Abstract
Few studies have addressed the expression profiles associated with progression of pancreatic cancer to advanced disease. Towards this end, we performed expression profiling of a series of normal pancreas, pancreatitis and cancer tissues representing early stage resected pancreatic cancers (stages pT2/T3), late stage unresectable cancers (stage pT4) and matched metastases to a variety of organ sites. Microarray data was analyzed using linear modeling of microarray data (LIMMA), and differentially expressed genes were subjected to Gene Set Enrichment Analysis (GSEA). While robust differences were found in primary cancers as compared to normal pancreatic tissues, no differences were found between primary cancers and metastases, whether using matched or unmatched samples. When resected pancreatic cancers were specifically compared to advanced pancreatic cancers, significant differences in gene expression were found associated with growth at the primary site. These differentially expressed genes were most prominent in gene classes that related to MAPK and Wnt pathway, metabolism, immune regulation, cell-cell and cell-matrix interactions within the infiltrating carcinoma. One candidate upregulated gene (MXI1) was validated as having increased expression in advanced stage (T4) carcinomas by real-time PCR (p<0.05) and immunolabeling (p<0.003). We conclude that in addition to the robust changes in expression that accompany pancreatic carcinogenesis additional specific changes occur in association with growth at the primary site. By contrast, metastatic spread is not accompanied by reproducible changes in gene expression. These findings add to our understanding of pancreatic cancer and offer new topics for investigation into the aggressive nature of this deadly tumor type.
Keywords: MAX-interacting protein 1, c-Myc, MAP kinase, microarray, metastasis, pancreas, carcinoma, oncogene, autopsy
Introduction
Despite several advances in our basic understanding and the clinical management of pancreatic cancer, most patients diagnosed with pancreatic cancer in the United Stages will die from this disease [1]. The late stage at which this neoplasm is usually detected, its aggressiveness, and the lack of effective systemic therapies all contribute to the poor survival rate. Approximately 80% of patients with pancreatic cancer are not candidates for surgical resection because they have either locally advanced or metastatic disease at the time of diagnosis; of those patients who undergo surgical resection, 70% will develop recurrent or metastatic disease within 1 year [2]. Despite this sobering reality, few studies are designed to understand advanced pancreatic cancers and their metastases in a comprehensive manner.
Genetic alterations of numerous genes have been identified that are fundamental to pancreatic carcinogenesis involving diverse and overlapping cellular pathways such as the cell cycle regulation, apoptosis or mitogenic signaling (reviewed in [3]). However, those genes or cellular pathways that specifically modulate metastasis formation of pancreatic cancers are not well understood. Nonetheless, a variety of studies have shown evidence for a role of proteolytic enzyme expression [4, 5], TGF-beta [6–9], Hedgehog [10] and E-cadherin signaling [11], glycoprotein expression [12, 13], decreased cell adhesion and enhanced motility [14–16] in increased metastatic ability of pancreatic cancer. By contrast, fewer studies have used unbiased methods such as whole genome expression profiling to understand the features associated with the metastatic phenotype of pancreatic cancer. Nonetheless, evidence for a gene signature associated with lymph node metastasis of pancreatic cancer has been shown [17].
An improved understanding of pancreatic cancer metastasis may afford novel strategies for intervention and treatment. Towards this goal, we have previously reported the utility of a rapid autopsy approach to the collection of high quality tissues from patients who have succumbed to metastatic pancreatic cancer [18]. We now report our analyses of the expression profiles of a series of resected pancreatic cancers and advanced stage unresectable pancreatic adenocarcinoma in an effort to begin characterization of the gene signatures associated with progression of this tumor type.
Materials and Methods
Clinical Specimens and Cell lines
Samples of pancreatic carcinoma or chronic pancreatitis were collected from resection specimens at Johns Hopkins Hospital. All samples were harvested within 1 hour from areas free of gross hemorrhage or necrosis, and were immediately snap frozen in liquid nitrogen. Samples of advanced stage and/or metastatic pancreatic carcinoma were collected from participants of the Johns Hopkins Gastrointestinal Cancer Rapid Medical Donation Program (GICRMDP) as previously described [18]. For all tissue samples used, a frozen section was prepared to confirm the diagnosis of normal tissue, chronic pancreatitis or pancreatic cancer. For cancer tissue samples, all non-neoplastic tissue (i.e. adjacent normal liver, normal lung, normal pancreas) was removed from the specimen before macrodissection of the neoplastic cells from frozen sections prepared for each sample. Only cancer tissues in which we achieved a neoplastic cellularity of at least 50% were used. Archival samples of primary and metastatic pancreatic cancer from an additional 34 patients were also obtained from the Pathology Files of The Johns Hopkins Hospital as previously described [19]. The collection and use of all fresh frozen and paraffin-embedded surgical and autopsy samples for use in this project was approved by the Johns Hopkins Institutional Review Board. The two immortalized normal pancreatic ductal epithelium cell lines HPDE and HPNE were also used and prepared as previously described [20].
