Abstract
Microsatellite instability (MSI) due to defects in DNA mismatch repair genes may be involved in the development of a subset of human ovarian carcinomas. The role of one such gene, hMSH6, in ovarian cancer is not well documented. We investigated the expression of hMSH6 protein in different histotypes of ovarian carcinoma and the associations between loss of hMSH6 protein and tumor grade, disease stage, familial history of cancer and patient survival. We stained an ovarian carcinoma tissue microarray consisting of formalin-fixed, paraffin-embedded tissue samples from 322 patients with an anti-hMSH6 antibody and scored the results semiquantitatively as negative or positive. Twelve cases were excluded owing to loss of cores during staining. Absence of hMSH6 protein was noted in 20 of 230 serous carcinomas (8.7%), in 7 of 16 clear cell carcinomas (43.7%), in 4 of 34 endometrioid carcinomas (11.7%), in 1 of 14 malignant mixed Müllerian tumors, 2 of 6 mucinous carcinomas, 0 of 2 transitional cell carcinomas and in 0 of 8 undifferentiated carcinomas. Loss of hMSH6 protein was not associated with survival, patient age, tumor grade, or disease stage but was associated with clear cell, mucinous and endometrioid carcinoma histology (P<0.007). These findings indicate that loss of hMSH6 expression in ovarian carcinoma is more common in certain histologic subtypes, particularly in clear cell, endometrioid, and mucinous carcinoma, suggesting that loss of hMSH6 function may participate in the pathogenesis of these subtypes of cancer. Loss of hMSH6 expression did not predict survival and was not associated with disease stage, tumor grade, patient age or family history of cancer.
Keywords: hMSH6, microsatellite instability, ovarian carcinoma, tissue microarrays
Introduction
The presence of microsatellite instability (MSI), changes in the patterns of polymorphic di- or trinucleotide repeat segments distributed throughout the genome, reflect abnormalities in DNA mismatch repair genes (e.g., hMLH1, hMSH2, hPMS1, hPMS2, or hMSH6) and impair a cell's ability to repair errors produced during DNA replication. Germline mutations in these genes (most often hMLH1 and hMSH2), and the resultant increase in MSI levels, have been associated with the colonic, endometrial, or gastric carcinomas that develop in individuals with hereditary nonpolyposis colon carcinoma (HNPCC) [1]. Young women with HNPCC are also at higher risk of developing ovarian carcinoma [2], an observation that provides indirect evidence that MSI may have a role in the genesis of ovarian cancer.
Most primary ovarian carcinomas are of four morphologic types: serous, mucinous, endometrioid, and clear cell. Several studies indicate that different histologic types of ovarian adenocarcinomas probably represent distinct disease entities that involve different molecular pathways [3]. For example, high-grade serous adenocarcinomas often have p53 gene mutations, whereas K-ras activation is more common in serous tumors of low malignant potential, low-grade serous carcinomas, or mucinous adenocarcinomas [4, 5]. Understanding the molecular basis of each morphologic type and its biological behavior is important and will eventually lead to the development of more specific and effective treatments for ovarian cancer [6].
The first evidence that mutations in hMSH6 could be involved in the development of colorectal cancer came from the description of such mutations in two cell lines, HCT-15 and MT1, derived from tumors displaying MSI, primarily as mononucleotide repeats [7]. Germline mutations of hMSH6 were then reported in two Japanese families with atypical HNPCC that lacked mutations in hMSH2 or hMLH1; one of these families experienced a predominance of endometrial and ovarian carcinomas [8, 9]. The microsatellite sequences found in the hMSH6 gene may predispose microsatellite-unstable tumors to replication errors. Currently, the frequency of hMSH6 expression in ovarian carcinoma and how this correlates to the clinical setting is largely unknown. Our goal was to study the frequency of the expression of hMSH6 by immunoperoxidase technique in tissue microarrays in a large cohort of patients with primary ovarian carcinoma and correlate these findings with various clinicopathologic variables including survival.
