Abstract
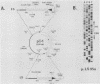
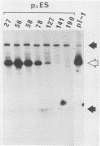
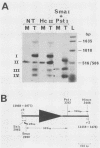
RmI, a chimeric DNA molecule containing polyomavirus (Py) and mouse sequences, generates unit-length Py DNA via intramolecular recombination between two directly repeated viral sequences of 182 base pairs (S repeats). To analyze the contribution of the S repeats in this process, we produced mutants of RmI carrying deletions in either one or both S repeats and tested them for their ability to recombine in mouse 3T6 cells. Mutant DNAs were found to yield unit-length Py DNA as long as they carried a minimal internal homology of 40 to 50 base pairs. Unlike RmI itself, however, the mutants also gave rise to nonhomologous recombination products. These results suggest that when the generation of homologous products is hampered by a limiting homology, nonhomologous products may arise instead of homologous ones. Therefore, the initial step(s) in the mechanisms yielding the two kinds of products could be identical.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayares D., Chekuri L., Song K. Y., Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):709–719. doi: 10.1101/sqb.1979.043.01.079. [DOI] [PubMed] [Google Scholar]
- Bourgaux P., Gendron D., Bourgaux-Ramoisy D. Preferred crossover sites on polyomavirus DNA. J Virol. 1990 May;64(5):2327–2336. doi: 10.1128/jvi.64.5.2327-2336.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgaux P., Sylla B. S., Chartrand P. Excision of polyoma virus DNA from that of a transformed mouse cell: identification of a hybrid molecule with direct and inverted repeat sequences at the viral-cellular joints. Virology. 1982 Oct 15;122(1):84–97. doi: 10.1016/0042-6822(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Effect of insertions, deletions, and double-strand breaks on homologous recombination in mouse L cells. Mol Cell Biol. 1985 Apr;5(4):684–691. doi: 10.1128/mcb.5.4.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillette S., Chartrand P. Intermolecular recombination assay for mammalian cells that produces recombinants carrying both homologous and nonhomologous junctions. Mol Cell Biol. 1987 Jun;7(6):2248–2255. doi: 10.1128/mcb.7.6.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Joffe S., Seidman M. M. Recombination and deletion of sequences in shuttle vector plasmids in mammalian cells. Mol Cell Biol. 1985 Sep;5(9):2265–2271. doi: 10.1128/mcb.5.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni V., Dailey L., Valle G. D., Basilico C. Requirements for excision and amplification of integrated viral DNA molecules in polyoma virus-transformed cells. J Virol. 1982 Aug;43(2):617–628. doi: 10.1128/jvi.43.2.617-628.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning A. M., Talmud P., Humphries S. E. Errors in the polymerase chain reaction. Nucleic Acids Res. 1988 Nov 11;16(21):10393–10393. doi: 10.1093/nar/16.21.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güssow D., Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res. 1989 May 25;17(10):4000–4000. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Huberdeau D., Sylla B. S., Herring-Gillam E., Bourgaux-Ramoisy D., Bourgaux P. Alternative excision products originating from a single integration of polyomavirus DNA. Mol Cell Biol. 1985 Oct;5(10):2608–2612. doi: 10.1128/mcb.5.10.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Smithies O. Recombinant fragment assay for gene targetting based on the polymerase chain reaction. Nucleic Acids Res. 1988 Sep 26;16(18):8887–8903. doi: 10.1093/nar/16.18.8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Schmidtke J., Rösler U. Polymerase chain reaction: replication errors and reliability of gene diagnosis. Nucleic Acids Res. 1989 Mar 25;17(6):2197–2201. doi: 10.1093/nar/17.6.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Piché A., Bourgaux P. Resolution of a polyomavirus-mouse hybrid replicon: release of genomic viral DNA. J Virol. 1987 Mar;61(3):840–844. doi: 10.1128/jvi.61.3.840-844.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piché A., Bourgaux P. Resolution of a polyomavirus-mouse hybrid replicon: viral function required for recombination. J Virol. 1987 Mar;61(3):845–850. doi: 10.1128/jvi.61.3.845-850.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Gauss P., Doherty D. H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982 Nov;31(1):25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Recombination between poly[d(GT).d(CA)] sequences in simian virus 40-infected cultured cells. Mol Cell Biol. 1985 Jun;5(6):1247–1259. doi: 10.1128/mcb.5.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D. J., Milman G. Short-term, high-efficiency expression of transfected DNA. Mol Cell Biol. 1984 Aug;4(8):1641–1643. doi: 10.1128/mcb.4.8.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla B. S., Allard D., Roy G., Bourgaux-Ramoisy D., Bourgaux P. A mouse DNA sequence that mediates integration and excision of polyoma virus DNA. Gene. 1984 Sep;29(3):343–350. doi: 10.1016/0378-1119(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Sylla B. S., Huberdeau D., Bourgaux-Ramoisy D., Bourgaux P. Site-specific excision of integrated polyoma DNA. Cell. 1984 Jun;37(2):661–667. doi: 10.1016/0092-8674(84)90398-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]