Abstract
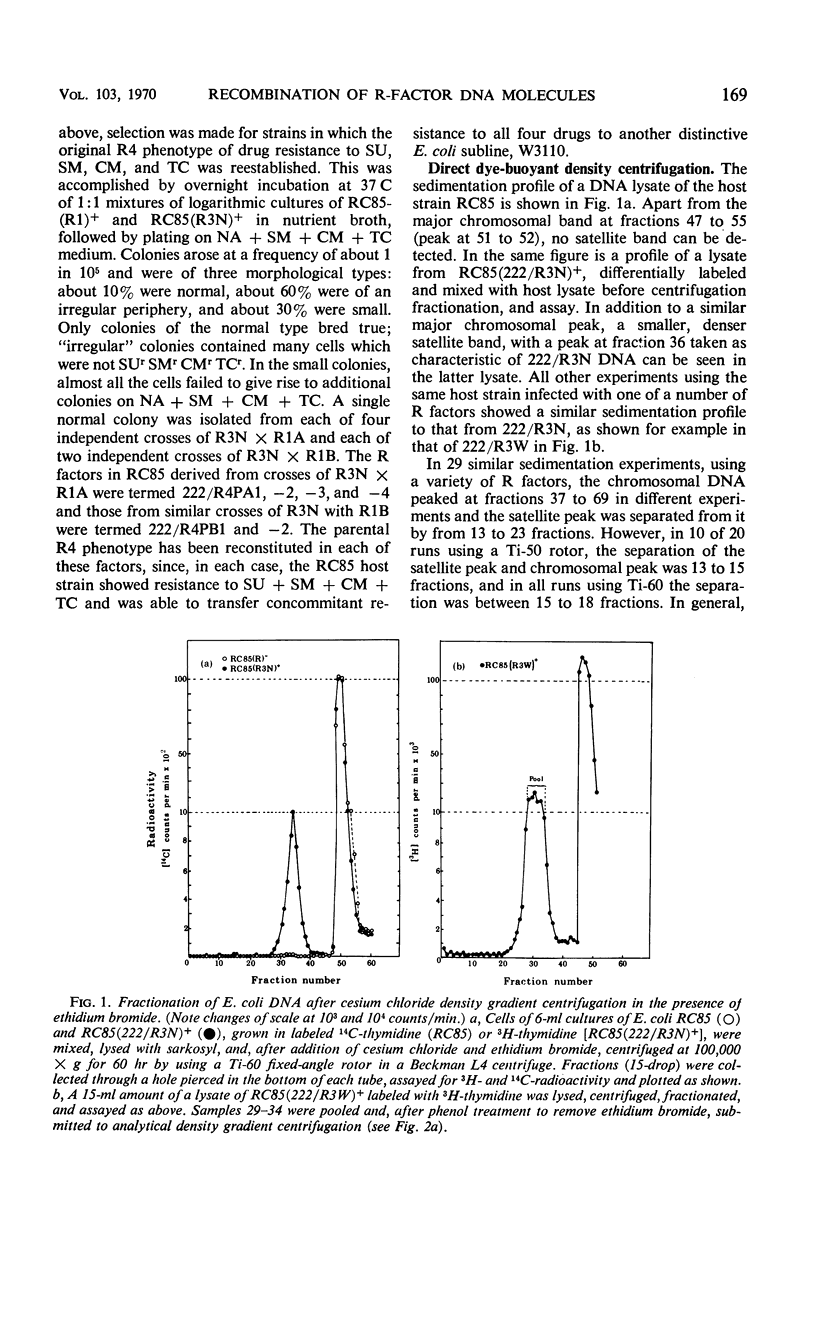
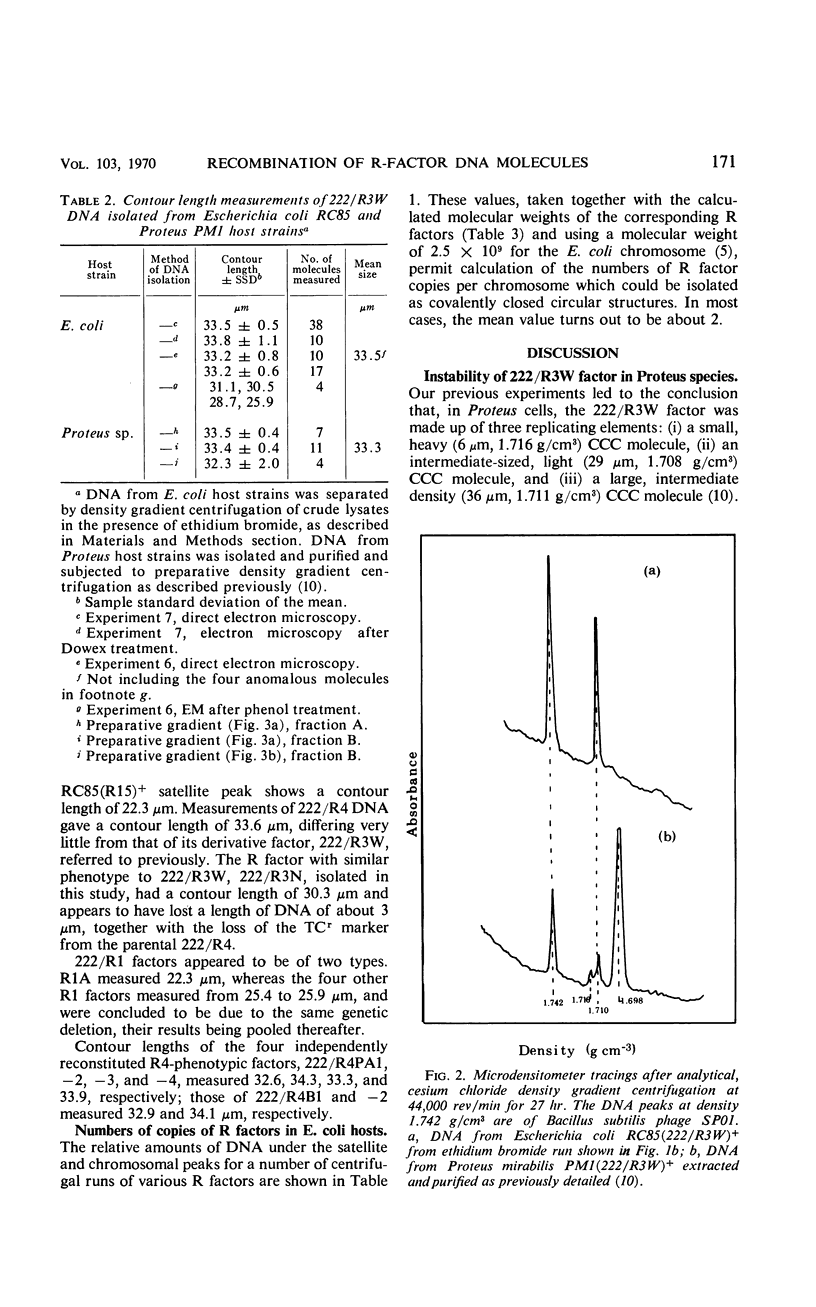
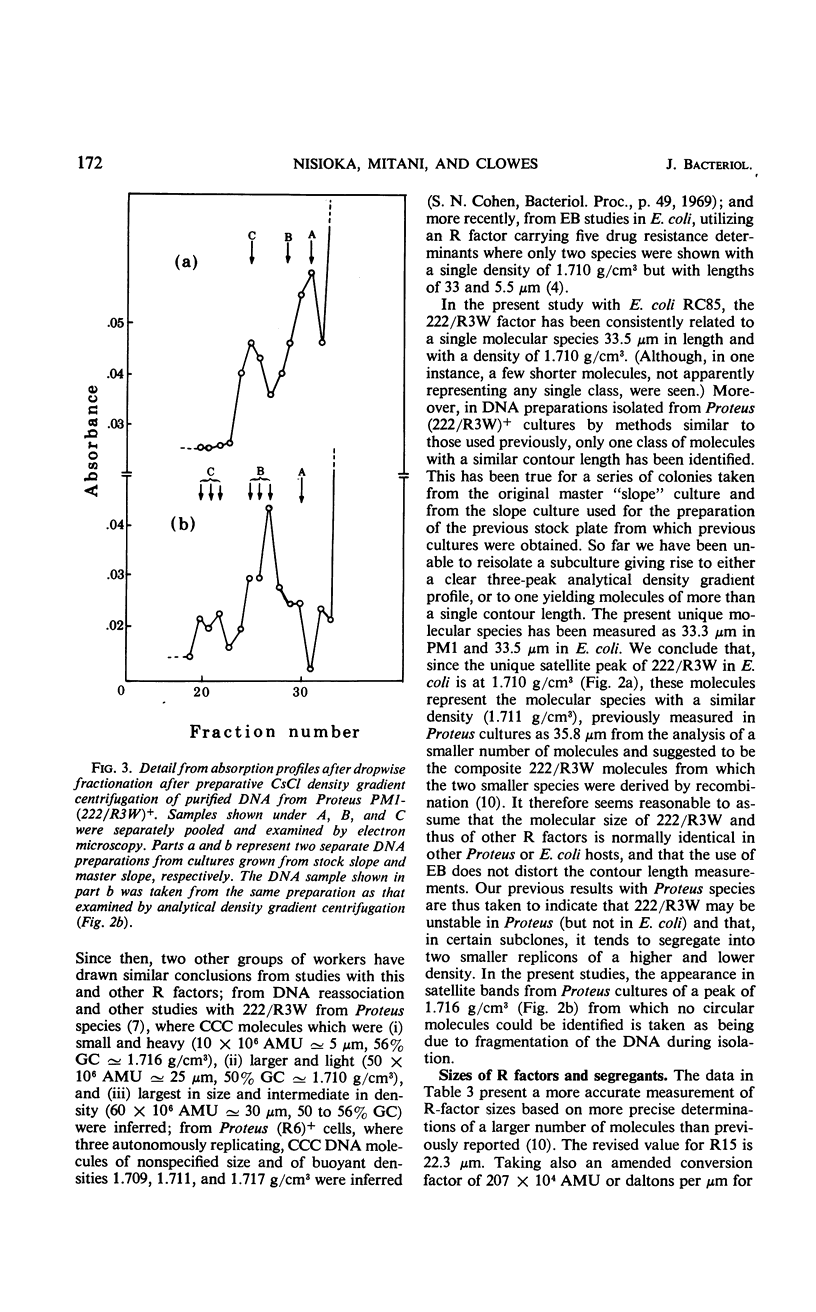
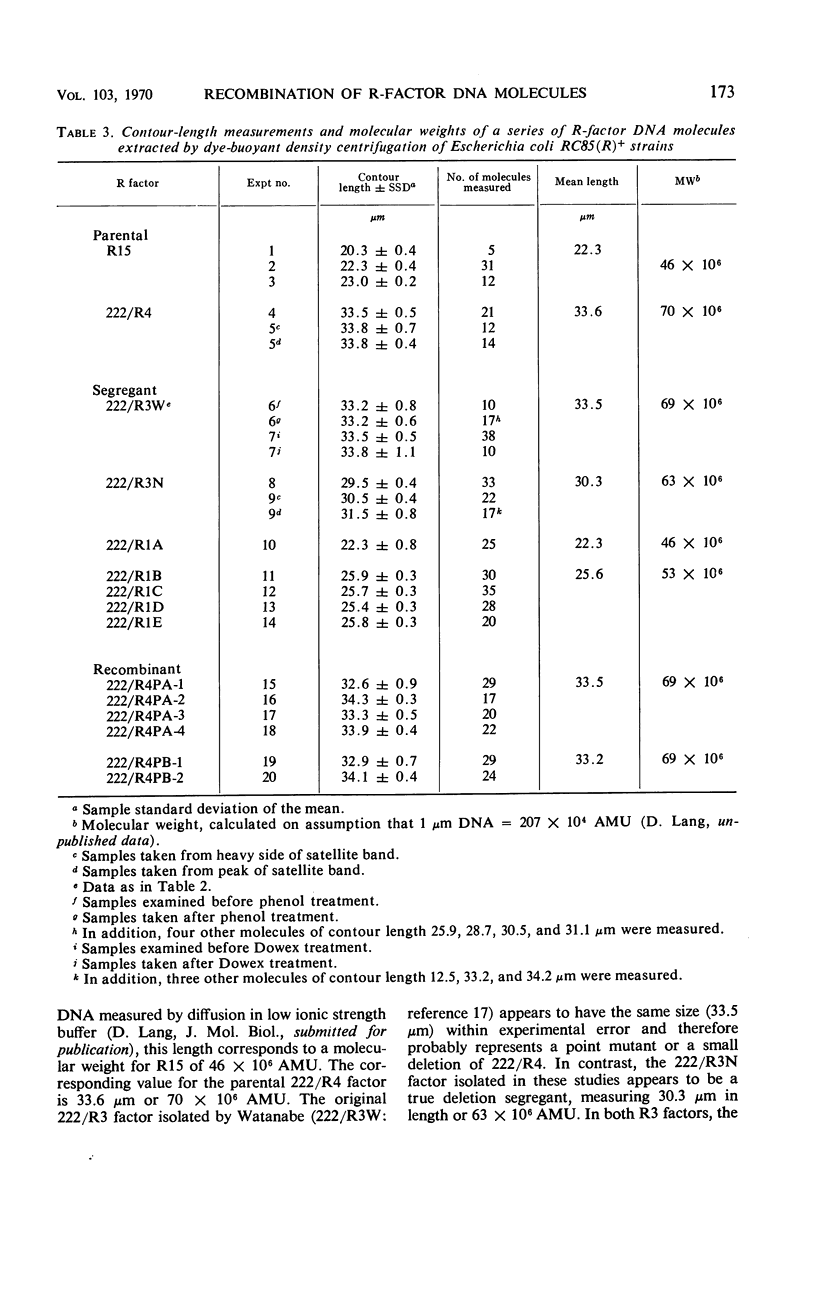
Three previously studied R factors were used: 222/R4, controlling transmissible resistance to sulfonamide, streptomycin, chloromycetin, and tetracycline (SUr SMr CMr TCr); 222/R3, a derivative of 222/R4 (now termed 222/R3W) having lost TCr; and R15, controlling infectious resistance to SU and SM only. Two types of derivative R factors were