Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignancies with high mortality, but its underlying molecular mechanisms remain not well understood. High-throughput, proteomic techniques targeting unique biological molecules may provide novel insights into HCC pathogenesis and prognosis. In this study, we systemically investigated tissue biomarkers of HCC by using surface-enhanced laser desorption and ionization time-of-flight mass spectrometry (SELDI-TOF-MS) technique. Proteomic spectra were generated from fresh tissues (26 HCC and 18 control cirrhotic liver) and analyzed by using Biomarker Wizard System. A total of 16 differential proteomic peaks were detected between HCC and cirrhotic liver tissues, and 11 between moderately and highly differentiated HCCs. The expression pattern of one proteomic peak was validated by immunohistochemistry. These molecules are potential candidate biomarkers for early diagnosis of and targeted therapy for HCC.
Keywords: Hepatocellular carcinoma, proteomic profiling, biomarker, mass spectrometry, immunohistochemistry
Introduction
Hepatocellular carcinoma (HCC) ranks fifth in prevalence worldwide among all malignancies and causes about 1 million deaths annually [1]. A rise in incidence and mortality from HCC has recently been observed in most industrialized countries. The etiology of HCC includes chronic viral hepatitis, alcohol abuse, metabolic disorders or other environmental agents. Among them, hepatitis B virus (HBV) infection is the major cause of HCC in China.
Pathologic features, such as tumor size and number, status of capsule or venous invasion, presence or absence of satellite nodules, and advanced pTNM stage are the best established risk factors for recurrence and prognosis in patients with HCC. Recent molecular research has identified many tumor biological molecules as the potential prognostic markers (biomarkers).
Considering the high fatality of HCC, early diagnosis is the key to increase the cure rate. At present, alpha fetoprotein (AFP) is the only serological marker available, but its diagnostic accuracy is unsatisfactory because of low sensitivity despite reliable specificity [2, 3]. It is, therefore, essential to identify new sensitive and specific biomarkers for early diagnosis of HCC.
The traditional technique used to discover new cancer-associated proteins is the two dimensional gel electrophoresis (2-DE) [4]. This technique allows high-resolution separation of proteins, good reproducibility and sufficient sensitivity, but it is neither a large-scale nor a high throughput technology. Nevertheless, in combination with mass spectrometry (MS), 2-DE can be a powerful method for separation and subsequent identification of proteins of interest. In particular, matrix assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF) is commonly used for the accurate measurement of the molecular masses of peptides derived from in-gel digested proteins and subsequent identification by peptide fingerprinting [5–7]. In contrast, the direct analysis of non-pre-fractionated biological samples is beyond the scope of traditional MALDI-TOF, limited by the complexity of the analyte, the presence of buffer components like salts or detergents as well as of non-protein components such as lipids and carbohydrates. Surface-enhanced laser desorption/ionization (SELDI) is a proteomic high-throughput technique that uses chromatographic surfaces able to retain proteins depending on their physicochemical properties followed by direct analysis via time of light mass spectrometry (TOF-MS) [8]. A number of studies using the ProteinChip technology have been carried out for proteomic profiling of biological fluids, including serums [9–11]. Because this technique demands only a small amount of samples, it is also ideal for small biopsies or microdissected tissues. Microdissected tissue material free from contaminating and unwanted tissue components is extremely important for the production of clean data for biomarker identification in cancer diagnostics and in elucidating clonal heterogeneity of tumors. Melle et al [12] showed that the detection of differentially expressed proteins was only possible in pure microdissected samples. Laser-based microdissection has previously been combined with ProteinChip technology to identify protein markers in other cancers [13–15].
In the present study, we analyzed the proteomic profiles in 26 HCC and 18 cirrhotic liver tissues using ProteinChip Technology. We identified the series of unique proteins by combined proteomic informatics and SELDI technique, and confirmed the identity of some protein molecules by immunohistochemistry.
Materials and Methods
Sample Collection and Clinical Data
Fresh tissues of HCC were collected from Beijing 306 Hospital, Armed Police General Hospital and Beijing Friendship Hospital, China. After surgical resection, the adjacent tissues of cancer were cut and immediately snap frozen in liquid nitrogen and stored at −80°C. A total of 26 HCC samples were collected for this study, including 3 cases of poorly differentiated, 13 moderately differentiated, and 10 well differentiated tumors. In addition, 18 cirrhotic liver tissue samples were also collected at the time of surgery. All patients had a history of HBV-related hepatitis. Table 1 summarizes the clinicopathological data of these patients. Formalin-fixed, paraffin-embedded tissue sections of the above cases were prepared for immunohistochemical staining.
Table 1.