RNA Extractions and Oligonucleotide Array Hybridization
Total RNA was isolated from cell lines or frozen tissues using the RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer's instructions and quantified using a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, Delaware). Only samples with yields ≥100 μg/uL total RNA and 260/230 absorbance ratios ≥ 1.9 were subjected to a second screen of RNA integrity with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). High quality samples of RNA were prepared for hybridization to U133plus2.0 microarrays according to the protocol described in the Affymetrix GeneChip® Expression Analysis Manual (Santa Clara, CA), using 3-10 μg of high quality total RNA as starting material. Microarray images were processed with Microarray Analysis Suite 5.0 (Affymetrix). All samples that demonstrated characteristics of high-quality cRNA (3′/5′ ratio of probe sets for glyceraldehyde-3-phosphate dehydrogenase <4.0) were subjected to subsequent statistical analysis.
Statistical Analyses of Microarray Data
The raw data for microarray results were normalized using the methods described by Irizarry et al [21]. Genes with expression levels below the detection limits of the Affymetrix platform and that therefore generated an absent call based on a proprietary algorithm developed by Affymetrix in all experiments were eliminated from analysis. Inter-array comparisons and determinations of false discovery rates (FDR) for each comparison were performed using the Bioconductor package ‘Limma’ [22]. Genes with p values ≤ 0.001 and with FDR values ≤ 0.30 were deemed potentially significant and selected for further study. For this study, analysis was performed with the following settings: two-class response type and log2 transformation of data. GO categories and KEGG pathways were tested using a variation on Gene Set Enrichment analysis (GSEA) [23] that is implemented in ‘Limma’ by use of a Wilcoxon test to examine whether genes in a gene category are more differentially expressed than the remaining genes. GSEA was performed using the March 2005 build of gene set collections.
Quantitative Real-Time PCR Amplification
Total RNA was extracted from tissue samples and aliquots of 1μg were reverse-transcribed to cDNA in a 20 μL final volume using the SuperScript II Reverse Transcriptase kit (Invitrogen Inc, CA). For quantitative PCR of differentially expressed genes, Taqman Gene Expression Assays were obtained from Applied Biosystems (Foster City, CA). Details of all assays used are available upon request. All reactions were performed in triplicate in the same run according to the manufacturers' instructions using 1μL of total cDNA per reaction. Negative controls in which cDNA was replaced with an equal volume of water were included in each PCR reaction to rule out contamination. Real-time quantitative RT-PCR analysis was performed using an automated sequence detection instrument (7300 Real Time PCR System, Applied Biosystems). Relative expression of mRNA in each sample was calculated using the comparative CT method as compared to the endogenous reference gene beta-GUS [24].
Immunohistochemistry and Analysis of Data
Immunolabeling was performed as described in detail in previous publications [25, 26]. The primary antibody used was goat polyclonal anti-human Mxi1 (Calbiochem, San Diego, CA #PC725) at a 1:25 dilution that was incubated at room temperature for 2 hours. Scoring of immunolabeling patterns were performed by two of the authors (D.C. and C.I.D.) at a two-headed microscope. Scoring was accomplished by independent evaluation of labeling intensity and labeling percentage within the tissue. For labeling intensity, 0 corresponded to no labeling, 1+ to weak positive labeling (labeling most convincingly seen at 10x or greater), 2+ to unequivocal positive labeling (labeling convincingly seen at 4x) and 3+ to intense positive labeling. The intensity value and the percent positive cells were multiplied to generate a Histology Score (H-score) with H = % positive cells X intensity for each tissue that was used for subsequent statistical analysis.
Statistics
All summary values are expressed as a mean ± standard deviation (SD) unless otherwise indicated. For parametric distributions a Student's T test was used, and for frequency distributions a Chi-squared test was used with modification by the Fishers exact test to account for frequency values less than 5. P values ≤0.05 were considered statistically significant.
Results
Samples and RNA Integrity
A total of 60 neoplastic samples were collected corresponding to 19 primary carcinomas and 41 samples of metastatic carcinoma to liver, lung, peritoneum or lymph node. Seven of 19 primary carcinomas were obtained from surgical resection specimens, and twelve of 19 primary carcinomas and all 41 metastases were obtained from rapid autopsy participants of the Johns Hopkins GICRMDP [18]. In addition, eight non-neoplastic tissues were collected to include three samples of chronic pancreatitis and five samples of normal bulk pancreas. Two immortalized normal pancreatic ductal epithelium cell lines (HPDE, HPNE) were also used.
