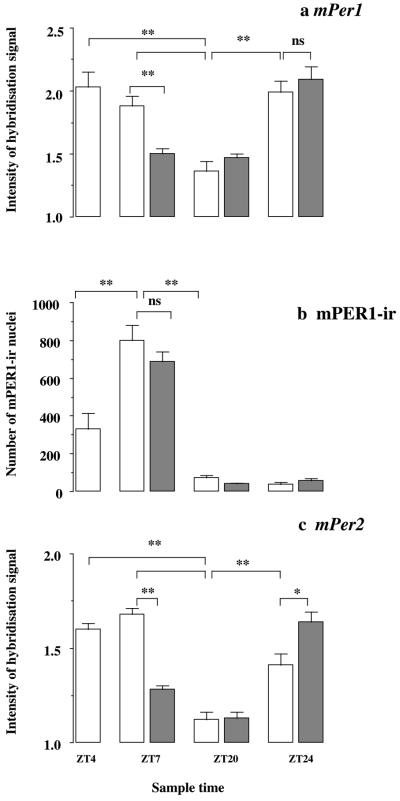
Figure 4.
Resetting by a nonphotic behavioral cue causes acute suppression of mPer signals, but not mPER1-ir in the SCN, shifting the endogenous mPer2 mRNA cycle. (a) Relative intensity (mean + SEM) of the mPer1 hybridization signal in the SCN exhibits a circadian rhythm in home-cage controls (open bars) and animals confined to a novel wheel (shaded bars) (ANOVA time effect F = 33.4, P < 0.01). Overall, there was no difference between treatment groups (F = 0.9, n.s.), but a highly significant interaction between treatment and time (ANOVA interaction F = 8.7, P < 0.01), arising from a significant suppression of the hybridization signal at ZT 7, after running in the novel wheel (n = 4–7 per group, n = 35). **P < 0.01 by posthoc Bonferroni and Dunnett's t test. (b) The abundance of mPER1-ir nuclei in the SCN exhibits a highly significant circadian rhythm in both groups of animals (ANOVA time effect F = 159, P < 0.01), with a peak during the subjective day, but no treatment effect (F = 1.0, n.s.) nor interaction (F = 1.1, n.s.). **P < 0.01 by posthoc Dunnett's t test. (c) Relative intensity (mean + SEM) of the mPer2 hybridization signal in the SCN exhibits a circadian rhythm in home-cage controls (open bars) and animals confined to a novel wheel (shaded bars) (ANOVA time effect F = 70.7, P < 0.01). Overall, there was no difference between treatment groups (F = 2.9, n.s.), but a highly significant interaction between treatment and time (ANOVA interaction F = 45.5, P < 0.01), arising from a significant suppression of the hybridization signal in the treatment group at ZT 7 and a significant elevation in this group at ZT 24. **P < 0.01, *P < 0.05 by posthoc Bonferroni and Dunnett's t test.
The abundance of mPER1-ir nuclei in the SCN also exhibited a significant daily cycle, phase-delayed relative to the mRNA rhythm, comparable to that in the first experiment. Levels rose between ZT 4 and ZT 7 in the subjective day, and were low in late subjective night (ZT 20, 24) (ANOVA time effect F = 159, P < 0.01, Fig. 4b). However, there was no significant treatment effect (F = 1.0, n.s.) nor interaction between time and treatment (F = 1.1, n.s.). Critically, there was no significant change in mPER1-ir in the SCN of animals confined to a running wheel for 3 hr and sampled at ZT 7. This highlights the role of mRNA rather than protein as an early point of regulation by nonphotic, behavioral resetting pathways.
Turning to mPer2, its rhythm of expression in the SCN of home-cage controls was comparable to that of mPer1, with a pronounced peak at ZT 7 (ANOVA time effect F = 70.7, P < 0.01, Fig. 3 g and i; Fig. 4c). However, the cycle appeared to be delayed relative to mPer1, insofar as levels showed a trend to increase, rather than decline, between ZT 4 and ZT 7; the intensity of signal at ZT 24 was significantly below peak levels, i.e., was still on the rising phase of the cycle. Overall, there was no difference between treatment groups (F = 2.9, n.s.), but there was a highly significant interaction between treatment and time (ANOVA interaction F = 45.5, P < 0.01), which arose from a significant suppression of hybridization signal in the animals confined to a novel wheel and sampled immediately after at ZT 7 (Fig. 3 h and j). In addition, there was a significant elevation in the group sampled at ZT 24 (Fig. 4c), indicative of an earlier rise caused by an advance of the subsequent endogenous cycle.