Clinicopathological data of patients with HCC and posthepatitic cirrhosis
| Tissue type | Number |
|---|---|
| HCC | 26 |
| Mean age (age range) | 55.5 (34–68) |
| Sex | |
| Male | 25 |
| Female | 1 |
| Subtype | |
| Poorly differentiated | 3 |
| Moderately differentiated | 13 |
| Well differentiated | 10 |
| Tumor size | |
| >2 cm | 15 |
| <2 cm | 11 |
| AFP status | |
| Positive | 15 |
| Negative | 11 |
| Cirrhotic liver | 18 |
| Mean age (age range) | 51 (30–70) |
| Sex | |
| Male | 15 |
| Female | 3 |
Preparation of Samples for SELDI-TOF-MS Analysis
Approximately 100 mg of fresh tissue (HCC or cirrhotic liver) was prepared for analysis using SELDI-TOF-MS as described by Scarlett et al [16]. Briefly, the tissue was solubilized via pestle homogenization in 0.5 mL of U9 buffer (9.5M/L urea/2% CHAPS/1% DTT, 50 mM/L Tris-HCl, pH9.0) on ice, then added to a QiaShredder spin column (Qiagen, Hilden, Germany) and centrifuged (13,000 rpm 4°C for 10 min) to remove insoluble material. The tissue homogenate was further centrifuged (40,000 rpm, 15°C for 2 h) and stored in 50 µL aliquots at −80°C. Before experiment, the protein level of the lysate was determined to be 2 to 10 g/L by Coomassie brilliant blue method. The tissue homogenate was defrosted on ice, and 10 μL sample was added to 20 μL U9 buffer and vortexed 30 min in iced bath. Then 360 μL binding buffer (100 mmoL/L NaAc, pH4.0) was added into it, admixed even, standing for 30 min.
The protein lysates from HCC and control cirrhotic liver tissue were analyzed on a weak cation exchange arrays (CM10; Ciphergen Biosystems, Inc., Fremont, CA) as described elsewhere [17]. In brief, weak cation exchanger arrays were pre-equilibrated with binding buffer (100 mmoL/L NaAc, pH4.0) for 5 minutes at room temperature twice. The binding buffer was then removed and 100 μL of sample mixture was added to each spot. The arrays were then incubated in a humidified chamber at room temperature for 1 hour, washed 2 times in binding buffer for 5 minutes each time, rinsed once in water, and allowed to air-dry. Each spot was treated with 0.5 μL of sinapinic acid (sinapinic acid, 50% saturated in 50% acetonitrile/0.5% TFA), and allowed to air dry. The protein chip arrays were then analyzed using the Ciphergen Protein Biological System IIc and ProteinChip Reader (Ciphergen Biosystems).
SELDI-TOF-MS Analysis
Mass spectra were setting for each sample with a laser intensity setting of 185. The laser was optimized for data collection between 2 and 20kD and the highest detection molecular weight is 200kD, with detector sensitivity set at 9. Considering the groundmass peaks, the peaks <1 kD were deflected away from the detector. The instrument was rectified by ALL-IN-ONE protein standard molecule (Ciphergen, USA) everyday. The mass deviation of system is ≤0.1%. All spectra were analyzed using Ciphergen ProteinChip Software Version 3.1 (Ciphergen Biosystems).
Immunohistochemistry
The sections were stained immunohisto-chemically by the PV-6000 method and bax antibody (Zymed, USA). Sections of 4 μm formalin-fixed paraffin-embedded HCC and cirrhotic liver tissue were air-dried for approximately 30 min at 65°C. For immunostaining, the sections were deparaffinized, rehydrated and washed with phosphate-buffered saline (PBS, pH 7.4) (3×3 min). Antigen was retrieved in 0.01 mol/L citrate buffer solution, pH 6.0 using a pressure cooker. To inhibit endogenous peroxidase activity, slides were sealed in 3% H2O2 for 10 min. Then slides were sealed with 1:15 cow serum 37°C for 15 min. Subsequently, they were incubated overnight at 4°C in a humidity chamber with the corresponding primary anti-bax antibody (Maixin, China). Secondary antibodies, anti-mouse IgG was applied for 30 min, visualized with diaminobenzidine (DAB), rinsed and soaked in PBS for 3–5 min, thrice after each step. They were counterstained with Mayer hematoxylin. The immunostaining results were graded as −, + or ++ when less than 10%, 10–30% and more than 30% of the hepatic parenchymal cells were positive [18].
Statistical Analysis
Statistical treatment of data was performed using Ciphergen ProteinChip Software Version 3.1 (Ciphergen Biosystems). A P value <0.01 was considered statistically significant. The data was analyzed and modeled using Biomarker patterns system.
