Abstract
HOX and three amino acid loop extension (TALE) proteins cooperate to induce transformation in mouse leukemia models, and are dysregulated in a variety of human leukemias. Despite decades of research, the mechanism of action for Hox proteins in embryogenesis and hematopoiesis remains unclear. Recent studies on the roles of Hoxa9 and Meis1 in leukemia has led to a wealth of new data, but their molecular mechanisms of action and synergy remain obscure. Advances in genome-wide technologies offer new avenues for understanding how homeodomain-containing transcription factors exert their programs in normal and neoplastic development.
Keywords: HOX, leukemia
Introduction
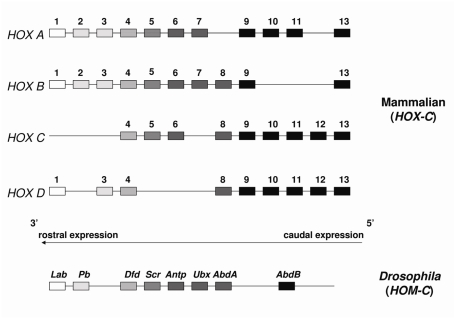
HOX proteins are an evolutionarily conserved family of homeodomain-containing DNA binding transcription factors with significant homology to the HOM-C factors in Drosophila melanogaster that are involved in the specification of segmental body pattern during development [1, 2]. In mammals, 39 HOX genes are arranged in four paralogous clusters (A-D) that map to four different chromosomes. The genes are arranged contiguously within each locus so that their 3′ to 5′ position parallels their temporal and spatial distribution; in the embryo 3′ factors are expressed anteriorly and 5′ factors are expressed posteriorly (Figure 1).
Figure 1.
Organization of the clustered mammalian HOX genes. HOX genes form linear arrays on 4 different chromosomes and exhibit significant homology to the Drosophila HOM-C clustered homeotic genes.
The homeodomain is a DNA-binding motif consisting of 60 amino acids configured in a helix-turn-helix motif with three alpha helices connected by short loops. Whereas the first two helices are anti-parallel, the C-terminal helix is longer, resting perpendicularly to the others, and binding directly to DNA in the major groove [3]. The N-terminus of the homeodomain is unstructured and interacts with the adjacent minor groove of DNA. Hox protein homeodomains share considerable homology and by themselves are unlikely to contribute significant DNA sequence specificity beyond a conserved TTNAT motif, which has been defined primarily through in vitro binding assays [4–6]. Additional target selectivity in vivo is likely conferred by associated proteins including TALE (three amino acid loop extension) cofactors such as Meis1 [5–7].
HOX Proteins in Hematopoiesis
Whereas body pattern development occurs only once at the beginning of life, hematopoietic maturation continues indefinitely, allowing for life-long renewal of blood and immune cells. Hox proteins are important regulators of hematopoiesis. Hox a, b, and c cluster genes are expressed early in hematopoiesis and are generally downregulated during differentiation [8–10]. Mouse models provide evidence of Hox protein involvement at various stages of hematopoiesis. MLL (mixed lineage leukemia)-null murine embryonic stem cells exhibit downregulation of many Hox a, b, and c cluster genes with significant impairment of normal hematopoiesis [11]. Hoxa7 appears to be involved in erythropoiesis and megakaryopoiesis [12]. Hoxb4 is a potent stimulator of hematopoietic stem cell (HSC) expansion [13–16]. However, Hoxb4 is not necessary for normal hematopoiesis as Hoxb4 deficient mice show only a mild proliferative defect in HSCs [17]. Hoxb3 deficiency causes impaired B lymphopoiesis [18]. Hoxa10 also appears to be involved in lymphopoiesis, erythropoiesis and megakaryopoiesis [19, 20]. Notably, Hoxa9 is the most highly expressed Hox gene in the HSC compartment. It is expressed in early hematopoietic progenitors (HPs), downregulated during differentiation and is important in HSC expansion [13, 21]. Among single Hox knockouts, Hoxa9 deficiency yields the most severe phenotype; however even in these animals, multilineage hematopoietic defects are mild (Table 1) [22, 23]. The most significant impairment exhibited by Hoxa9-null HSCs is the poor repopulation in bone marrow transplantation experiments [24]. Compound Hox knockouts show similarly mild hematopoietic phenotypes, most likely because of considerable redundancy of function among and between paralog groups [25].
Table 1.