When using tissues derived from different sources (i.e. resection specimens versus autopsy), it is conceivable that differences in gene expression detected by sensitive microarray analyses may simply reflect differences in RNA quality. To control for this possibility, all samples with adequate yields and purity were subjected to electrophoretic analysis on an Agilent Bioanalyzer. Despite seemingly high quality of many samples (both resected and autopsy tissues) based on yields and absorbance ratios, many showed evidence of significant RNA degradation by electropherogram. These were also excluded, leaving 36 high quality samples of the original 60 for microarray analysis corresponding to five surgically resected primary carcinomas (stage pT2 or pT3), five autopsy derived primary carcinomas (all pT4), 20 metastatic pancreatic carcinomas representing seven liver metastases, five lung metastases, five peritoneal metastases and three lymph node metastases, and two samples each of chronic pancreatitis, normal pancreas, and normal duct epithelium. Among these samples, no significant difference was found for the source of tissue (surgery versus autopsy) and GAPDH 3′/5′ ratios (1.46±0.42 versus 1.73±0.27, p=0.19). The clinicopathologic features of these patients whose RNA samples were ultimately used for microarray analysis are shown in Table 1.
Table 1.
Patient and sample characteristics used for microarray analysis
| Samples used for Microarray Analysisc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Age/Gender | Sample Origin | Tumor Location | Tumor Differentiation | Pathologic Stage at Time of Sampling | Primary Carcinoma Diameter (cm) | Gross Metastatic Disease | PR | LV | LU | LN | PE |
| S8 | 57F | Resection | Head | Poor | pT2N1bM0 | 3.5 | No | + | ||||
| S3 | 72M | Resection | Head | Poor | pT3N1bM0 | 1.7 | No | + | ||||
| S4 | 59M | Resection | Head | Poor | pT3N1bM0 | 2.5 | No | + | ||||
| S5 | 74F | Resection | Tail | Poor | pT3N0M0 | 4.6 | No | + | ||||
| S9 | 71F | Resection | Head | Moderate/Poor | pT2N1bM0 | 2.5 | No | + | ||||
| A2 | 63M | Autopsy | Body | Poor | pT4 | 7.0b | Yes | + | + | |||
| A3 | 68F | Autopsy | Body | Moderate | pT4 | naa | Yes | + | + | |||
| A6 | 57M | Autopsy | Head | Poor | pT4 | 5.0 | Yes | + | + | + | + | |
| A10 | 60M | Autopsy | Head | Poor | pT4 | 10.0 | Yes | + | + | + | + | |
| A13 | 60M | Autopsy | Body | Poor | pT4 | 15.0 | Yes | + | + | + | ||
| A17 | 50F | Autopsy | Head | Moderate/Poor | pT4 | 4.0 | Yes | + | + | |||
| A21 | 69M | Autopsy | naa | Moderate | pT4 | 3.0b | Yes | + | + | |||
| A22 | 52M | Autopsy | Body | Moderate | pT4 | 4.5 | Yes | + | + | + | ||
| A26 | 50M | Autopsy | Tail | Moderate/Poor | pT4 | naa | Yes | + | + | |||
| A28 | 73M | Autopsy | Head | Moderate | pT4 | naa | Yes | + | ||||
The primary carcinoma of these two patients was previously resected, thus detailed information was not available.
The RNA quality of the primary carcinoma in these two patients was suboptimal for microarray evaluation.
PR, primary carcinoma; LV, liver metastasis; LU, lung metastasis; LN, lymph node metastasis; PE, peritoneal metastasis.
Expression Profiles Associated with Pancreatic Carcinogenesis
To determine the changes in gene expression associated with pancreatic carcinogenesis in our sample set, we compared the 10 primary pancreatic cancers (both surgically resected and autopsy) to six non-neoplastic samples represented by two samples of bulk normal pancreas, two samples of chronic pancreatitis and two samples of normal duct epithelium. This analysis identified 8352 probesets with an estimated FDR of ≤0.30 indicating those genes whose expression was significantly different in cancer versus normal samples and with a false discovery rate of 30%. Of these, 171 probesets had an estimated FDR of ≤0.05 and 825 probesets had an estimated FDR of ≤0.10. The subset of known genes with a FDR of ≤0.05 within the larger group of 8352 probesets encoded proteins known to play a role in pancreatic carcinogenesis and behavior, including members of the Notch signaling pathway (NOTCH1, HES1) [27, 28], the WNT signaling pathway (WNT5A and WNT5B) [29], the MAPK signaling pathway (DUSP6) [30], angiogenesis (VEGF) [31, 32], and cell-cell contacts (CLDN4, MUC1) [12, 26, 33] among others (Supplementary Table 1). These genes not only confirm the many gene expression profiling studies that have implicating these gene products in pancreatic cancer, but also validate our sample set and methodological approach as well in identifying gene expression of biologic significance.