Materials and Methods
Patients
Subjects were 322 patients with primary epithelial ovarian cancer who had undergone initial surgery at The University of Texas M. D. Anderson Cancer Center between 1990 and 2000 and for whom tissue samples and medical records were available. Tumors of low malignant potential, nonepithelial ovarian carcinomas, and benign lesions were excluded. Follow-up was updated through June 2005 by reviewing medical records and the U.S. Social Security Index. Demographic and survival data were entered into a comprehensive database for linking with histopathologic data (described below). Histopathologic diagnoses were based on World Health Organization (WHO) criteria [10] and grade based on Gynecologic Oncology Group [11] criteria. Serous carcinomas were graded according to a two-tier (low-grade and high-grade) system proposed by Malpica et al [12]. For statistical analysis grade 2 and grade 3 endometrioid ovarian carcinomas were grouped as “high grade” and grade 1 tumors as “low grade”. Disease was staged according to the International Federation of Gynecology and Obstetrics (FIGO) system [13–17]. Disease-specific survival time (overall survival) was reported as time since diagnosis or treatment, and only deaths from ovarian cancer were counted. The use of tissue blocks and chart review was approved by the appropriate institutional review boards at M. D. Anderson Cancer Center.
Construction of Tissue Microarrays
Tissue microarray blocks were constructed as previously described [18]. Tumor samples were arranged randomly. For each case, two replicate 1-mm core-diameter samples were collected, and each was placed on a separate recipient block. The final tissue microarray consisted of 4 blocks, the first two (1a and b) containing duplicates of 164 spots and the second two (2a and b) containing duplicates of 158 spots, with samples spaced 0.5 mm apart. Five-micrometer sections from each block were obtained and stained with H&E to confirm the presence of tumor and to assess tumor histology.
Sample tracking was based on coordinate positions for each tissue spot in the block; the spots were transferred onto tissue microarray slides for staining. This sample tracking system was linked to a Microsoft Access (version 97) database containing demographic, clinicopathologic, and survival data, thereby allowing rapid links between histologic data and clinical features. The array was read according to the given tissue microarray map; each core was scored individually, and results are presented as the means of the two replicate core samples.
Immunohistochemical Analysis
Tissue microarray slides were subjected to immunohistochemical staining as follows. After initial deparaffinization, endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide. Deparaffinized sections were microwaved in 10 mM citrate buffer (pH 6.0) to unmask the epitopes. The slides were then incubated with anti-hMSH6 antibody (BD Transduction Laboratories) in a 1:300 dilution for 1 hour at room temperature, followed by incubation with biotin-labeled secondary antibody (1:40) for 20 minutes and then with a 1:40 solution of streptavidin:peroxidase for 20 minutes. Slides were then stained for 5 minutes with 0.05% 3',3-diaminobenzidine tetrahydrochloride that had been freshly prepared in 0.05 M Tris buffer at pH 7.6 containing 0.024% H2O2 and then counterstained with hematoxylin, dehydrated, and mounted. All of the dilutions of antibody, biotin-labeled secondary antibody, and streptavidin-peroxidase were made in phosphate-buffered saline (pH 7.4) containing 1% bovine serum albumin. The primary antibody was raised against clone 44 of the MSH6 gene. It is highly specific and sensitive immunohistochemically, which led to an extensive usage investigating microsatellite instability in different organs and systems [30, 31, 37, 38]. Colon carcinoma tissue sections obtained form our archives were used as a positive control. Negative controls were treatments in which the primary antibody was replaced with phosphate-buffered saline. All controls gave satisfactory results.
The immunostained slides were reviewed by 2 pathologists (J.Z., D.R) who followed the tissue microarray map to record a score for each sample. Each reviewer was blinded as to the other's assessment and to the clinicopathologic information; scoring discrepancies were resolved by a third pathologist (J.L.). Staining for hMSH6 was graded as follows: Negative (no cells stained), weakly positive (less than 25% of cells stained), or strongly positive (more than 26% cells stained). For statistical analysis patients exhibiting weak and strong positive staining were grouped together. Means of the results from the two replicate core samples from each tumor specimen were considered for each case.
Statistical Analyses
Differences in proportions were evaluated by the χ2 or Fisher's exact tests as appropriate. Disease-specific survival rates were calculated by the method of Kaplan and Meier and compared by using log-rank tests. A Cox proportional hazards regression model was used for the multivariate analysis of survival. SAS Language Reference, Version 8 (1999) was used for the statistical analyses (SAS Institute Inc., Cary, NC). Results were considered statistically significant at the P < 0.05 level.