Results
Proteomic Profiling of Tumor and Non-tumor Tissues
Compared the proteomic profiling of the tumor with the non-tumor group, there existed 16 stable differential expression proteins. Among them, 7 were up-regulated, and 9 were down-regulated (Table 2, Figures 1, 2 and 3). By enquiring ExPasy protein database (http://www.expasy.org), we inferred the candidate proteins based on the definite molecular mass. As search standards, the molecular mass was set as m/z±0.1%, PI 9.0±5.0 (Table 3). Some protein or peptide molecule had only one retrieval result, so the result was inferred as the protein with the same molecular mass. Others had more than one results. Based on their histological origin and biological functions, we made an initial screen and determined the candidate proteins (Table 3, highlighted in blue).
Table 2.
Differential expression of proteins detected in HCC
| Theoretical MW (Da) | Expression | P values |
|---|---|---|
| 2273 | ↓ | 0.00771 |
| 2641 | ↓ | 0.00254 |
| 4524 | ↓ | 0.00655 |
| 4674 | ↓ | 0.00011 |
| 4974 | ↑ | 0.00711 |
| 6758 | ↓ | 0.00836 |
| 7219 | ↓ | 0.00017 |
| 7431 | ↓ | 0.00771 |
| 7576 | ↑ | 0.00555 |
| 7678 | ↑ | 0.00212 |
| 9846 | ↓ | 0.00001 |
| 10747 | ↓ | 0.00026 |
| 15137 | ↑ | 0.00469 |
| 15341 | ↑ | 0.00510 |
| 31037 | ↑ | 0.00362 |
| 31748 | ↑ | 0.00161 |
↑ and ↓ indicate increased and decreased expression in HCC as compared to control cirrhotic liver, respectively.
Figure 1.
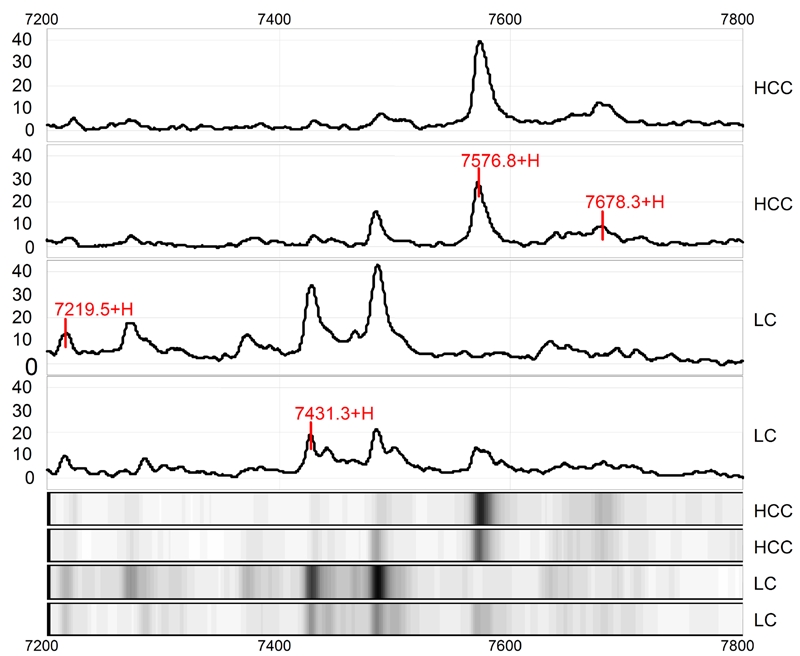
Protein finger printing of hepatocellular carcinoma (HCC) and control cirrhotic liver (LC) in range from 7200 to 7800Da. The protein peaks 7219, 7431, 7576 and 7678Da are randomly extracted from HCC protein finger printing. Among them, 7219 and 7431Da proteins are down-regulated and 7576 while 7678Da proteins are up-regulated in HCC as compared to LC. The next 4 tracts are simulated electropherograms of the above-mentioned results.
Figure 2.