Expression Profiles Associated with Metastatic Disease
As the ultimate purpose of our study was to understand the expression profiles associated with pancreatic cancer progression, we next compared the 10 primary carcinomas to the 20 metastatic carcinomas from a variety of target sites. No robust differences in gene expression were found, with an estimated false discovery rate of 1.0 for all genes in this comparison, indicating the observed differences are no greater than expected by chance. To determine if the lack of detectable changes in gene expression was due to the use of unmatched samples, we repeated this comparison using only the five primary carcinomas for which their 11 matched distant metastases were available. Similar to our finding using the entire subset of primary and metastatic carcinoma samples, no remarkable differences in gene expression were found for these matched primary and metastatic carcinomas, with all estimated false discovery rates ≥0.81. It is important to note that these findings do not rule out the presence of gene expression of biologic significance in pancreatic cancer metastasis; rather, it indicates that only a few genes with potential differences in expression exist within the range of false discovery. Finally, we determined if changes in gene expression associated with progression could best be detected when each patient's primary carcinoma was individually compared to their own matched metastases. Several genes showed fold-change differences in each matched primary and metastatic carcinoma, although each gene list was largely unique to each matched pair (Supplementary Table 2). Nonetheless, two genes were present in four of five matched pairs (PCK1 and SFRP2) and four genes were present in three of five matched pairs (COL10A1, FBXO32, MFAP5, and PDGFD). In addition, 31 genes were present in two of five matched pairs that included MMP9, a gene well described as playing a role in metastatic ability [34]. Thus, unlike pancreatic carcinogenesis that is associated with numerous robust and reproducible changes in gene expression, few differences exist among primary infiltrating carcinomas and their matched distant metastases, at least at the level of mRNA expression.
Expression Profiles Associated with Advanced Tumor Stage
We next compared the five surgically resected primary carcinomas (pathologic stage pT2/pT3) to the five primary carcinomas with co-existent gross metastatic disease (pathologic stage pT4) that were obtained at autopsy. This comparison identified 242 probesets (137 upregulated and 105 downregulated) in the pT4 carcinomas as compared to the pT2/pT3 carcinomas corresponding to 173 known genes (Supplementary Table 3). To determine the biologic significance of those genes or pathways most represented in this gene set, we performed Gene Set Enrichment Analysis (GSEA) [23]. GSEA revealed gene expression associated with advanced stage carcinomas was related to a variety of pathways including MAPK signaling, T and B cell receptor signaling, cell adhesion, cell adhesion and Wnt signaling (Table 2).
Table 2.
Top 20 KEGG pathways enriched in advanced stage pancreatic carcinoma microarray data
| KEGG IDa | Pathway | P value | Gene(s)a |
|---|---|---|---|
| hsa03010 | Ribosome | 0 | RPS5, RPS26 |
| hsa04010 | MAPK signaling pathway | 0 | FOSB, TAOK1, EGFR, PRKAB2 |
| hsa04360 | Axon Guidance | 0 | - |
| hsa04510 | Focal adhesion | 0 | EGF, PTEN, THBS1, VWF, EGFR |
| hsa04514 | Cell adhesion molecules (CAMs) | 0 | HLA-DOA, HLA-DQB1 |
| hsa04520 | Adherens junction | 0 | WASL, EGFR |
| hsa04530 | Tight junction | 0 | PPP2R1B |
| hsa04612 | Antigen processing and presentation | 0 | HLA-DOA, HLA-DQB 1 |
| hsa04640 | Hematopoietic cell lineage | 0 | HLA-DRB4, HLA-DRB5 |
| hsa04660 | T cell receptor signaling pathway | 0 | FOSB |
| hsa04662 | B cell receptor signaling pathway | 0 | FOSB |
| hsa04664 | Fc epsilon RI signaling pathway | 0 | - |
| hsa04810 | Regulation of actin cytoskeleton | 0 | EGF, WASL, EGFR |
| gamma-Hexachlorocyclohexane | - | ||
| hsa00361 | degradation | 0.001 | |
| hsa00460 | Cyanoamino acid metabolism | 0.001 | - |
| hsa00561 | Glycerophospholipid metabolism | 0.001 | PPAPDC1B |
| hsa04120 | Ubiquitin mediated proteolysis | 0.001 | UBE2D4 |
| hsa04310 | Wnt signaling pathway | 0.001 | THEM4(CTBP1), PPP2R1B |
| hsa04670 | Leukocyte transendothelial migration | 0.001 | - |
| hsa04910 | Insulin signaling pathway | 0.001 | PRKAB2, SOCS3 |
Pathway identifiers as listed on the KEGG Pathway Finder at T1D Base (http://www.t1dbase.org). The genes shown in the right-most column are representative genes that were used for KEGG pathway analysis.