isolated from 222/R4 by serial subculture in Salmonella species. One derivative, termed 222/R1, lost resistance to SU, SM, and CM, and the other, termed 222/R3N, lost only TCr. Each factor was transferred to a standard Escherichia coli K-12 host. Recombinant factors of 222/R4 phenotype were isolated by selection after mixed culture of E. coli (222/R1)+ and (222/R3N)+ strains. Density-gradient equilibrium centrifugation of lysates of E. coli R+ hosts in the presence of ethidium bromide separated R-factor deoxyribonucleic acid (DNA) as a heavy satellite peak which was subjected to electron microscopy or analytical density gradient centrifugation. Each DNA comprised a unimolecular species of circular DNA. The contour of R15 measured 22.3 μm [equivalent to 46 × 106 atomic mass units (AMU)], and that of 222/R4 measured 33.6 μm (70 × 106 AMU). 222/R3W appeared to be a point mutant or small deletion of 222/R4 with an almost identical size, whereas 222/R3N had lost a DNA segment of about 3 μm, and measured 30.3 μm or 63 × 106 AMU. The 222/R1 factors also appeared to have arisen by loss of DNA from 222/R4, 222/R1A being 22.3 μm or 46 × 106 AMU, whereas all other 222/R1 factors appeared to be duplicates, measuring 25.6 μm or 53 × 106 AMU. The DNA from six recombinant factors of R4 phenotype was indistinguishable in size and configuration from the parental 222/R4. In most cases, the number of R-factor copies (present as covalently closed circular molecules) per copy of the E. coli chromosome was less than 2, ranging from 1.2 to 3.3.