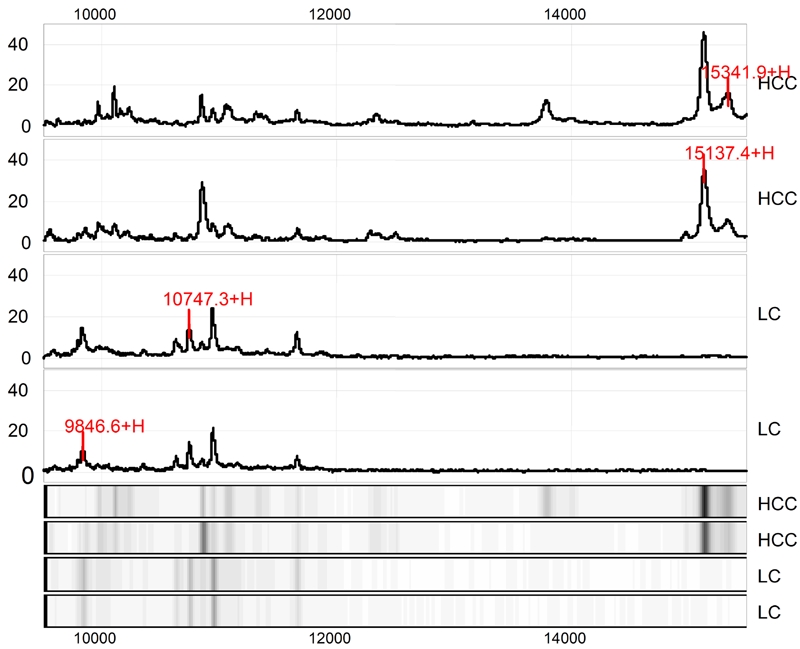
Protein finger printing of hepatocellular carcinoma (HCC) and control cirrhotic liver (LC) in range from 9500 to 16000Da. The protein peaks 9846, 10747, 15137 and 15341Da are randomly extracted from HCC protein finger printing. Among them, 9846Da and 10747Da proteins are down-regulated while 15137Da and 15147Da proteins are up-regulated in HCC as compared to LC. The next 4 tracts are simulated electropherograms of the above-mentioned results.
Figure 3.
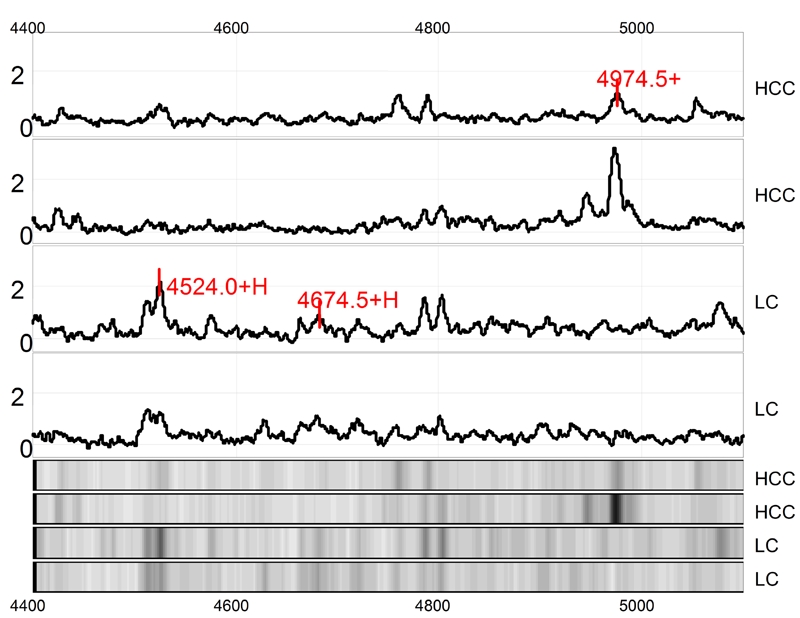
Protein finger printing of hepatocellular carcinoma (HCC) and control cirrhotic liver (LC) in range from 4400 to 5200 Da. The protein peaks 4524, 4674 and 4974Da were randomly extracted form HCC protein finger printing. Among them, 4524 and 4674Da proteins are down-regulated while 4974 Da protein is up-regulated in HCC as compared to LC. The next 4 tracts are simulated electropherograms of the above-mentioned results.
Table 3.
Candidate protein (peptide) biomarkers in HCC
| Theoretical MW (Da) | Candidate protein (peptide) inferred |
|---|---|
| 9846.428 | Anaphase-promoting complex subunit 11 (Q9NYG5) |
| Cyclin-dependent kinases regulatory subunit 2(P33552) | |
| Pro-MCH-like protein 2 (Q9BQD1) | |
| Protein transport protein S61 subunit beta (P60468) | |
| Caspase-9 subunit p10 (P55211) | |
| 4674.091 | BAX protein, cytoplasmic isoform gamma (Q07815) |
| 7219.294 | Platelet basic protein precursor (P02775) |
| 10747.82 | Cholecystokinins precursor (P06307) |
| NADH-ubiquinone oxidoreductase chain 4L (P03901) | |
| Protein S100-A5 (P33763) | |
| 31748.55 | Nit protein 2 (Q86X76) |
| 7678.362 | S-adenosylmethionine decarboxylase proenzyme (P17707) |
| Platelet factor 4 variant precursor (P10720) | |
| 2641.05 | Endothelin-3 precursor (P14138) |
| 31037.76 | Enoyl-coenzyme A hydratase 1, short-chain (Q58EZ5) |
| 15137.76 | Vascular endothelial growth factor (VEGF) isoform |
| 15341.82 | Growth/differentiation factor 9 precursor (O60383) |
| Beta-glucuronidase-like protein 1 (Q9NQQ0) | |
| Interleukin-2 precursor (P09105) | |
| Interleukin-2 precursor (P60568) | |
| 7576.037 | Dipeptidyl-peptidase 1 precursor (P53634) |
| 4524.928 | Orexigenic neuropeptide QRFP precursor (P83859) |
| 4974.046 | Beta-defensin 107A precursor (Q8IZN7) |
| Gastric inhibitory polypeptide precursor (P09681) | |
| 2273.905 | Prolactin-releasing peptide precursor (P81277) |
| 7431.591 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-T2 subunit |
| precursor(O14610) | |
| Growth-regulated protein alpha precursor (P09341) | |
| 6758.022 | Cytochrome c oxidase polypeptide VIIa-heart, mitochondrial precursor (P24310) |
Note: The number in parentheses is the serial number of the protein in ExPASY database.