To verify our findings, we first focused upon four candidate upregulated genes (MAX Interacting Protein 1 (MXI1); Protein Kinase, AMP-Activated, Noncatalytic, Beta-2 (PRKAB2); Protein-Tyrosine Phosphatase, Receptor-Type Gamma (PTPRG) and Avian Musculo-aponeurotic Fibrosarcoma Oncogene Homolog (MAF)) and four candidate downregulated genes (Tribbles, Drosophila Homolog of 1 (TRIB1); Epidermal Growth Factor Receptor (EGFR); C-Terminal Modulator Protein (THEM4/CTMP); and Suppressor of Cytokine Signaling 3 (SOCS3)) in advanced stage carcinomas using semi-quantitative real-time PCR of cDNAs prepared from the same carcinomas used for microarray analysis. All candidate upregulated genes were verified as having increased expression in advanced stage carcinomas compared to resectable carcinomas, ranging from 1.94 fold increased for MAF to 5.59 fold increased for MXI1 (data not shown). However, candidate down-regulated genes showed no difference in mRNA expression by quantitative PCR indicating these genes represent false positives in our statistical analysis of microarray data.
We next analyzed the four validated upregulated genes by qPCR in an independent set of ten pT1-T3 resected primary carcinomas and five matched pairs of pT4 stage primary carcinomas and their liver metastases. Consistent with our findings on microarray, MXI1 showed an 8.7 fold increase in gene expression in pT4 advanced stage primary carcinomas as compared to the levels found pT1-pT3 stage resected carcinomas, but no difference in gene expression was found specifically among the five advanced stage primary carcinomas and their matched liver metastases (Figure 1). By contrast, no significant differences in gene expression were found for MAF, PRKAB2 or PTPRG in either comparison.
Figure 1.

Verification of candidate upregulated genes in pancreatic cancers. Total mRNA from ten resected pancreatic cancers (stages pT1-pT3) and five advanced stage cancers with associated metastases (all pT4) was extracted and used to generate cDNAs for quantitative PCR of each gene. Ct values for each gene were normalized to that of beta-glucoronidase in the same sample and experiments were performed in triplicate. MXI1 is confirmed as having increased expression in pT4 carcinomas (p<0.05), whereas PRKAB2, PTPRG and MAF show no difference in expression.
To determine if increased MXI1 gene expression found by both microarrays and real-time PCR corresponds to an increase in protein expression as well, we performed immunolabeling for Mxi1 protein in an independent sampling of 13 pT2/pT3 paraffin embedded carcinomas and 10 pT4 paraffin-embedded carcinomas. Immunolabeling for Mxi1 was detected in the normal duct epithelium and in the neoplastic epithelium of all pancreatic cancers analyzed where it was localized to the nucleus although significant cytoplasmic labeling was also present (Figure 2). Labeling within normal ducts of the pancreas was confined to scattered cells within the duct whereas labeling was more diffuse within the epithelium of the carcinoma samples. However, significant heterogeneity in the intensity of labeling was found within individual pT2/T3 stage carcinomas whereas all pT4 stage carcinomas showed uniform labeling throughout the sections analyzed. Labeling was also present within the desmoplastic stroma of each carcinoma although labeling was also heterogeneous and of weak intensity compared to the epithelium of each case. When an H score was calculated specifically for the epithelium for each carcinoma, a significantly increased H score was found for the pT4 carcinomas compared to the pT2/pT3 carcinomas (279±27 versus 207±0.62, p<0.003) (Figure 3).
Figure 2.
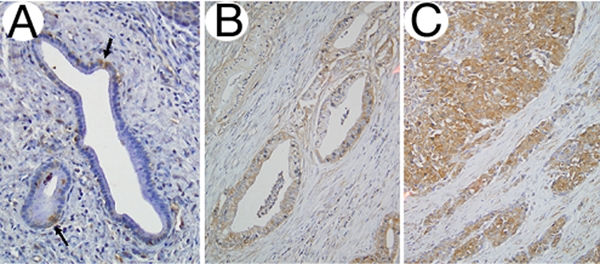
Mxi1 immunolabeling in pancreatic tissues. A. Normal pancreatic duct with scattered Mxi1 positive cells within the duct (indicated by arrows) with labeling present within the nucleus of positive cells. B. Mxi1 immunolabeling in a representative pT2 stage pancreatic cancer. The cancer glands are diffusely positive, although of less intensity than that of the representative pT4 stage pancreatic cancer shown in panel C.
Figure 3.

