Abstract
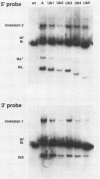
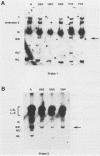
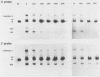
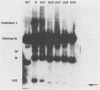
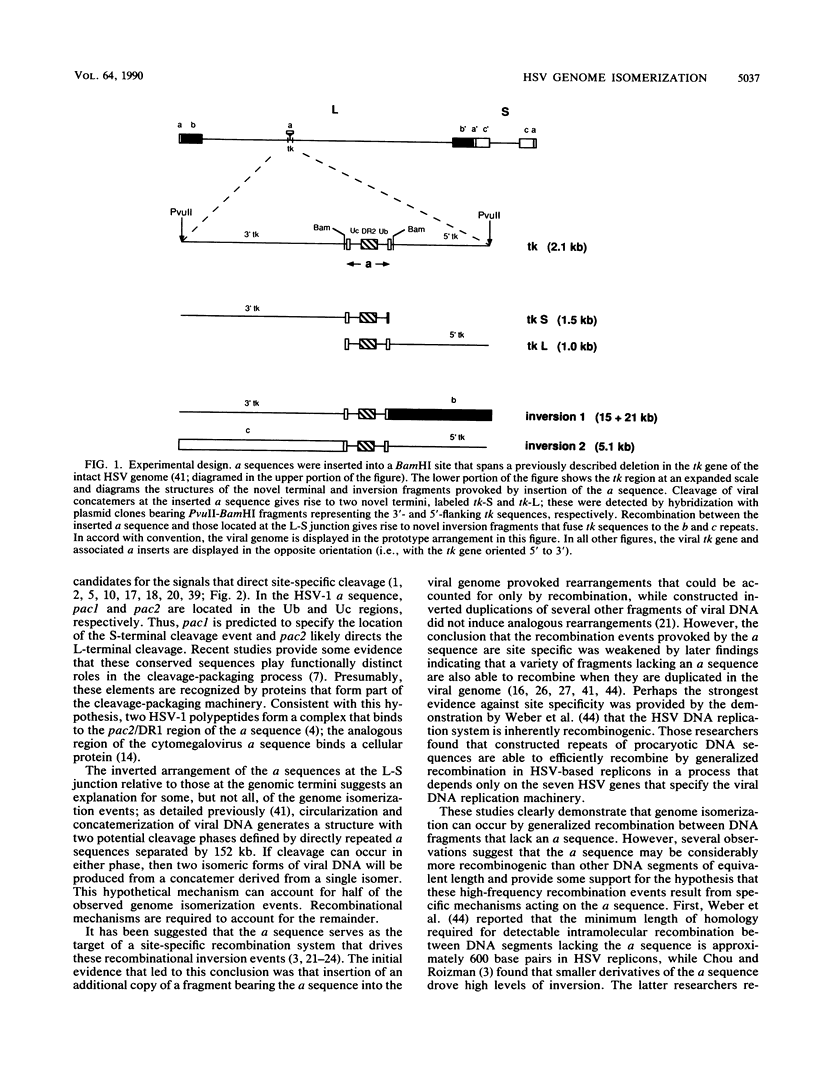
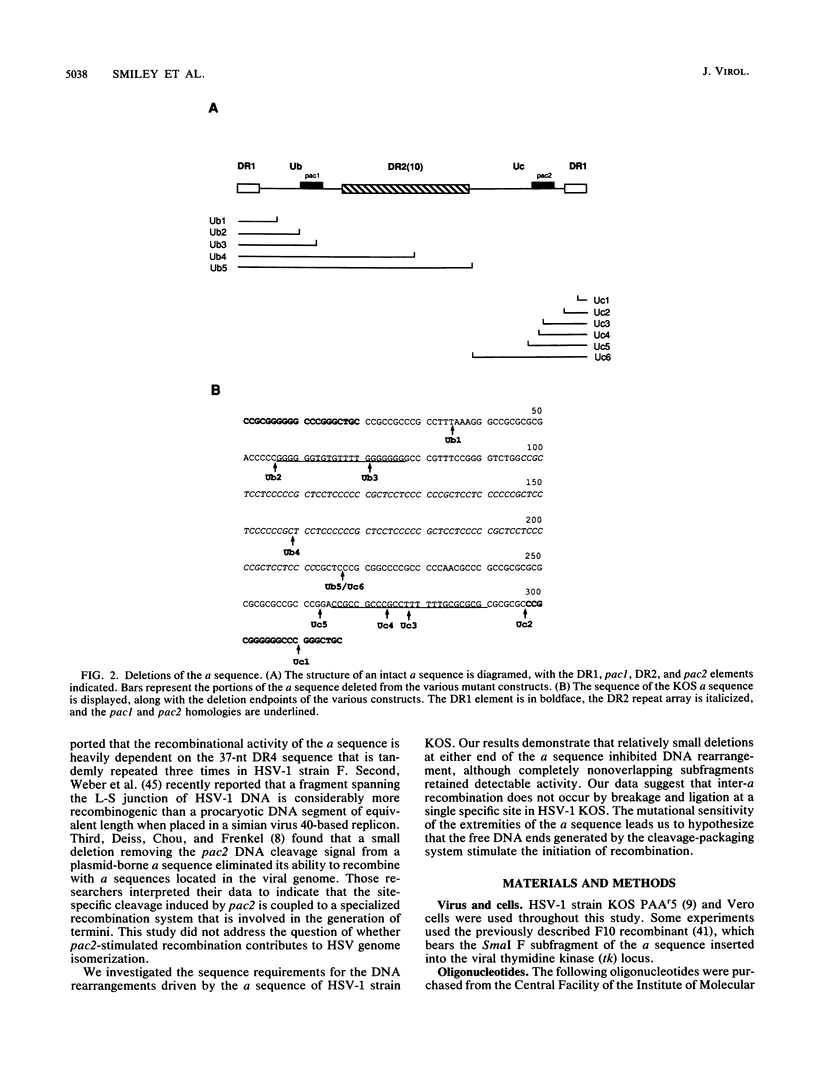
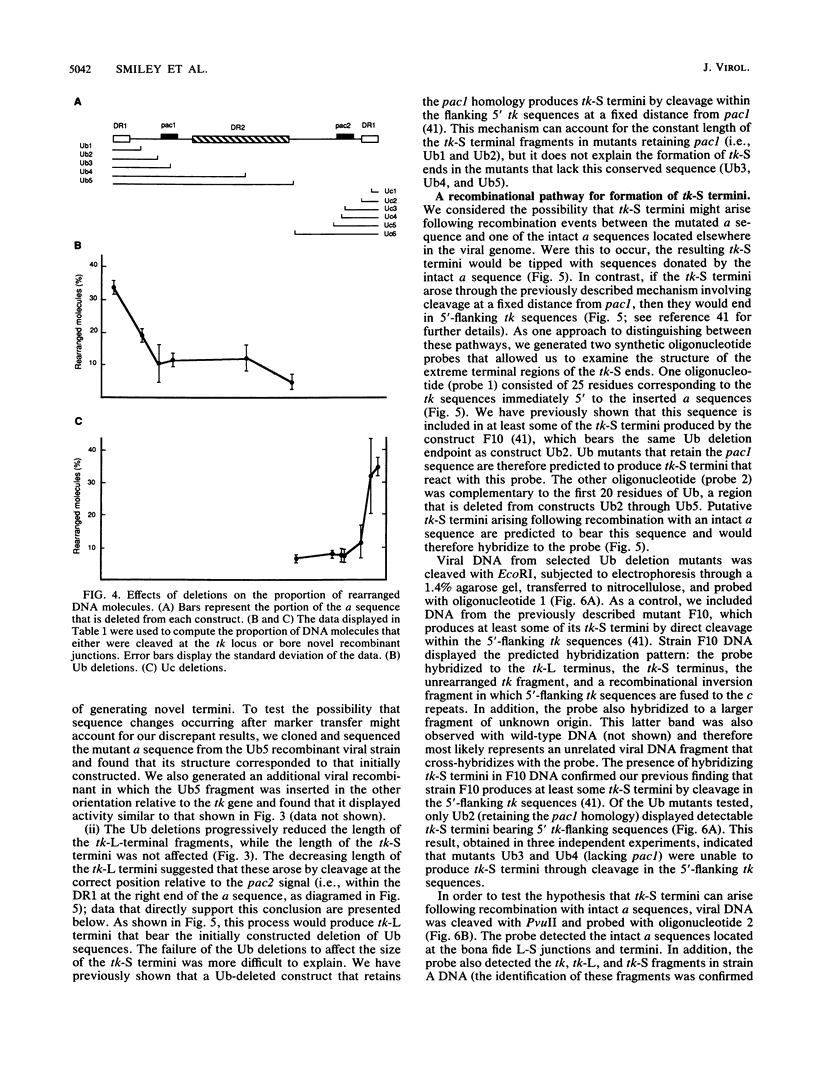
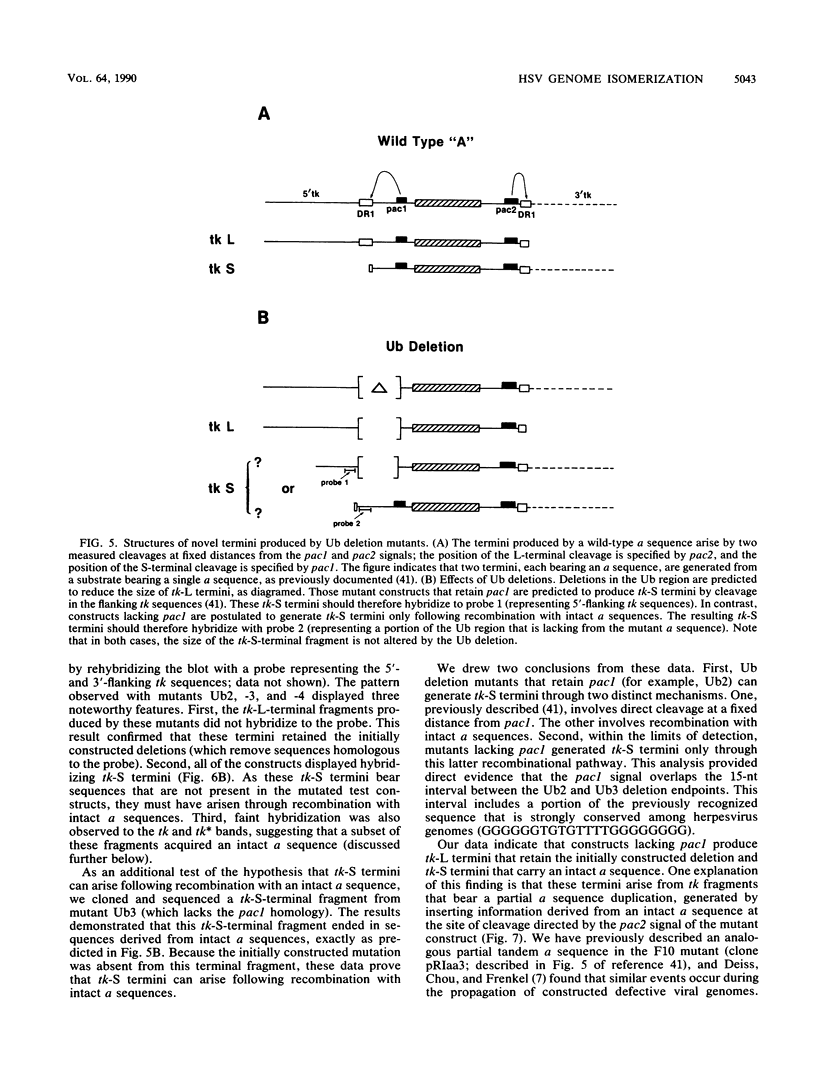
We investigated the sequence requirements for the site-specific DNA cleavages and recombinational genome isomerization events driven by the terminal repeat or a sequence of herpes simplex virus type 1 KOS DNA by inserting a series of mutated a sequences into the thymidine kinase locus in the intact viral genome. Our results indicate that sequences located at both extremities of the a sequence contribute to these events. Deletions entering from the Ub side of the a sequence progressively reduced the frequency of DNA rearrangements, and further deletion of the internal DR2 repeat array had an additional inhibitory effect. This deletion series allowed us to map the pac1 site-specific DNA cleavage signal specifying the S-terminal cleavage to a sequence that is conserved among herpesvirus genomes. Constructs lacking this signal were unable to directly specify the S-terminal cleavage event but retained a reduced ability to give rise to S termini following recombination with intact a sequences. Deletions entering from the Uc side demonstrated that the copy of direct repeat 1 located adjacent to the Uc region plays an important role in the DNA rearrangements induced