Proteomic Profiling of HCC with Different Clinicopathological Features
Based on the AFP immunostaining results, we divided the HCC cases into AFP-positive and AFP-negative, and analyzed their proteomic profiling separately. However, the data did not show any statistical differences of candidate proteins between the two groups.
By analyzing the proteomic profiling of 3 poorly differentiated, 13 moderately differentiated and 10 well differentiated HCC tissues, we found that there was no statistical difference between poorly differentiated and moderately differentiated tumors, and between poorly differentiated and well differentiated tumors. However, there existed 11 protein markers of statistical significance between moderately differentiated and well differentiated HCCs. Among them, 3 were low-expression and 8 were high-expression in moderately differentiated HCC. By enquiring ExPasy protein database, we preliminarily determined the candidate proteins of definite molecular mass. As search standards, the molecular mass is set as m/z ±0.1% and PI 9.0±5.0. Considering their histological origin and biological function, we made an initial screen of candidate proteins (peptides) (Table 4).
Table 4.
Candidate protein (peptide) biomarkers between moderately and well differentiated HCC
| Theoretical MW (Da) | Expression | P value | Candidate protein (peptide) inferred |
|---|---|---|---|
| 10945.197 | ↓ | 0.00919 | 60S ribosomal protein L37 (P61927) |
| 11675.955 | ↓ | 0.00034 | Glycoprotein hormone alpha-2 precursor |
| 11893.256 | ↓ | 0.00309 | (Q96T91) |
| 2553.324 | ↑ | 0.00541 | Caspase-2 precursor (P42575) |
| 2911.205 | ↑ | 0.00707 | Endothelin-2 precursor (P20800) |
| 3099.609 | ↑ | 0.00919 | Orexin precursor (O43612) |
| 6186.219 | ↑ | 0.00541 | Neuropeptide B precursor (Q8NG41) |
| 6554.271 | ↑ | 0.00410 | 60S ribosomal protein L40 (P62987) |
| 9627.260 | ↑ | 0.00541 | Surfactant-associated protein G (Q6UW10) |
| 7155.589 | ↑ | 0.00919 | Cytochrome c oxidase polypeptide VIa-liver, mitochondrial precursor (P12074) |
| FXYD domain-containing ion transport regulator 3 precursor (Q14802) |
↑ and ↓ indicate up-regulation and down-regulation of expression in moderately differentiated HCC tissue as compared to well differentiated HCC. The number in parenthesis is the serial number of the protein in ExPASY database.
Immunohistochemistry of Bax
To confirm the identified biomarkers, we chose bax protein (4674 Da in proteomic profiling) and examined its expression in all hepatic tissue samples by immunohistochemistry. In HCC group, most cases (18 of 26, 69.2%) were negative, 2 were weakly positive (7.6%) and 6 were positive (23.1%). In well differentiated HCC, 2 of 10 cases were positive (20%), 1 was weakly positive (10%), and 7 were negative (70%). In moderately differentiated HCC, 4 of 13 cases were positive (30.7%), 1 was weakly positive (7.7%), and 8 were negative (61.5%). All poorly differentiated HCC was negative (100%) (Figure 4A). In contrast, all 18 liver tissues with cirrhosis were positive for bax (17 strongly positive and 1 weakly positive) (Figure 4B).
Figure 4.