Discussion
The current studies demonstrate circadian cycling of mPer1 and mPer2 gene expression in the SCN of the Syrian hamster, and reveal a phase relationship between mPer1 mRNA and mPER1 protein, consistent with a transcriptional negative-feedback oscillator. Moreover, the study demonstrates an acute sensitivity of mPer1 and mPer2 mRNA levels (but not mPER1 protein) to a potent behavioral cue, which resets the circadian clock by nonphotic pathways. The sensitivity of mPer products to both light and behavioral resetting stimuli underlines their proximity to the core circadian oscillator, and identifies the regulation of mRNA levels as a common mechanism for the resetting of the mammalian clockwork by diverse cues.
The spontaneous cycling of mPer products in the hamster SCN, with a phase lag between the mRNA hybridization signal and nuclear mPER1-ir, is consistent with the transcriptional negative-feedback model for the circadian clock (3) proposed from studies that revealed a similar delayed phase relationship for the gene products in Drosophila and mice (11). The nuclear localization of endogenous mPER1-ir, which was also reported in the mouse (11), is consistent with nuclear localization of recombinant protein when transfected into cells lines (14, 15, 24), and is a necessary feature if the endogenous protein is to participate in a transcriptional cycle. The nature of such a cycle is inevitably complex, with interactions between the positive regulators of E-box-mediated transcription, CLOCK/BMAL (25), and the negative trans-acting factors, mPER1, mPER2, mPER3, CRY1, and CRY2 (14, 15, 24). Moreover, temporal differences in the expression of these components, such as the phase delay of mPer2 relative to mPer1, described here in the hamster and previously in the mouse (9, 10), will add a further degree of complexity.
In Drosophila, resetting to light involves inactivation of the TIMELESS protein, a dimerization partner of PERIOD (26). Mammalian TIMELESS is not regulated by light (11), resetting being mediated by changes in mPer mRNA. Nonphotic resetting in mammals could, however, potentially involve changes in mPER proteins. The current study failed to detect an early response of mPER1 protein to nonphotic cues, although it cannot be excluded that examination with a finer temporal resolution might reveal subtle effects on protein levels. Given that the SCN oscillator is reset by the stimulus, a phase advance in the mPER1 rhythm must take place on the subsequent cycle. The current study did not examine the rising phase of mPER1-ir to test this prediction. It also remains to be determined whether other clock elements that are insensitive to light, such as mPer3 (10, 28) or clock (25), respond to nonphotic cues. Indeed, acute suppression of mPer transcription might be achieved by interference of CLOCK/BMAL1-mediated drive to the mPer genes.
The present demonstration of the acute sensitivity of both mPer1 and mPer2 to a nonphotic behavioral cue highlights the proximity of these genes to the core oscillation, rather than a more peripheral role as, for example, their involvement in light input pathways. In addition, it provides a molecular explanation for behavioral responses observed in the whole organism. In vivo, phase shifts to light (28) as well as to nonphotic cues (18, 19, 29) are completed within a single circadian cycle. The suppression of mPer expression immediately after the end of running in the novel wheel can be interpreted as evidence of the clock being rapidly driven to a new phase, and so provides a molecular correlate of the behavioral studies. Moreover, the earlier rise in mPer2 levels on the next cycle is further evidence that resetting is completed within a single cycle. The apparent absence of such an effect on mPer1 levels may be a reflection of the earlier phasing of the mPer1 rhythm, in combination with the limited number of time points sampled. Samples directed at the midpoint of the spontaneous rise rather than to its peak might reveal an advance to mPer1 and to mPer2 as well. A similar approach might also be expected to reveal an advance to the mPER1-ir rhythm.
A clear distinction has been established between the signaling pathways that mediate photic and nonphotic resetting. Photic induction of mPer in the SCN is probably mediated by glutamatergic retinal afferents, acting through a signaling cascade based on increased intracellular calcium and activation of the transcription factors CREB (27, 30) and ERK (31). In contrast, nonphotic resetting, through confinement to a novel wheel or scheduled arousal, requires neuropeptide Y (NPY)-ergic innervation of the SCN (16, 32, 33); there is a strong prediction from the current work that resetting by local infusion of NPY will be accompanied by a rapid suppression of the expression of mPer1 and mPer2 in the SCN. The potential for negative regulation of the transcriptional apparatus of the SCN by nonphotic cues has been demonstrated by the reported suppression of cFOS-ir in the SCN of hamsters subjected to a novel wheel (34), although the role played by this and other immediate-early gene products in the regulation of mPer is not yet known. The current identification of mPer genes as targets for contrasting resetting cues (light and behavioral inputs) suggests that novel therapeutic agents for manipulation of clock-related disorders could be identified by examining their actions on the expression of these genes in the SCN. Furthermore, the most appropriate time for the use of such agents, i.e., their phase dependence, could be predicted from the regulation of their molecular targets, either positive or negative.
In conclusion, the present paper shows that nonphotic resetting of the mammalian clock is associated with the acute suppression of the putative clock genes mPer1 and mPer2 in the SCN and is a necessary element in their definition as core elements of the circadian mechanism. Moreover, it reveals a molecular target for the convergent but opposing actions of environmental and behavioral stimuli that regulate circadian time, and thus offers an integrative explanation for circadian entrainment.