Characteristics of Hoxa9 and Meis1 in mouse models.
| Experimental model | Hoxa9 | Meis1 |
|---|---|---|
| Loss of expression (null mice) | Viable, morphologically normal, normal number of HSCs [22] | Fatal by E14.5 – bleeding from amegakaryopoiesis |
| Mild leucopenia/blunted G-CSF response [22] | Additional defects in angiogenesis, eye development | |
| Decreased number of committed myeloid and lymphoid progenitors [22, 23] | Reduction in myeloerythroid colony-forming units | |
| BMT repopulation defect with decreased HSC proliferation [24] | Heterozygotes viable; no phenotypic defects [26] | |
| Enforced expression (HSC transduction) | Immortalization, differentiation block [100] | Pro-apoptotic |
| Non-transforming when transplanted [60] | Does not promote immortalization | |
| Lack of expression in MLL-fusion cells | Can be tolerated [71] | Required for transformation [63] |
TALE Proteins – HOX Partners and Beyond
TALE proteins are homeodomain-containing transcription factors that are thought to enhance DNA binding specificity of Hox proteins through heterodimerization [5, 7]. Within this category, subgroups include the Pbx and Meis families, which have been shown to assemble in trimeric complexes composed of Hox, Pbx and Meis subunits upon a single DNA sequence [6]. Although the crystal structure of a Hoxa9/Pbx heterodimer bound to DNA has been reported [4], no structural data is available for a Hoxa9-Meis1 dimer or Pbx-containing trimer.
Like Hox proteins, TALE proteins are involved in embryogenesis as well as hematopoiesis [26]. Meis1 expression during hematopoiesis parallels Hox expression, with levels high in HPs and increasingly downregulated with further differentiation [10]. Meis1 plays an important role in limb and eye development, as evidenced by the severe phenotypes seen in Meis1-null embryos (Table 1). However, lethality results from its hematopoietic contribution, as embryos succumb from amegakaryopoiesis by day E14 [26]. This broader phenotype compared to Hox knockout animals may result from hetero-dimerization of Meis1 with several Hox proteins; alternatively the phenotype may be the result of non-Hox related contributions to embryogenesis. A recent study revealed linkage of the neurological disorder Restless Legs Syndrome to the MEIS1 locus, underscoring the broad scope of physiologic processes impacted by TALE factors [27].
HOX and TALE Proteins in MLL-Rearranged Leukemias
Acute leukemias with rearrangements of the MLL gene (11q23) have an intermediate to poor prognosis compared to cytogenetically favorable types [28–31]. MLL rearrangements comprise 5–6% of all acute myeloid leukemias and about 20% of acute lymphoblastic leukemias [32, 33]. In development and hematopoiesis, wild-type MLL activates Hox gene transcription by methylation of histone H3 at lysine 4 through its intrinsic histone methyltransferase activity [34]. Additionally, fusion proteins involving MLL deregulate Hoxa9 and Meis1 expression [34–37]. Acute lymphoblastic leukemias with MLL rearrangements show high expression of HOXA7 and HOXA9 as well as MEIS1 [38–40]. These findings suggest that HOXA9 and MEIS1 are key mediators of transformation by MLL rearrangements.
Clinical Evidence for HOX Involvement in Other Leukemias
HOX genes are overexpressed or rearranged in a variety of experimental and human leukemias [38–45]. In acute lymphoblastic leukemias, HOXA9 overexpression appears to be largely limited to those with MLL translocations, whereas a large fraction of myeloid leukemias show dysregulated HOXA9 expression [38]. HOXA9 is associated with refractory AML and poor prognosis [46]. High level HOXA9 expression is implicated in T-cell ALL cases, including subtypes harboring MLL, CALM-AF10 t(10;11), and TCRβ-HOXA locus (inv(7)(p15q34)) rearrangements [47, 48]. A recent study identified a correlation between HOXA9 expression in T-ALL and a prognostically distinct subgroup with relatively primitive phenotype [49]. Given that the prevalence of leukemia increases with age, it is interesting that HOXA9 levels tend to be higher in marrow from elderly donors [50]. Direct involvement of HOX proteins in oncogenic fusions with NUP98, such as NUP98-HOXA9 t(7;11) and NUP98-HOXD13 t(2;11)(q31;p15) in AML has been well reviewed [51, 52].
Like HOXA9, MEIS1 expression appears to correlate with poor prognosis in AML. Two patient series showed decreased MEIS1 expression in AML patients with good outcome [53, 54].