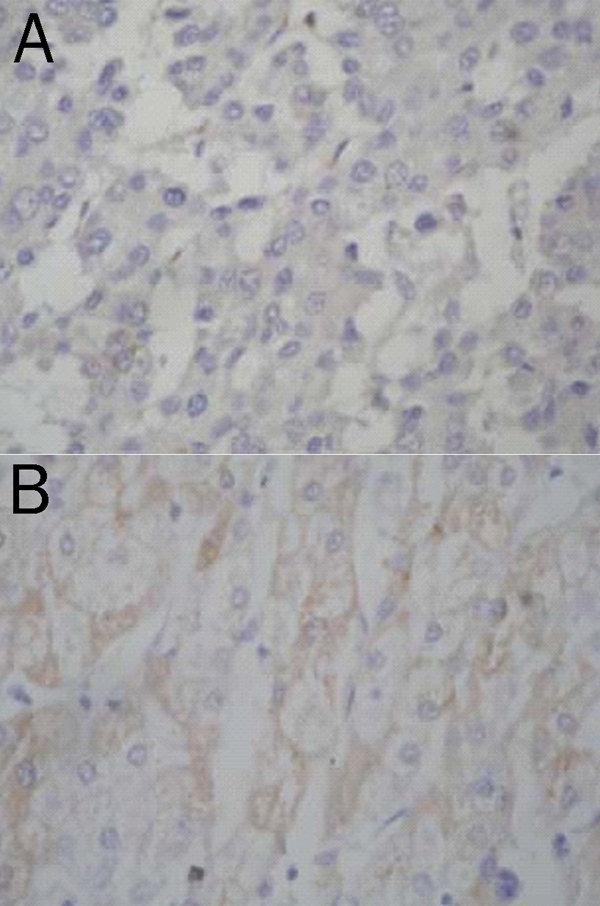
Expression of Bax in hepatocellular carcinoma (A) and non-tumor hepatic cells (B). (Immunostain for bax, × 400)
Then we used Biomarker Patterns System to analyze the protein profiling. 4674Da was designated as a standard marker of this group, and the results showed that this protein peak was low in 18 of 26 HCC and high in the remaining 8 HCC tissues. In all 18 liver tissues with cirrhosis, the 4674Da protein peak was high. These findings were consistent with the immunohistochemical results.
The difference in the expression level of bax between HCC and hepatic cirrhosis was statistically significant (P<0.05). There was no significant correlation between bax expression and age (P=0.17), sex (P =0.09) or the degree of HCC differentiation (P =0.07).
Discussion
The molecular mechanisms of HCC are not well understood, although aneuploidy and multiple genetic alterations are often present. For example, mutations of AFP, p53 and TrACP have been reported in HCC, while c-myc and cyclin D1 are frequently overexpressed [19–21]. In China, HBV infection is the major cause of HCC. However, these findings do not piece together a clear portrait of HCC and none of the biological markers have been proven to be specific enough for diagnosis and prediction of prognosis. One reason might be that HCC is a very heterogeneous tumor entity [22] and multiple changes at the cellular and subcellular level occur during tumor development and progression.
To address this problem, novel automated and high throughput molecular methods have been used. One of the most promising proteomic tools for the detection of new cancer biomarkers is the ProteinChip technology [23, 24]. Preliminary studies have demonstrated the ability of SELDI-TOF-MS proteomic profiling to differentiate several cancer types from nonmalignant diseases [23, 25–26]. By comparing the proteins present in samples from diseased tissue with those present in samples from normal tissue, it is possible to identify changes in expression of proteins that potentially may be related to tumor progression, invasion and metastasis, and prognosis.
Based on our pilot study, we used weak cation exchange arrays (CM10) instead of hydrophobic protein chip or strong anionic exchanger chip in order to capture more differential proteins. There are 16 candidate protein markers of statistical significance in HCC. The most differential protein peaks are 4674Da, 7219Da and 9846Da. By searching for protein database, we could infer that they might be Bax protein, cytoplasmic isoform gamma, platelet basic protein precursor and caspase-9 subunit p10, respectively. Bax is a cell apoptotic enhancing factor. It promotes apoptosis by inducing release of caspases and activating cytochrome C [27]. Caspase-9 subunit p10 is critical in the apoptotic pathway, reduction in its expression in HCC may be important in the progression from cirrhosis to HCC.
11 proteins are differentially expressed between moderately differentiated and well differentiated HCCs. Among those, 11893Da is down-regulated in moderately differentiated HCC. 11893Da may be caspase-2 precursor, an early factor of apoptosis cascade reaction. 2553Da may represent endothelin precursor, a polypeptide with strong activity of shrinking blood vessel [28]. This study demonstrated for the first time that it exists in HCC.
This study did not discover differential protein markers between moderately differentiated and poorly differentiated HCC tissues, nor between well differentiated and poorly differentiated HCC tissues. It is possible that the number of poorly differentiated HCC samples is too small to have any statistical significance.
By SELDI-TOF-MS, we could get the similar molecular mass of protein and peptide to those obtained by other proteomic methods, but we did not know what kind of protein they were. The convenient method is searching for database well established by the international society. But some proteins were modified in the biosystem, such as glycosylation and methylation. So it is very difficult to determine a protein based only on its molecular weight, and the most precise method is still protein purification and identification.