Summary of Mxi1 immunolabeling in pancreatic cancers. Shown are the mean histology (H) scores for the ten pT2/T3 and five pT4 pancreatic carcinomas immunolabeled for Mix1 protein. Mix1 labeling is consistently greater in the advanced stage carcinomas (p<0.003).
Discussion
Several studies have reported the gene expression profiles of infiltrating pancreatic cancers from surgical resection specimens [17, 35–42], but the expression profiles of advanced stage pancreatic cancers associated with locally destructive behavior and/or metastatic spread have not. This is largely due to the fact that advanced pancreatic cancer is not a surgical disease, thus beyond diagnosis tissue samples are often not available for research. We now report one of the first studies of advanced stage pancreatic cancer using tissues obtained from rapid autopsy of patients with end stage pancreatic cancer.
Our comparisons among primary carcinomas and their matched metastases failed to show commonly deregulated genes. This is consistent with that shown in breast cancers [43, 44] although in the study by Ramaswamy et al [43] a 17 gene metastasis signature was found in the primary carcinomas that had matched metastases, whereas no such signature was found in the current study. While this may be a reflection of our sample sizes, another possibility is that our experimental findings support the common perception that all pancreatic cancers are metastatic, even at the time of surgical resection [45]. Similar profiles among matched primary and metastatic cancers have also been suggested as evidence for development of the metastatic phenotype during carcinogenesis time of tumor formation [43, 46] although the intriguing proposal that metastatic ability is programmed in normal tissues has also been put forth [47–49]. Thus, as consistent and robust changes in gene expression have been found in association with pancreatic carcinogenesis but not metastasis formation further study of the expression profiles of pancreatic cancer and correlation to outcomes is warranted to address this possibility.
While the expression profiles of advanced stage disease were found to be highly similar to that of resected cancers, our data nonetheless suggest that additional changes in gene expression may occur within the primary site that correlates with progression to advanced stage disease. One of the changes found and validated was increased expression of MXI1, a member of the MAD family of transcriptional repressors that negatively regulates c-Myc [50]. Amplification and overexpression of c-Myc has been well described in pancreatic cancer where it acts as a central regulator of gene transcription and predisposes cells to apoptosis under nutrient, growth factor or oxygen deprivation conditions [51, 52]. Thus, while c-Myc amplification and/or overexpression may promote pancreatic carcinogenesis, up-regulation of Mxi1 may occur during disease progression to refine c-Myc oncogenic signals and to protect cancer cells from c-Myc induced apoptosis [52]. To state that this refinement causes metastasis is unlikely. Rather, with continued growth of the neoplasm within an increasingly tenuous microenvironment, such as from small cancers confined to the pancreas to bulky unresectable cancers, subclones may constantly evolve with the neoplasm to maintain a positive net growth by modulation of oncogenic signaling. Indeed, the findings of Graeber et al support this possibility, as they have shown selection of clonal variants with diminished apoptotic potential in solid human tumors due to hypoxic environments [47].
Our findings from gene set enrichment analyses provide additional insight into the expression profiles associated with pancreatic cancer progression, such as the MAPK and WNT signaling pathways. Consistent with this finding, deregulation of the MAPK and Wnt signaling pathways have been implicated in epithelial-mesenchymal transition and in the metastasis of a variety of tumor types, including pancreatic cancer [53–58]. MAPK signaling is also a central component of the oxidative stress response, consistent with ongoing MAPK pathway deregulation with tumor growth at the primary site [59]. We also found evidence suggestive of immune deregulation in tumor progression. This finding is particularly intriguing considering the emerging role of immune-boosting therapies in the treatment of pancreatic cancer [60] and supports further investigations in this area particularly as the immune system relates to tumor progression.
In summary, we provide examples of the expression profiles associated with pancreatic cancer progression and offer possibilities for further investigation into the mechanisms underlying the aggressive behavior of this disease.