Role of HOX and TALE Proteins in Experimental Models of Transformation
Enforced retroviral expression of certain Hox genes (for example Hoxa9, Hoxa10, Hoxb3, or Hoxb6) in murine bone marrow cells results in differentiation block with continuous self-renewal in culture [21, 55–58]. The stage at which maturation arrest occurs varies with the particular Hox protein that is overexpressed; for example, Hoxa9 overexpression causes arrest with an early myelomonocytic phenotype [59]. Transplantation of Hoxa9-transduced bone marrow into mice does not result in a fully penetrant leukemia; instead, transformation to leukemia is seen in a small subset of animals after prolonged latency, suggesting that additional “hits” are required [56, 57, 60]. Co-expression of Meis1 greatly increases the transformation efficiency of Hoxa9; HSCs transduced with both Hoxa9 and Meis1 confer a rapidly fatal leukemia in transplanted animals [60]. Meis1 has also been shown to accelerate Nup98-Hox fusion mediated leukemogenesis [61, 62]. These findings imply synergy between Hox genes and Meis1 in leukemic transformation.
The role of Pbx proteins in transformation process is unclear. MLL fusion proteins have been shown to upregulate Pbx3 expression [36, 63]. In addition, joint reduction of Pbx2 and Pbx3 levels interferes with MLL-driven leukemogenesis [63]. Furthermore, the Pbx interaction domain of Meis1 is required for MLL-fusion mediated transformation and for promoting leukemia in Hoxa9-immortalized cells [63, 64]. Hoxa9 also requires its Pbx-interaction domain for its myeloid expansion phenotype [65].
Despite its critical role in promoting leukemic transformation when expressed with Hox proteins, enforced expression of Meis1 in hematopoietic progenitors does not lead to immortalization and is in fact pro-apoptotic [66]. Two groups have shown that addition of a VP16 transactivating domain to the N-terminus of Meis1 results in a chimeric factor capable of immortalizing HPs; however VP16-Meis1 leukemias show much longer latency than seen with Hoxa9+Meis1 coexpression [67, 68]. The implications of this observation are discussed later in the review.
Cooperation between HOX and TALE Proteins in Leukemia
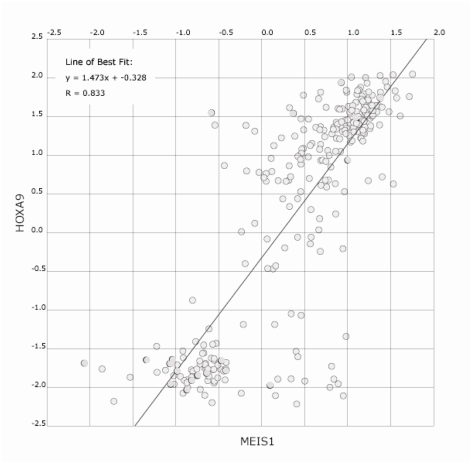
In addition to their parallel up-regulation by MLL fusion proteins, several lines of evidence indicate a strong synergy between Hox and TALE proteins. In the BXH2 murine retroviral mutagenesis model of AML, Hoxa9 and Meis1 loci are nearly always activated in tandem through independent viral integrations [69]. In addition, analysis of human AML samples by microarray (www.oncomine.org) shows strong correlation between MEIS1 and HOXA9 levels (Figure 2) [70]. The most convincing evidence comes from the phenotype of murine HSCs coexpressing Hoxa9 and Meis1 when compared to overexpression of either gene alone: as discussed above, the combination is potently leukemogenic [60, 64]. Since Meis1 has the opposite phenotype when expressed by itself (Table 1), the nature of synergy between Hoxa9 and Meis1 is unlikely to be simply additive.
Figure 2.
HOXA9 and MEIS1 expression levels in AML patient samples. Expression data from 285 leukemias [104] was analyzed using the co-expression module at oncomine.org and shows the correlation between HOXA9 and MEIS1 expression [70]. MLL-fusion cases represent only 7% of cases in this study [104].
Are HOXA9 and MEIS1 Dispensible in Leukemogenesis?