Results
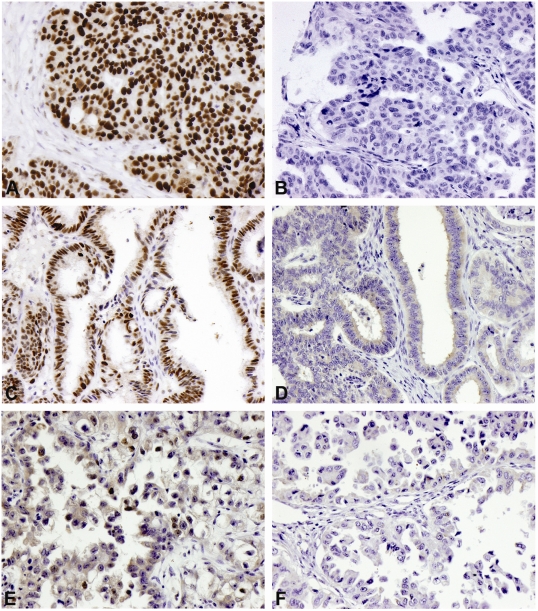
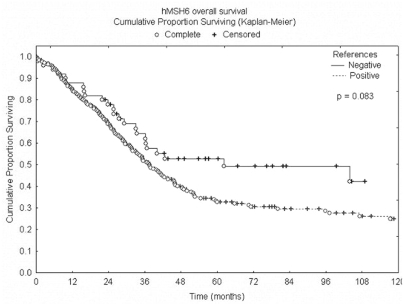
A total of 310 cases were analyzed, as 12 were lost during the staining process. Examples of positive and negative stains of different histotypes are shown in Figure 1. hMSH6 protein was not expressed in 34 cases (11%). Loss of expression was proportionally more frequent in clear cell carcinomas (43.8%), mucinous carcinomas (33.3%) and endometrioid carcinomas (11.8%) compared to the other histotypes (Table 1). The high rate of absence of hMSH6 protein expression among clear cell carcinoma was statistically different compared to serous carcinoma (p = 0.0001) and endometrioid carcinoma (p = 0.02) (Table 2). No correlations were found between hMSH6 expression and patient age, tumor grade, disease stage, or family history of cancer (Table 1). The 14 hMSH6 negative cases with familial history of cancer were divided as follows: 5 had a history of breast carcinoma, 1 of colorectal and the rest of other types of cancer related history. Overall survival seemed to have been better among patients whose tumors did not express hMSH6 protein, but this apparent difference was not statistically significant (p=0.083) (Figure 2).
Figure 1.
Representative pictures of immunostains using antibody against hMSH6: serous carcinoma positive (A) and negative (B), endometrioid carcinoma positive (C) and negative (D), and clear cell carcinoma positive (E) and negative (F) (200×).
Table 1.
hMSH6 staining according to clinical characteristics
| hMSH6 staining results | |||
|---|---|---|---|
| Negative | Positive | Total | |
| Tumor Histotype | |||
| Serous adenocarcinoma | 20(8.7%) | 210(91.3%) | 230 |
| Endometrioid adenocarcinoma | 4(11.8%) | 30(88.2%) | 34 |
| Malignant mixed Mullerian tumor | 1(7.1%) | 13(92.9%) | 14 |
| Clear cell carcinoma | 7(43.8%) | 9(56.3%) | 16 |
| Poorly differentiated carcinoma | 0(0%) | 8(100%) | 8 |
| Mucinous adenocarcinoma | 2(33.3%) | 4(66.7%) | 6 |
| Transitional cell carcinoma | 0(0%) | 2(100%) | 2 |
| Disease State | |||
| I | 5(16.1%) | 26(83.9%) | 31 |
| II | 5(20.0%) | 20(80.0%) | 25 |
| III | 19(9.7%) | 176(90.3%) | 195 |
| IV | 5(8.5%) | 54(91.5%) | 59 |
| P value | 0.30 | ||
| Tumor Grade | |||
| Low | 1(9.1%) | 10(90.9%) | 11 |
| High | 33(11.1%) | 266(89.0%) | 299 |
| P value | 0.83 | ||
| Patient Age (mean)(years) | 55.1 | 59.2 | |
| P value | 0.64 | ||
| Family History of Cancer | |||
| Yes | 14(11.2%) | 111(88.8%) | 125 |
| No | 20(10.9%) | 163(89.1%) | 183 |
| Unknown | 0(0%) | 2(100%) | 2 |
| P value | 0.91 | ||
| Total | 34(11%) | 276(89%) | 310 |
Table 2.