Lin et al [29] used SELDI-TOF-MS technique to analyze the protein profile of follicular lymphoma with frozen tissue samples and found two differential proteins, 32.5kD and 11.2kD. The authors then combined proteomic informatics with immunohistochemistry and Western blot, confirmed that the two proteins were cyclin D3 and caspase-3, respectively. By a similar way, Ranganathan et al identified transthyretin, cystatin C and neuroendocrine protein 7B2 in cerebrospinal fluid of amyotrophic lateral sclerosis patients [30].
In this study, we combined ProteinChip technique with proteomic informatics to try to define candidate proteins in HCC tissues. According to its molecular weight and biological function, we assumed that 4674Da protein was bax, and used immunohisto-chemical technique to validate it. As a result, we found that the expression pattern of bax in the formalin-fixed paraffin-embedded tissue of HCC and non-tumor liver control was similar to 4674Da peaks in protein profiling.
Bax is the most important apoptosis-promoting protein. Bax induces release of cytochrome C, and activates caspase-3, resulting in nuclear fragmentation [18]. Down-regulation of bax expression in HCC may promote growth and progression of HCC cells.
In conclusion, we show in this study that ProteinChip technology can be used to define proteomic signatures specific for HCC and hepatic cirrhosis. We also show that the combination of ProteinChip, proteomic informatics and immunohistochemical technique is an easier approach to identify the candidate proteins. Nevertheless, additional studies are needed to validate these proteomic signatures by analyzing additional tissues or by examining sera from patients with HCC and hepatic cirrhosis. In addition, identifying the proteins behind this signature and analyzing their biological function will aid our understanding of their relationship to the pathogenesis and progression of HCC.
References
- 1.Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN 2000: Cancer Incidence, Mortality and Prevalence Worldwide. Lyon: IARC Scientific Publications, IARC Press; 2001. [Google Scholar]
- 2.Qin LX, Tang ZY. The prognostic significance of clinical and pathological features in hepatocellular carcinoma. World J Gastroenterol. 2002;8:193–199. doi: 10.3748/wjg.v8.i2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giannelli G, Antonaci S. New frontiers in biomarkers for hepatocellular carcinoma. Dig Liver Dis. 2006;38:854–859. doi: 10.1016/j.dld.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Klose J, Kobalz U. Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis. 1995;16:1034–1059. doi: 10.1002/elps.11501601175. [DOI] [PubMed] [Google Scholar]
- 5.Siuzdak G. Mass Spectrometry for Biotechnology. San Diego: Academic Press; 1996. [Google Scholar]
- 6.Loo JA, Brown J, Critchley G, Mitchell C, Andrews PC, Ogorzalek Loo RR. High sensitivity mass spectrometric methods for obtaining intact molecular weights from gel-separated proteins. Electrophoresis. 1999;20:743–748. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<743::AID-ELPS743>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Lahm HW, Langen H. Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis. 2000;21:2105–2114. doi: 10.1002/1522-2683(20000601)21:11<2105::AID-ELPS2105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Tang N, Tornatore P, Weinberger SR. Current developments in SELDI affinity technology. Mass Spectrom Rev. 2004;23:34–44. doi: 10.1002/mas.10066. [DOI] [PubMed] [Google Scholar]
- 9.Paradis V, Degos F, Dargere D, Pham N, Belghiti J, Degott C, Janeau JL, Bezeaud A, Delforge D, Cubizolles M, Laurendeau I, Bedossa P. Identification of a new marker of hepatocellular carcinoma by serum proteomic profiling of patients with chronic liver diseases. Hepatology. 2005;41:40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Dang TA, Shen J, Perlaky L, Hicks J, Murray J, Meyer W, Chintagumpala M, Lau CC, Man TK. Identification of a plasma proteomic signature to distinguish pediatric osteo-sarcoma from benign osteochondroma. Proteomics. 2006;6:3426–3435. doi: 10.1002/pmic.200500472. [DOI] [PubMed] [Google Scholar]