Studies using HSCs purified from Meis1-null embryos prior to demise indicate that Meis1 is necessary for MLL-fusion protein mediated transformation [63]. This was found to be the case for 12 different MLL fusion proteins (but not for the unrelated translocation E2A-HLF), and was linked to both induction and maintenance of leukemic proliferation. The report also showed acceleration of MLL-fusion leukemias through addition of ectopically expressed Meis1. The combination of these two findings lends strong support to Meis1 dose dependency, since the baseline Meis1 concentration induced by MLL-fusion expression is necessary, but higher level expression of Meis1 has a potentiating effect [63].
The necessity of Hoxa9 for MLL fusion protein mediated leukemogenesis is less clear-cut. Initial data offers conflicting results, as a single group found loss of transformation by MLL-ENL in Hoxa7- or Hoxa9-null mice but retention of leukemic potential with MLL-GAS7 expression in a Hoxa9-/- background [12, 37]. At the same time, others found Hoxa9 to be dispensible in generation of leukemias from transgenically expressed MLL-AF9; in fact, Hoxa9-null HSCs exhibited less myeloid differentiation than wild-type counterparts [71]. These observations may be due to experimental design: in the MLL-ENL study, Lin- cells (HSCs) were chosen for retroviral transduction, while the MLL-GAS7 report used c-Kit-selected precursors which include common myeloid progenitors [the MLL-AF9 study involved a germline transgene and did not require a selection step] [72]. One potential explanation is that Hoxa9 freezes cells in a particular stage of differentiation that permits MLL-fusion mediated transformation. Kit-selected cells represent a spectrum between HSCs and later myeloid progenitors, while Lin- HSCs are homogeneous. Indeed, these observations have been attributed to the concept that the committed myeloid progenitor is the “leukemic stem cell” for MLL fusions [73].
More recent data suggests that continued HOXA9 expression is essential for maintenance of immortalization by MLL-fusion proteins; shRNA knockdown of HOXA9 in a t(9;11) human leukemia cell line (MOLM-14) inhibits proliferation as early as 48 hours after transduction [74]. In addition HOXA9 reduction reverses the in vivo transformation phenotype in mice transplanted with SEMK2, a t(4;11) leukemia line. Whereas proliferation/viability of non-MLL-rearranged leukemic cell lines is also affected by HOXA9 knockdown, there is a statistically significant difference in magnitude when compared to human AML cells with MLL translocations. Not surprisingly, the greatest effects are seen in leukemic cells with the highest baseline expression levels of HOXA9 [74]. These findings suggest an ongoing role for HOXA9 in AML cells, rather than a “hit-and-run” contribution to leukemogenesis.
Why is HOXA9 so Commonly Implicated in Leukemogenesis?
Although HOXA9 is the most commonly deregulated HOX factor in leukemia, abundant evidence suggests that HOX proteins play redundant roles in leukemogenesis. The ability of MLL fusion proteins to transform in a Hoxa9-null background (discussed above) points to this: In addition, Hoxb6, Hoxa7, Hoxa10, and others show similar properties in HSC immortalization/transformation studies [12, 56, 57, 67]. Gene expression profiling of MLL-fusion mouse models shows overexpression of Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10, and a subset of human leukemias show coordinated upregulation of HOXA5-10 along with HOXA9 [38, 75]. However, it is HOXA9 that emerges as the single most prognostic gene in clinical leukemias, and Hoxa9 and Hoxa7 are the most common insertion sites (along with Meis1) in BXH2 insertion-mediated mouse leukemias [46, 69]. Although viable, Hoxa9-null mice have a more profound hematopoietic phenotype than other characterized Hox knockouts [52]. Additionally, comparison of NPM1-mutated and MLL-rearranged human leukemias shows a shared HOX dysregulation profile including the posterior HOXA cluster and MEIS1, while other HOX genes show differential regulation [42]. Finally, t(7;11)/NUP98-HOXA9 is the most common recurring cytogenetic abnormality involving a HOX protein [76, 77]. The precise state of myeloid differentiation specified by HOXA9 may be a key contributor to its relative leukemogenic potential. In this context it may be relevant that HOXA9 is the most highly predictive transcript for AML vs ALL [38]. Overall, although current data supports considerable redundancy among HOX paralogs, human and mouse studies indicate a particularly important role for HOXA9 in leukemogenesis.
Which HOXA9 and MEIS1 Targets are Essential for Leukemogenesis?