P values for the correlation between different histotypes
| TCCa | MC | PDC | CCC | MMMT | EC | |
|---|---|---|---|---|---|---|
| SC | 0.4 | 0.18 | 0.82 | 0.0001 | 0.77 | 0.79 |
| EC | 0.52 | 0.45 | 0.72 | 0.02 | 0.96 | |
| MMMT | 0.24 | 0.41 | 0.77 | 0.06 | ||
| CCC | 0.66 | 0.96 | 0.08 | |||
| PDC | 1 | 0.32 | ||||
| MC | 1 |
TCC, transitional cell carcinoma; SC, serous carcinoma; EC, endometrioid carcinoma; MMMT, malignant mixed Müllerian tumor; CCC, clear cell carcinoma; PDC, poorly differentiated carcinoma; MC, mucinous carcinoma
Figure 2.
Kaplan-Meier overall survival curves according to hMSH6 expression in ovarian carcinoma.
Discussion
In the present study we found that hMSH6 protein expression was absent in 11% of the cases. This loss of expression was associated with some of the ovarian carcinoma histotypes (clear cell, mucinous and endometrioid) and with a trend to better survival. We could not demonstrate any significant correlation with any of the other known clinical variables related to ovarian carcinoma prognosis or family history of cancer.
The occurrence of MSI in ovarian carcinoma remains unclear. Increasing evidence suggests that MSI, mainly at hMSH2 and hMLH1, might be associated with certain histologic types of ovarian carcinoma, being more common in endometrioid, mucinous, and clear cell types than in serous carcinoma [19–21, 22]. Previous studies have reported that 10% to 17% of ovarian carcinomas display MSI, but if the analyses are limited to endometrioid ovarian cancer, then the prevalence of MSI increases to 30% to 50% of cases [19, 20, 23–25]. Also, MSI was frequently found in 38% of mucinous adenocarcinoma and 14% of clear cell carcinomas [21, 26]. Consistently, most of the studies report a relatively low frequency of MSI in ovarian serous carcinomas (0–13%) [19–21, 27]. In contrast, some studies could not find such association with ovarian carcinoma histotypes [24]. In most of these studies MSI was assessed using different kinds of microsatellite markers and with the demonstration of only one locus, which is no longer considered adequate for MSI positive criteria. Currently, an MSI criterion was standardized with the use of 2 mono-nucleotide repeats (BAT26 and BAT25), and 3 dinucleotide repeats (D5S346, D2S123 and D17S250) known as the National Cancer Institute panel [28]. Subsequently this panel was adapted for use in ovarian cancer [29]. This may partially explain the controversial frequency of MSI observed in these studies. The use of PCR-based methods to detect MSI is relatively expensive and time consuming, which may limit the number of cases that can be analyzed. Immunohistochemical analysis offers an alternative method for assessment of MSI status and has proved to be both highly sensitive and highly specific for identifying MSI as a result of the inactivation of the hMSH1, hMSH2, and hMSH6 genes [26]. Moreover, the use of tissue microarrays provides the ability to assess hundreds of cases under identical testing conditions [18].
Only two reports have analyzed the frequency of mutations in the hMSH6 gene in ovarian carcinomas but only in the clear cell and endometrioid histotype. The first report from Gras et al describes a low frequency of hMSH6 mutation in a mixed group of patients with clear cell and endometrioid carcinoma [22]. The second report corresponds to our previous study of pure endometrioid ovarian carcinomas where we found that the genetic alterations in the polynucleotide tracts of the mismatch repair genes hMSH6 is infrequent [30]. To our knowledge no reports have been published on any clinical associations between ovarian cancer and hMSH6 gene expression. In the present study we found that lack of hMSH6 expression was more common in clear cell carcinoma, endometrioid carcinoma, and mucinous carcinoma than in serous carcinoma, an observation that represents further evidence that genetic derangements may play a role in ovarian carcinoma histogenesis.
The frequency of hMSH6 germline mutations has been evaluated in various population-based studies of sporadic colorectal cancer, familial non-HNPCC, classical HNPCC, early-onset colorectal cancer, and colorectal tumors with low levels of MSI [31, 32, 33–36]. Abnormalities of hMSH6 in association with HNPCC may indicate an increased risk of having an atypical HNPCC phenotype, characterized by late-onset colorectal cancer and frequent extracolonic tumors, particularly endometrial cancers [31–36]. In the present study we found that only 14 cases had a documented familial history of cancer in those lacking hMSH6 protein expression, suggesting that the lack of hMSH6 expression does not correlate with a familial history of cancer.
In our present study we found a trend towards better survival among patients whose tumors did not express hMSH6. However, this correlation did not reach statistical significance. The importance of these findings is still a matter of speculation.