by the a sequence: mutants lacking this sequence displayed a reduced frequency of novel terminal and recombinational inversion fragments, and further deletions of the Uc region had a relatively minor additional effect. By using a construct in which site-specific cleavage was directed to heterologous DNA sequences, we found that the recombination events leading to genome segment inversion did not occur at the sites of DNA cleavage used by the cleavage-packaging machinery. This observation, coupled with the finding that completely nonoverlapping portions of the a sequence retained detectable recombinational activity, suggests that inter-a recombination does not occur by cleavage-ligation at a single specific site in herpes simplex virus type 1 strain KOS. The mutational sensitivity of the extremities of the a sequence leads us to hypothesize that the site-specific DNA breaks induced by the cleavage-packaging system stimulate the initiation of recombination.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albrecht M., Darai G., Flügel R. M. Analysis of the genomic termini of tupaia herpesvirus DNA by restriction mapping and nucleotide sequencing. J Virol. 1985 Nov;56(2):466–474. doi: 10.1128/jvi.56.2.466-474.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankier A. T., Dietrich W., Baer R., Barrell B. G., Colbère-Garapin F., Fleckenstein B., Bodemer W. Terminal repetitive sequences in herpesvirus saimiri virion DNA. J Virol. 1985 Jul;55(1):133–139. doi: 10.1128/jvi.55.1.133-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989 Mar;63(3):1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984 Nov;65(Pt 11):1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss L. P., Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986 Mar;57(3):933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Coen D. M., Fisher B. L., Weisslitz M., Randall S., Almy R. E., Gelep P. T., Schaffer P. A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984 Jan 15;132(1):26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Ludwig H., Buhk H. J. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J Virol. 1988 Apr;62(4):1355–1363. doi: 10.1128/jvi.62.4.1355-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerner H. C., Maichle I. B., Ott A., Schröder C. H. Origin of two different classes of defective HSV-1 Angelotti DNA. Nucleic Acids Res. 1979 Apr;6(4):1467–1478. doi: 10.1093/nar/6.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G. W., Mocarski E. S. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J Virol. 1989 Nov;63(11):4715–4728. doi: 10.1128/jvi.63.11.4715-4728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J. R., Spector D. H. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology. 1988 Jan;162(1):98–107. doi: 10.1016/0042-6822(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Matsuo T., Heller M., Petti L., O'Shiro E., Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984 Dec 14;226(4680):1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Liu A. C., Spaete R. R. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain). J Gen Virol. 1987 Aug;68(Pt 8):2223–2230. doi: 10.1099/0022-1317-68-8-2223. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Lee G. T., Spear P. G. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985 Feb;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Spear P. G. Enhanced rate of conversion or recombination of markers within a region of unique sequence in the herpes simplex virus genome. J Virol. 1986 May;58(2):704–708. doi: 10.1128/jvi.58.2.704-708.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The organization of the herpes simplex virus genomes. Annu Rev Genet. 1979;13:25–57. doi: 10.1146/annurev.ge.13.120179.000325. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. The alpha sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985 Jun;54(3):817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C., Davison A. J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983 Dec 10;11(23):8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Spector D. H. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986 Sep;59(3):591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Summers W. C. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J Virol. 1978 Aug;27(2):374–387. doi: 10.1128/jvi.27.2.374-387.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Levine M., Glorioso J. C. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990 Jan;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]