Supplementary Material
Acknowledgments
This work was supported by NIH grant CA106610-01 to Christine A. Iacobuzio-Donahue), the Family of George Rubis, Sigma Beta Sorority, the Jeff Zgonina Foundation for Pancreatic Cancer Research and the Skip Viragh Foundation.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, Sauter PK, Coleman J, Hruban RH, Lillemoe KD. Resected adenocarcinoma of the pancreas-616 patients: Results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567–579. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 3.Hansel DE, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Annu Rev Genomics Hum Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- 4.Schneiderhan W, Diaz F, Fundel M, Zhou S, Siech M, Hasel C, Moller P, Gschwend JE, Seufferlein T, Gress T, Adler G, Bachem MG. Pancreatic stellate cells are an important source of mmp-2 in human pancreatic cancer and accelerate tumor progression in a murine xenograft model and cam assay. J Cell Sci. 2007;120:512–519. doi: 10.1242/jcs.03347. [DOI] [PubMed] [Google Scholar]
- 5.Ellenrieder V, Hendler SF, Ruhland C, Boeck W, Adler G, Gress TM. Tgf-beta-induced invasiveness of pancreatic cancer cells is mediated by matrix metalloproteinase-2 and the urokinase plasminogen activator system. Int J Cancer. 2001;93:204–211. doi: 10.1002/ijc.1330. [DOI] [PubMed] [Google Scholar]
- 6.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, Adler G, Gress TM. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal trans-differentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- 7.Jungert K, Buck A, von Wichert G, Adler G, Konig A, Buchholz M, Gress TM, Ellenrieder V. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 2007;67:1563–1570. doi: 10.1158/0008-5472.CAN-06-1670. [DOI] [PubMed] [Google Scholar]
- 8.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, Gabrielson KL, Matsui W, Maitra A. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: A new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin T, Wang C, Liu T, Zhao G, Zha Y, Yang M. Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J Surg Res. 2007;141:196–203. doi: 10.1016/j.jss.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, Goto M, Yonezawa S, Hollingsworth MA. Rna interference suppression of muc1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res. 2006;12:2976–2987. doi: 10.1158/1078-0432.CCR-05-1197. [DOI] [PubMed] [Google Scholar]
- 13.Kayed H, Kleeff J, Keleg S, Felix K, Giese T, Berger MR, Buchler MW, Friess H. Effects of bone sialoprotein on pancreatic cancer cell growth, invasion and metastasis. Cancer Lett. 2007;245:171–183. doi: 10.1016/j.canlet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Sawai H, Okada Y, Funahashi H, Matsuo Y, Takahashi H, Takeyama H, Manabe T. Integrin-linked kinase activity is associated with interleukin-1 alpha-induced progressive behavior of pancreatic cancer and poor patient survival. Oncogene. 2006;25:3237–3246. doi: 10.1038/sj.onc.1209356. [DOI] [PubMed] [Google Scholar]
- 15.Sawai H, Okada Y, Funahashi H, Matsuo Y, Takahashi H, Takeyama H, Manabe T. Interleukin-1alpha enhances the aggressive behavior of pancreatic cancer cells by regulating the alpha6beta1-integrin and urokinase plasminogen activator receptor expression. BMC Cell Biol. 2006;7:8. doi: 10.1186/1471-2121-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu CS, Xue B, Tu C, Feng ZH, Shi YH, Miao Y, Wen CJ. Nrage suppresses metastasis of melanoma and pancreatic cancer in vitro and in vivo. Cancer Lett. 2007;250:268–275. doi: 10.1016/j.canlet.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Kim HN, Choi DW, Lee KT, Lee JK, Heo JS, Choi SH, Paik SW, Rhee JC, Lowe AW. Gene expression profiling in lymph node-positive and lymph node-negative pancreatic cancer. Pancreas. 2007;34:325–334. doi: 10.1097/MPA.0b013e3180317b01. [DOI] [PubMed] [Google Scholar]
- 18.Embuscado EE, Laheru D, Ricci F, Yun KJ, de Boom Witzel S, Seigel A, Flickinger K, Hidalgo M, Bova GS, Iacobuzio-Donahue CA. Immortalizing the complexity of cancer metastasis: Genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–554. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin W, Yun KJ, Ricci F, Zahurak M, Qiu W, Su GH, Yeo CJ, Hruban RH, Kern SE, Iacobuzio-Donahue CA. Map2k4/mkk4 expression in pancreatic cancer: Genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 20.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Wettenhall JM, Smyth GK. Limmagui: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Mudali SV, Fu B, Lakkur SS, Luo M, Embuscado EE, Iacobuzio-Donahue CA. Patterns of epha2 protein expression in primary and metastatic pancreatic carcinoma and correlation with genetic status. Clin Exp Metastasis. 2006;23:357–365. doi: 10.1007/s10585-006-9045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: Support for use as a therapeutic target. Am J Clin Pathol. 2004;121:226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, Schmid RM, Leach SD. Notch mediates tgf alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 29.Ripka S, Konig A, Buchholz M, Wagner M, Sipos B, Kloppel G, Downward J, Gress T, Michl P. Wnt5a–target of cutl1 and potent modulator of tumor cell migration and invasion in pancreatic cancer. Carcinogenesis. 2007;28:1178–1187. doi: 10.1093/carcin/bgl255. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. Potential tumor suppressive pathway involving dusp6/mkp-3 in pancreatic cancer. Am J Pathol. 2003;162:1807–1815. doi: 10.1016/S0002-9440(10)64315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang AD, Camp ER, Fan F, Shen L, Gray MJ, Liu W, Somcio R, Bauer TW, Wu Y, Hicklin DJ, Ellis LM. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66:46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 32.Tang RF, Wang SX, Peng L, Wang SX, Zhang M, Li ZF, Zhang ZM, Xiao Y, Zhang FR. Expression of vascular endothelial growth factors a and c in human pancreatic cancer. World J Gastroenterol. 2006;12:280–286. doi: 10.3748/wjg.v12.i2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Lohr M, Leder G, Iwamura T, Adler G, Gress TM. Claudin-4 expression decreases invasive-ness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–6271. [PubMed] [Google Scholar]