Full text
PDF




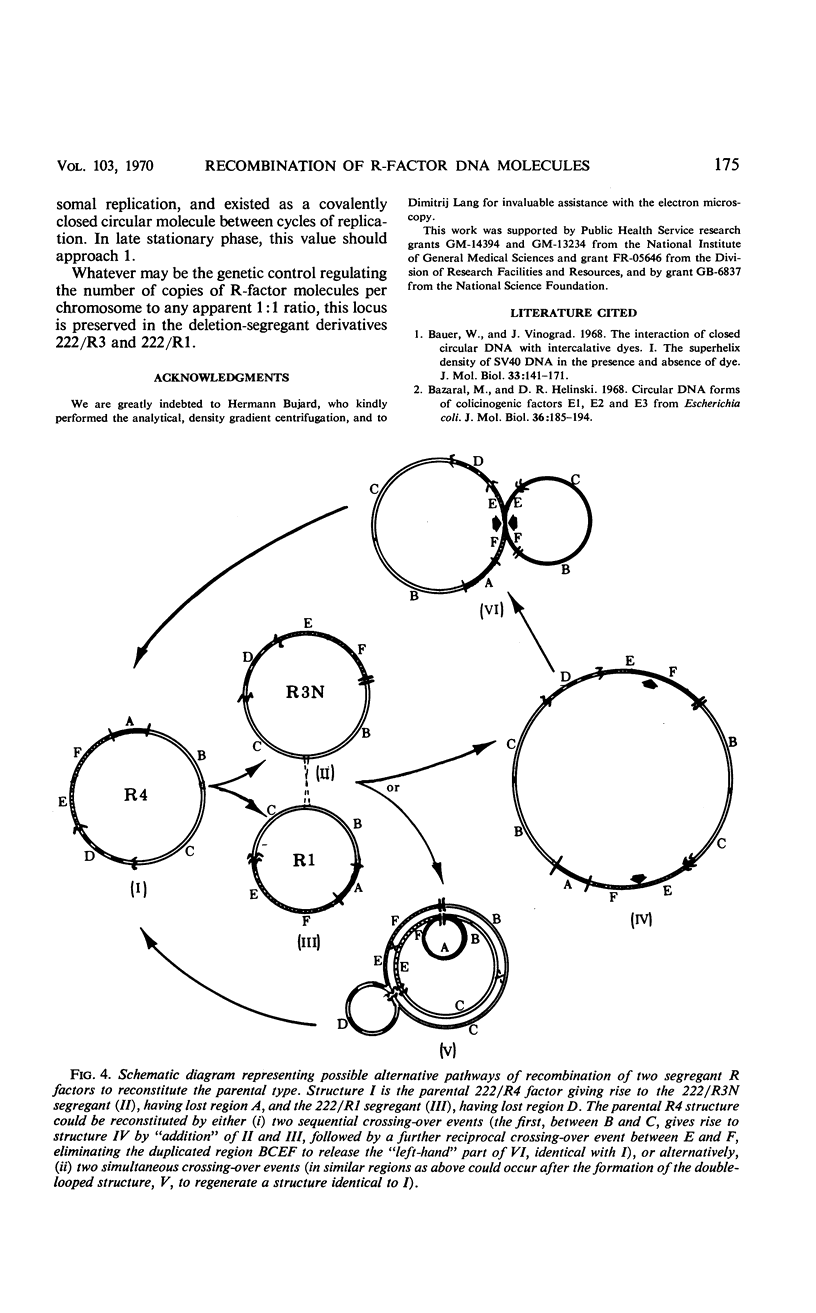
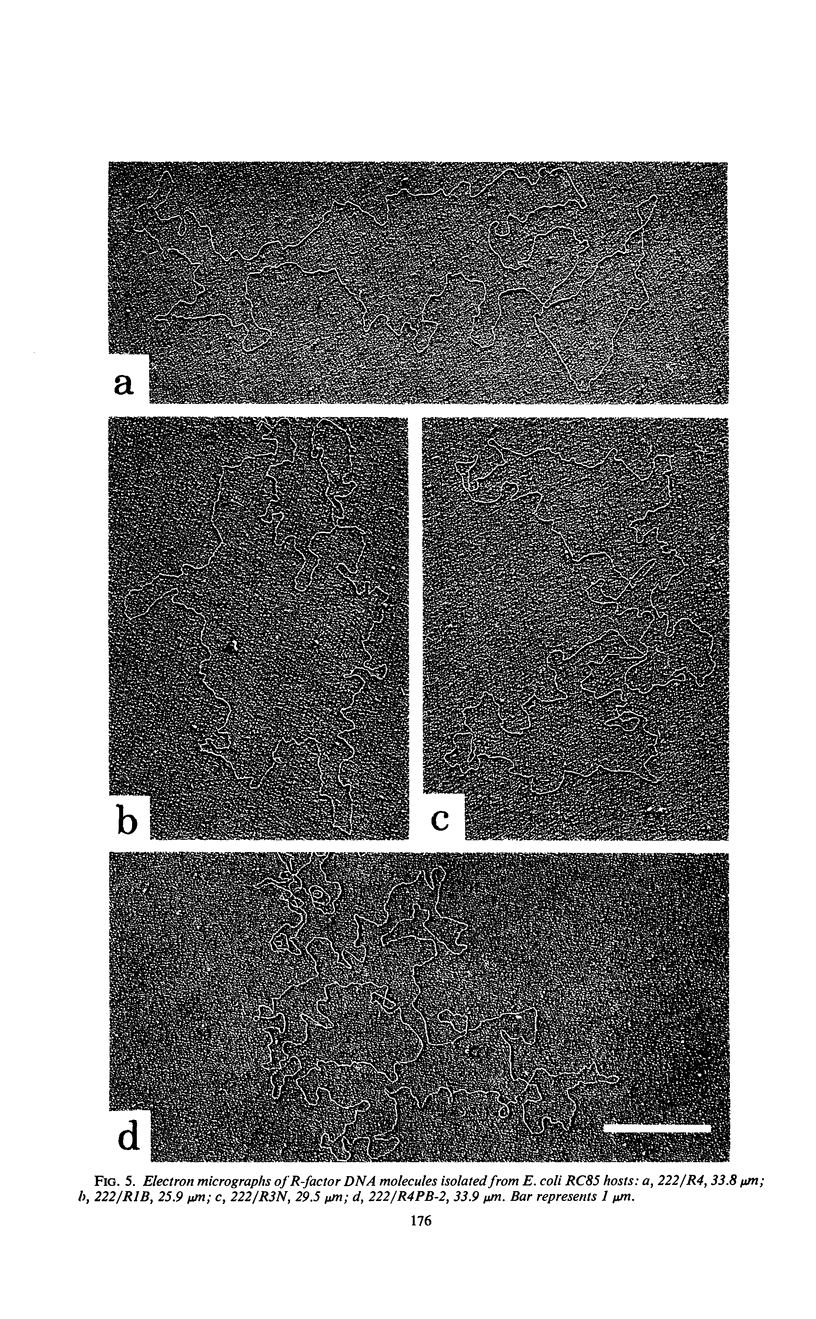

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Multiple molecular species of circular R-factor DNA isolated from Escherichia coli. Nature. 1969 Dec 27;224(5226):1273–1277. doi: 10.1038/2241273a0. [DOI] [PubMed] [Google Scholar]
- Cooper S., Helmstetter C. E. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- Falkow S., Citarella R. V., Wohlhieter J. A. The molecular nature of R-factors. J Mol Biol. 1966 May;17(1):102–116. doi: 10.1016/s0022-2836(66)80097-9. [DOI] [PubMed] [Google Scholar]
- Hirota Y. THE EFFECT OF ACRIDINE DYES ON MATING TYPE FACTORS IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1960 Jan;46(1):57–64. doi: 10.1073/pnas.46.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Nisioka T., Mitani M., Clowes R. Composite circular forms of R factor deoxyribonucleic acid molecules. J Bacteriol. 1969 Jan;97(1):376–385. doi: 10.1128/jb.97.1.376-385.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Rownd R. Replication of a bacterial episome under relaxed control. J Mol Biol. 1969 Sep 28;44(3):387–402. doi: 10.1016/0022-2836(69)90368-4. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- WATANABE T., FUKASAWA T. Episome-mediated transfer of drug resistance in Enterobacteriaceae. II. Elimination of resistance factors with acridine dyes. J Bacteriol. 1961 May;81:679–683. doi: 10.1128/jb.81.5.679-683.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T., LYANG K. W. Episome-mediated transfer of drug resistance in Enterobacteriaceae. V. Spontaneous segregation and recombination of resistance factors in Salmonella typhimurium. J Bacteriol. 1962 Sep;84:422–430. doi: 10.1128/jb.84.3.422-430.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]