- 11.Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ, Ismail T, Wakelam MJ, Johnson PJ, Martin A. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94:1898–1905. doi: 10.1038/sj.bjc.6603188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melle C, Ernst G, Schimmel B, Bleul A, Thieme H, Kaufmann R, Mothes H, Settmacher U, Claussen U, Halbhuber KJ, Von Eggeling F. Discovery and identification of R-defensins as low abundant, tumor-derived serum markers in colorectal cancer. Gastroenterology. 2005;129:66–73. doi: 10.1053/j.gastro.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Cheung PK, Woolcock B, Adomat H, Sutcliffe M, Bainbridge TC, Jones EC, Webber D, Kinahan T, Sadar M, Gleave ME, Vielkind J. Proteomic profiling of microdissected prostate tissue links growth differentiation factor 15 to prostate carcinogenesis. Cancer Res. 2004;64:5929–5933. doi: 10.1158/0008-5472.CAN-04-1216. [DOI] [PubMed] [Google Scholar]
- 14.Melle C, Ernst G, Schimmel B, Bleul A, Kaufmann R, Hommann M, Richter KK, Daffner W, Settmacher U, Claussen U, von Eggeling F. Characterization of pepsinogen C as a potential biomarker for gastric cancer using a histoproteomic approach. J Proteome Res. 2005;4:1799–1804. doi: 10.1021/pr050123o. [DOI] [PubMed] [Google Scholar]
- 15.Melle C, Bogumil R, Ernst G, Schimmel B, Bleul A, von Eggeling F. Detection and identification of heat shock protein 10 as a biomarker in colorectal cancer by proteomic profiling. Proteomics. 2006;6:2600–2608. doi: 10.1002/pmic.200500427. [DOI] [PubMed] [Google Scholar]
- 16.Scarlett CJ, Smith RC, Saxby A, Nielsen A, Samra JS, Wilson SR, Baxter RC. Proteomic classification of pancreatic adenocarcinoma tissue using protein chip technology. Gastroenterology. 2006;130:1670–1678. doi: 10.1053/j.gastro.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Melle C, Ernst G, Schimmel B, Bleul A, Koscielny S, Wiesner A, Bogumil R, Moller U, Osterloh D, Halbguber KJ, von Eggeling F. Biomarker discovery and identification in laser microdissected head and neck squamous cell carcinoma with ProteinChip(R) technology, two-dimensional gel electrophoresis, tandem mass spectrometry and immunohistochemistry. Mol Cell Proteomics. 2003;2:443–452. doi: 10.1074/mcp.M300033-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Krajewski S, Blomqvist C, Franssila K, Krajewska M, Wasenius VM, Niskanen E, Nordling S, Reed JC. Reduced expression of proapoptotic gene bax is associated with poor response rates to combination chemotherapy and shorter survival in woman with metastatic breast adenocarcinoma. Cancer Res. 1995;55:4471–4478. [PubMed] [Google Scholar]
- 19.Nhieu JT, Renard CA, Wei Y, Cherqui D, Zafrani ES, Buendia MA. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol. 1999;155:703–710. doi: 10.1016/s0002-9440(10)65168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng SY, Lai PL, Hsu HC. Amplification of the c-myc gene in human hepatocellular carcinoma: biologic significance. J Formos Med Assoc. 1993;92:866–870. [PubMed] [Google Scholar]
- 21.Nishida N, Fukuda Y, Komeda T, Kita R, Sando T, Furukawa M, Amenomori M, Shibagaki I, Nakao K, Ikenaga M, et al. Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res. 1994;54:3107–3110. [PubMed] [Google Scholar]
- 22.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 23.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 24.Rosty C, Christa L, Kuzdzal S, Baldwin WM, Zahurak ML, Carnot F, Chan DW, Canto M, Lillemoe KD, Cameron JL, Yeo CJ, Hruban RH, Goggins M. Identification of hepato-carcinoma-intestine-pancreas/pancreatitis-asoociated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- 25.Rai AJ, Zhang Z, Rosenzweig J, Shih IeM, Pham T, Fung ET, Sokoll LJ, Chan DW. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126:1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 26.Kozak KR, Amneus MW, Pusey SM, Su F, Luong MN, Luong SA, Reddy ST, Farias-Eisner R. Identification of biomarkers for ovarian cancer using strong anion-exchange ProteinChips: potential use in diagnosis and prognosis. Proc Natl Acad Sci USA. 2003;100:12343–12348. doi: 10.1073/pnas.2033602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 28.Ishibashi M, Fujita M, Nagai K, Kako M, Furue H, Haku E, Osamura Y, Yamaji T. Production and secretion of endothelin by hepatocellular carcinoma. J Clin Endocrinol Metab. 1993;76:378–383. doi: 10.1210/jcem.76.2.7679399. [DOI] [PubMed] [Google Scholar]
- 29.Lin Z, Jenson SD, Lim MS, Elenitoba-Johnson KS. Application of SELDI-TOF mass spectrometry for the identification of differentially expressed proteins in transformed follicular lymphoma. Mod Pathol. 2004;17:670–678. doi: 10.1038/modpathol.3800100. [DOI] [PubMed] [Google Scholar]
- 30.Ranganathan S, Williams E, Ganchev P, Gopalakrischnan V, Lacomis D, Urbinelli L, Newhall K, Cudkowicz ME, Brown RH, Jr, Browser R. Proteomic profiling of cerebrospinal fluid identifies biomarkers for amyotrophic lateral sclerosis. J Neurochem. 2005;95:1461–1471. doi: 10.1111/j.1471-4159.2005.03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