When ectopically expressed in hematopoietic progenitors, Hoxa9 induces two main phenotypic effects: differentiation arrest (at an early myeloid progenitor phase) and indefinite self-renewal (immortalization) [64]. The strongest support for the latter are the repopulation defects that are seen in Hoxa9-null mice (see Table 1) [24]. The long latency required to reach transformation in animals, and the elimination of this latency by coexpression of Meis1, imply that by themselves Hox proteins do not confer sufficient proliferative capacity for leukemogenesis. There is in vitro support for a Meis1 contribution to proliferation. Cells transduced with Hoxa9 and Meis1 outgrow Hoxa9-transduced cells in culture, and also allow for selective growth in particular cytokine combinations [64, 67]. The genes that mediate these effects are not well defined.
Putative downstream effectors of HOXA9 were first reported from microarray experiments using transient overexpression of HOXA9 in cultured cell lines in 2004 [78]. Cells included Jurkat T lymphoblasts, K562 lymphoblasts, and U937 promonocytes. These lines were established from human leukemias and are functionally independent of exogenous HOXA9 expression. Many identified targets were cell line-specific, but a small subset of genes (14/220) was found to correlate with HOXA9 expression in published AML tumor profiling data. In a second study from the same laboratory, HOXA9 was transiently overexpressed in human CD34+ cord blood cells. Surprisingly this resulted in a distinct target set with minimal overlap [79].
A second group of investigators published two reports measuring gene expression differences between cells immortalized with Hoxa9 and Hoxa9+Meis1 [64, 67]. Acute leukemias often arise as the result of activation of a nuclear protooncogene, which blocks differentiation, along with a cytoplasmic receptor tyrosine kinase, which promotes proliferation [80]. One Hoxa9/Meis1 target that fits this model is the tyrosine kinase Flt3, which is the most common signal transduction “hit” in AML [81] Increased expression of Flt3 is seen in Hoxa9/Meis1-transduced HPs when compared to HPs immortalized by Hoxa9 alone; in addition, chromatin immuno-precipitation (ChIP) demonstrated binding of the Flt3 promoter by both Hoxa9 and Meis1 [67]. However, HPs from Flt3-null mice can be transformed by Hoxa9+Meis1 with efficiency comparable to wild-type cells, so Flt3 cannot be considered the single, necessary mediator of leukemogenesis [82]. Wild-type Flt3 is insufficient for transformation of hematopoietic precursors on its own; Flt3 can promote leukemogenesis with Nup98-Hoxa9, but the latency in this setting is longer than with co-expression of Meis1 [83]. Other notable targets include the stem-cell associated surface marker CD34 and the proleukemic transcription factor Sox4.
A third laboratory in collaboration with our own took a different approach to identify targets of joint Hoxa9 and Meis1 regulation: inducible expression of MLL-ENL was used to immortalize hematopoietic progenitors; the effects of retroviral introduction of Hoxa9 and Meis1 were observed after withdrawal of MLL-ENL support [84]. The oncogene c-Myb was found to be necessary but not sufficient for transformation by Hoxa9+Meis1.
Several reports at the 2007 American Society of Hematology meeting highlighted Hoxa9 targets identified through RNA interference knock-down, including Bim and Mef2c [85, 86]. The latter factor is of interest as it is part of an 11-gene minimal “immediate” gene expression signal in MLL-AF9 transformed HSCs which includes Hoxa9 and Meis1 [87]. More recently, Insulin-like growth factor-1 receptor (IGF-1R) was shown to be upregulated in the setting of HOXA9 overexpression suggesting a possible downstream mechanism for HOXA9 induced cell proliferation [88].
Molecular Functions of HOXA9 and MEIS1 in Leukemia
Canonical transcription factors merge a sequence-specific DNA binding domain with an interaction motif capable of recruiting enzymatic activities. Although evidence for both functions has been described for individual HOX proteins, the mechanisms by which HOX proteins regulate transcription remain unclear. For example, interaction with CREB binding protein (CBP) has been demonstrated for multiple HOX proteins including HOXA9 [89]. Although the most straightforward explanation is that HOX proteins function by recruiting histone acetyltransferase activity, another report suggests that CBP prevents DNA binding by HOX proteins and that the HOX interaction inhibits CBP acetylase activity [90].