Conclusions
These results underscore the importance of identifying the correct HNPCC-associated tumors and genes toward the recognition of affected families that may develop ovarian carcinoma as well as appropriate clinical surveillance. We found negative hMSH6 protein expression in several histologic subtypes of ovarian carcinoma, particularly in clear cell, endometrioid, and mucinous carcinoma, suggesting that loss of hMSH6 function may participate in the genesis of these subtypes of cancer. However, loss of hMSH6 protein expression did not predict overall survival, and it was not associated with disease stage, tumor grade, patient age or family history of cancer.
References
- 1.Lynch HT, Smyrk T. Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer. 1996;78:1149–1167. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1149::AID-CNCR1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown GJ, St John DJ, Macrae FA, Aittomaki K. Cancer risk in young women at risk of hereditary nonpolyposis colorectal cancer: implications for gynecologic surveillance. Gynecol Oncol. 2001;80:346–349. doi: 10.1006/gyno.2000.6065. [DOI] [PubMed] [Google Scholar]
- 3.Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38:87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 4.Aunoble B, Sanches R, Didier E, Bignon YJ. Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer (review) Int J Oncol. 2000;16:567–576. doi: 10.3892/ijo.16.3.567. [DOI] [PubMed] [Google Scholar]
- 5.Fujita M, Enomoto T, Inoue M, Tanizawa O, Ozaki M, Rice JM, Nomura T. Alteration of the p53 tumor suppressor gene occurs independently of K-ras activation and more frequently in serous adenocarcinomas than in other common epithelial tumors of the human ovary. Jpn J Cancer Res. 1994;85:1247–1256. doi: 10.1111/j.1349-7006.1994.tb02937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 7.Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D'Arrigo A, Markowitz S, Willson JK, Kinzler KW, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 9.Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 10.Tavassoli FA. In: Tumours of the Breast and Female Genital Organs. Devilee P, editor. Lyon, France: WHO/IARC; 2003. [Google Scholar]
- 11.Benda JA, Zaino R. GOG Pathology Manual. Buffalo, NY: 1994. [Google Scholar]
- 12.Malpica A, Deavers MT, Lu K, Bodurka DC, Atkinson EN, Gershenson DM, Silva EG. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Scully RE. World Health Organization classification and nomenclature of ovarian cancer. Natl Cancer Inst Monogr. 1975;42:5–7. [PubMed] [Google Scholar]
- 14.Russell P. The pathological assessment of ovarian neoplasms. II: The proliferating ‘epithelial’ tumours. Pathology. 1979;11:251–282. doi: 10.3109/00313027909061951. [DOI] [PubMed] [Google Scholar]
- 15.Russell P. The pathological assessment of ovarian neoplasms. III: The malignant “epithelial” tumours. Pathology. 1979;11:493–532. doi: 10.3109/00313027909059027. [DOI] [PubMed] [Google Scholar]
- 16.Russell P. The pathological assessment of ovarian neoplasms. I: Introduction to the common ‘epithelial’ tumours and analysis of benign ‘epithelial’ tumours. Pathology. 1979;11:5–26. doi: 10.3109/00313027909063533. [DOI] [PubMed] [Google Scholar]
- 17.Shepherd JH. Revised FIGO staging for gynaecological cancer. Br J Obstet Gynaecol. 1989;96:889–892. doi: 10.1111/j.1471-0528.1989.tb03341.x. [DOI] [PubMed] [Google Scholar]
- 18.Rosen DG, Huang X, Deavers MT, Malpica A, Silva EG, Liu J. Validation of tissue microarray technology in ovarian carcinoma. Mod Pathol. 2004;17:790–797. doi: 10.1038/modpathol.3800120. [DOI] [PubMed] [Google Scholar]
- 19.King BL, Carcangiu ML, Carter D, Kiechle M, Pfisterer J, Pfleiderer A, Kacinski BM. Microsatellite instability in ovarian neoplasms. Br J Cancer. 1995;72:376–382. doi: 10.1038/bjc.1995.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita M, Enomoto T, Yoshino K, Nomura T, Buzard GS, Inoue M, Okudaira Y. Microsatellite instability and alterations in the hMSH2 gene in human ovarian cancer. Int J Cancer. 1995;64:361–366. doi: 10.1002/ijc.2910640602. [DOI] [PubMed] [Google Scholar]