- 34.Overall CM, Kleifeld O. Tumour microenvironment – opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 35.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: A comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 36.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 37.Missiaglia E, Blaveri E, Terris B, Wang YH, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Lemoine NR. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 38.Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, Kalthoff H, Kremer B, Kloppel G, Saeger HD. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA micro-arrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grutzmann R, Boriss H, Ammerpohl O, Luttges J, Kalthoff H, Schackert HK, Kloppel G, Saeger HD, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 40.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cdna microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 41.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 42.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cdna microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 44.Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ducreux M, Boige V, Malka D. Treatment of advanced pancreatic cancer. Semin Oncol. 2007;34:S25–30. doi: 10.1053/j.seminoncol.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 47.Hunter KW. Host genetics and tumour metastasis. Br J Cancer. 2004;90:752–755. doi: 10.1038/sj.bjc.6601590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malins DC, Gilman NK, Green VM, Wheeler TM, Barker EA, Anderson KM. A cancer DNA phenotype in healthy prostates, conserved in tumors and adjacent normal cells, implies a relationship to carcinogenesis. Proc Natl Acad Sci USA. 2005;102:19093–19096. doi: 10.1073/pnas.0509630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malins DC, Gilman NK, Green VM, Wheeler TM, Barker EA, Vinson MA, Sayeeduddin M, Hellstrom KE, Anderson KM. Metastatic cancer DNA phenotype identified in normal tissues surrounding metastasizing prostate carcinomas. Proc Natl Acad Sci USA. 2004;101:11428–11431. doi: 10.1073/pnas.0404572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurlin PJ, Huang J. The max-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 52.Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El-Deiry WS. Mxi1 is induced by hypoxia in a hif-1-dependent manner and protects cells from c-myc-induced apoptosis. Cancer Biol Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- 53.Tan X, Tamori Y, Egami H, Ishikawa S, Kurizaki T, Takai E, Hirota M, Ogawa M. Analysis of invasion-metastasis mechanism in pancreatic cancer: Involvement of tight junction transmembrane protein occludin and mek/erk signal transduction pathway in cancer cell dissociation. Oncol Rep. 2004;11:993–998. [PubMed] [Google Scholar]
- 54.Dissanayake SK, Wade M, Johnson CE, O'Connell MP, Leotlela PD, French AD, Shah KV, Hewitt KJ, Rosenthal DT, Indig FE, Jiang Y, Nickoloff BJ, Taub DD, Trent JM, Moon RT, Bittner M, Weeraratna AT. The wnt5a/protein kinase c pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo Y, Zi X, Koontz Z, Kim A, Xie J, Gorlick R, Holcombe RF, Hoang BH. Blocking wnt/lrp5 signaling by a soluble receptor modulates the epithelial to mesenchymal transition and suppresses met and metalloproteinases in osteosarcoma saos-2 cells. J Orthop Res. 2007;25:964–971. doi: 10.1002/jor.20356. [DOI] [PubMed] [Google Scholar]
- 56.Zhao CH, Bu XM, Zhang N. Hyper-methylation and aberrant expression of wnt antagonist secreted frizzled-related protein 1 in gastric cancer. World J Gastroenterol. 2007;13:2214–2217. doi: 10.3748/wjg.v13.i15.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendes O, Kim HT, Lungu G, Stoica G. Mmp2 role in breast cancer brain metastasis development and its regulation by TIMP2 and ERK1/2. Clin Exp Metastasis. 2007;24:341–351. doi: 10.1007/s10585-007-9071-0. [DOI] [PubMed] [Google Scholar]
- 58.Hulit J, Suyama K, Chung S, Keren R, Agiostratidou G, Shan W, Dong X, Williams TM, Lisanti MP, Knudsen K, Hazan RB. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007;67:3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 59.Wu JJ, Bennett AM. Essential role for mitogen-activated protein (map) kinase phosphatase-1 in stress-responsive map kinase and cell survival signaling. J Biol Chem. 2005;280:16461–16466. doi: 10.1074/jbc.M501762200. [DOI] [PubMed] [Google Scholar]
- 60.Laheru D, Jaffee EM. Immunotherapy for pancreatic cancer - science driving clinical progress. Nat Rev Cancer. 2005;5:459–467. doi: 10.1038/nrc1630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