Acknowledgments
We are grateful to J. Drew, N. White, and P. Salmon for excellent technical assistance. The anti-mPER1 serum was a gift from Dr. S. M. Reppert, Harvard Medical School, Boston, MA. Plasmids for production of riboprobes were gifts from Dr. Z. S. Sun (Baylor College of Medicine, Houston, TX) and Prof. H. Okamura (University of Kobe Medical School, Kobe, Japan). This work was supported by the Biotechnology and Biological Sciences Research Council of the U.K., with a project grant (Ref. S/09882) to M.H.H. and Prof. C. P. Kyriacou (Department of Genetics, University of Leicester, U.K.), and by a Medical Research Council (Canada) grant and a Gonville and Caius College (Cambridge, U.K.) Visiting Fellowship to N.M.
Abbreviations
- ir
immunoreactivity
- n.s.
not significant
- SCN
suprachiasmatic nuclei
- ZT
Zeitgeber time
References
- 1.Pittendrigh C S. Annu Rev Physiol. 1993;55:17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Aschoff J. Handbook of Behavioral Neurobiology. New York: Plenum; 1981. [Google Scholar]
- 3.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Klein D C, Moore R Y, Reppert S M. Suprachiasmatic Nucleus: The Mind's Clock. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 5.Welsh D K, Logothetis D E, Meister M, Reppert S M. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 6.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 7.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 8.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J J, Dunlap J C, et al. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 9.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 10.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 11.Hastings M H, Field M D, Maywood E S, Weaver D R, Reppert S M. J Neurosci. 1999;19(RC 11):1–7. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. Nature (London) 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 14.Sangoram A M, Saez L, Antoch M P, Gekakis N, Staknis D, Whiteley N, Fruechte E M, Vitaterna M H, Shimomura K, King D P, et al. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 15.Jin X, Shearman L P, Weaver D R, Zylka M J, De Vries G J, Reppert S M. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 16.Mrosovsky N. Biol Rev. 1996;71:343–372. doi: 10.1111/j.1469-185x.1996.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 17.Hastings M H, Best J D, Ebling F J P, Maywood E S, McNulty S, Schurov I, Selvage D, Sloper P, Smith K L. Prog Brain Res. 1996;111:147–174. doi: 10.1016/s0079-6123(08)60406-9. [DOI] [PubMed] [Google Scholar]
- 18.Mead S, Ebling F J P, Maywood E S, Humby T, Herbert J, Hastings M H. J Neurosci. 1992;12:2516–2522. doi: 10.1523/JNEUROSCI.12-07-02516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mrosovsky N. J Biol Rhythms. 1991;6:167–179. doi: 10.1177/074873049100600207. [DOI] [PubMed] [Google Scholar]
- 20.Hastings M H, Duffield G E, Smith E J D, Maywood E S, Ebling F J P. Chronobiol Int. 1998;15:425–445. doi: 10.3109/07420529808998700. [DOI] [PubMed] [Google Scholar]
- 21.Mrosovsky N, Salmon P A, Vrang N. Chronobiol Int. 1998;15:147–158. doi: 10.3109/07420529808998679. [DOI] [PubMed] [Google Scholar]
- 22.Mrosovsky N. Chronobiol Int. 1996;13:387–392. doi: 10.3109/07420529609012662. [DOI] [PubMed] [Google Scholar]
- 23.Maywood E S, Bittman E L, Ebling F J P, Barrett P, Morgan P J, Hastings M H. J Neuroendocrinol. 1995;7:215–223. doi: 10.1111/j.1365-2826.1995.tb00750.x. [DOI] [PubMed] [Google Scholar]
- 24.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 25.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 26.Myers M P, Wager-Smith K, Rothenfluh-Hilfiker A, Young M W. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 27.Best J D, Maywood E S, Smith K L, Hastings M H. J Neurosci. 1999;19:828–835. doi: 10.1523/JNEUROSCI.19-02-00828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, et al. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumova A, Illnerova H. Neurosci Lett. 1996;218:181–184. doi: 10.1016/s0304-3940(96)13159-1. [DOI] [PubMed] [Google Scholar]
- 30.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 31.Obrietan K, Impey S, Storm D R. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 32.Biello S M, Janik D, Mrosovsky N. Neuroscience. 1994;62:273–279. doi: 10.1016/0306-4522(94)90331-x. [DOI] [PubMed] [Google Scholar]
- 33.Maywood E S, Smith E, Hall S J, Hastings M H. Eur J Neurosci. 1997;9:1739–1747. doi: 10.1111/j.1460-9568.1997.tb01531.x. [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen J D, Janik D, Mrosovsky N. Brain Res Bull. 1998;47:367–376. doi: 10.1016/s0361-9230(98)00121-x. [DOI] [PubMed] [Google Scholar]