- 21.Ohwada M, Suzuki M, Saga Y, Sato I. DNA replication errors are frequent in mucinous cystadenocarcinoma of the ovary. Cancer Genet Cytogenet. 2000;117:61–65. doi: 10.1016/s0165-4608(99)00145-4. [DOI] [PubMed] [Google Scholar]
- 22.Gras E, Catasus L, Arguelles R, Moreno-Bueno G, Palacios J, Gamallo C, Matias-Guiu X, Prat J. Microsatellite instability, MLH-1 promoter hyper-methylation, and frameshift mutations at coding mononucleotide repeat microsatellites in ovarian tumors. Cancer. 2001;92:2829–2836. doi: 10.1002/1097-0142(20011201)92:11<2829::aid-cncr10094>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Krajinovic M, Richer C, Gorska-Flipot I, Gaboury L, Novakovic I, Labuda D, Sinnett D. Genomic loci susceptible to replication errors in cancer cells. Br J Cancer. 1998;78:981–985. doi: 10.1038/bjc.1998.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arzimanoglou II, Lallas T, Osborne M, Barber H, Gilbert F. Microsatellite instability differences between familial and sporadic ovarian cancers. Carcinogenesis. 1996;17:1799–1804. doi: 10.1093/carcin/17.9.1799. [DOI] [PubMed] [Google Scholar]
- 25.Diebold J. Molecular genetics of ovarian carcinomas. Histol Histopathol. 1999;14:269–277. doi: 10.14670/HH-14.269. [DOI] [PubMed] [Google Scholar]
- 26.Cai KQ, Albarracin C, Rosen D, Zhong R, Zheng W, Luthra R, Broaddus R, Liu J. Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum Pathol. 2004;35:552–559. doi: 10.1016/j.humpath.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Haas CJ, Diebold J, Hirschmann A, Rohrbach H, Schmid S, Lohrs U. Microsatellite analysis in serous tumors of the ovary. Int J Gynecol Pathol. 1999;18:158–162. doi: 10.1097/00004347-199904000-00010. [DOI] [PubMed] [Google Scholar]
- 28.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 29.Sood AK, Holmes R, Hendrix MJ, Buller RE. Application of the National Cancer Institute international criteria for determination of micro-satellite instability in ovarian cancer. Cancer Res. 2001;61:4371–4374. [PubMed] [Google Scholar]
- 30.Albarracin CT, Silva EG, Malpica A, Luthra R, Liu J. The role of hMSH3 and hMSH6 in ovarian endometrioid carcinoma and relationship with microsatellite instability phenotype. Oncol Rep. 2004;12:1217–1219. [PubMed] [Google Scholar]
- 31.Wijnen J, de Leeuw W, Vasen H, van der Klift H, Moller P, Stormorken A, Meijers-Heijboer H, Lindhout D, Menko F, Vossen S, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142–124. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, Kuismanen SA, Liu T, Chadwick RB, Johnson CK, Stevens MW, Richards SK, Meek JE, Gao X, Wright FA, et al. MSH6 and MSH3 are rarely involved in genetic predisposition to nonpolypotic colon cancer. Cancer Res. 2001;61:1619–1623. [PubMed] [Google Scholar]
- 33.Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 34.Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parc YR, Halling KC, Wang L, Christensen ER, Cunningham JM, French AJ, Burgart LJ, Price-Troska TL, Roche PC, Thibodeau SN. HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res. 2000;60:2225–2231. [PubMed] [Google Scholar]
- 36.Plaschke J, Kruppa C, Tischler R, Bocker T, Pistorius S, Dralle H, Ruschoff J, Saeger HD, Fishel R, Schackert HK. Sequence analysis of the mismatch repair gene hMSH6 in the germline of patients with familial and sporadic colorectal cancer. Int J Cancer. 2000;85:606–613. doi: 10.1002/(sici)1097-0215(20000301)85:5<606::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 37.Stefansson I, Akslen LA, MacDonald N, Ryan A, Das Soma, Jacobs IJ, Salvesen HB. Loss of hMSH2 and hMSH6 expression is frequent in sporadic endometrial carcinomas with microsatellite instability: A population-based study. Clin Cancer Res. 2002;8:134–143. [PubMed] [Google Scholar]
- 38.Hussein MR, Roggero E, Sudilovsky E, Tuthill R, Wood GS, Sudilovsky O. Alterations of mismatch repair protein expression in benign melanocytic nevi, melanocytic dysplastic nevi, and cutaneous malignant melanomas. Am J Dermatopathol. 2001;23:308–314. doi: 10.1097/00000372-200108000-00006. [DOI] [PubMed] [Google Scholar]