Moreover, it remains an open question whether HOX proteins act predominantly as activators, repressors, or both. It is likely that this is highly dependent on cellular and specific promoter context. It is appealing to propose that the unique synergy between HOXA9 and MEIS1 results from an altered property of the heterodimer. As mentioned above, addition of the VP16 activating moiety to Meis1 reverses its pro-death phenotype and promotes immortalization [67]. Notably, the Meis1-VP16 fusion retains cooperativity with Hoxa9, arguing against a straightforward “replacement” of Hoxa9 function by VP16. Hoxa9 shows transcriptional activation in reporter gene assays using a Hox-responsive enhancer element; this activity is retained when the N-terminus of Hoxa9 is replaced by VP16 [65]. In a more physiologic model employing the CYBB (gp91Phox) proximal promoter, HOXA9 was shown to activate transcription in a PBX-dependent manner; however addition of MEIS1B blocked transcriptional activation [91]. Occupancy experiments suggest that HOXA9 is not prevented from binding to DNA but that MEIS1 forms a tripartite complex with PBX1.
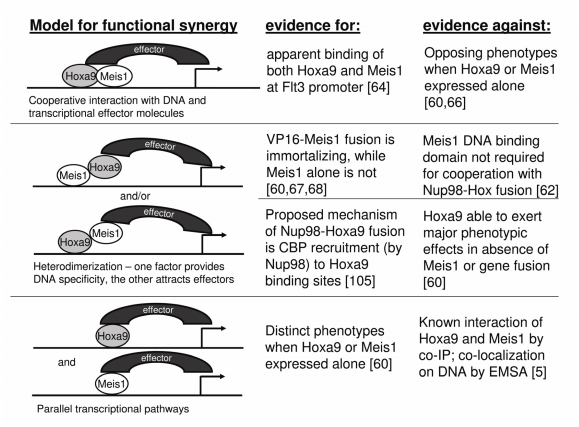
Another unresolved topic is the molecular mechanism for the synergy between HOXA9 and MEIS1 in transformation. Figure 3 illustrates possible models of cooperation, including data supporting and refuting each theory. For example, the finding that VP16-Meis1 has immortalizing activity in HPs, suggests that the DNA binding specificity of Meis1 is crucial to its function [64]. However, the homeodomain of Meis1 is not necessary for acceleration of transformation by the Nup98-Hoxd13 fusion protein, suggesting that Meis1 may play an important role independent of DNA binding [62].
Figure 3.
Possible modes of cooperation between Hoxa9 and Meis1. No single model is completely supported by current evidence; rather it is likely that targets of Hoxa9 and Meis1 are regulated by a combination of these mechanisms.
The contribution of PBX proteins to transformation and their role in HOXA9+MEIS1 transcriptional modulation is similarly unclear. TALE factors were traditionally thought to contribute to additional binding specificity rather than enzyme recruitment potential [7]. However, there is evidence for direct interaction between Pbx factors and chromatin remodeling machinery, and also between Pbx1 and histone deacetylase-containing corepressor complexes [92]. Additionally Pbx2 and Pbx3 appear to be involved in MLL-mediated transformation [63].
The solution to these unresolved questions is likely to come from more rigorous biochemical approaches to identifying HOX protein interactors as well as improved approaches to identify in vivo binding sites. The assumption that HOX proteins function as transcription factors is well supported, although there are intriguing reports of Hoxa9 modulating gene expression through potentiation of translational efficiency via eIF4E [93]. The Hoxa9-Pbx2-Meis1 trimer, characterized by electrophoretic mobility shift assays using “consensus” binding motifs, has not yet translated into a rigorous demonstration of triple occupancy at in vivo promoters using ChIP. In fact, the empiric nature of ChIP data is squandered when putative targets are selected or verified based on the presence of the TTNAT core (which mathematically recurs by chance within 300 bp, compared to an average ChIP DNA fragment size of 500 bp). Finally, the assumption of proximal promoter-mediated function, which has been repeatedly disproven for many transcription factors, continues to drive investigation of target gene regulation for HOX and other homeodomain families. With the advent of truly genomic screening capabilities, these entrenched constraints can be put to the test in an unbiased fashion.
Additional Insights from HOX Protein Expression in Non-Hematopoietic Tissues
In angiogenesis, Hoxa9 is a positive regulator of EphB4, which stimulates endothelial cell migration and tube forming activity [94]. A recent report demonstrates that Hoxa9 mediates TNFα induction of the proinflammatory marker E-selectin in endothelial cells [95]. As in hematopoiesis, Hoxa9 regulation in endothelial development is controlled by MLL [96]. The common hemangioblast theory would predict that the functions of Hoxa9 in hematopoiesis and angiogenesis may be quite similar, with possible overlap of target genes.
Expression of HOX proteins in other non-hematopoietic tissues and solid tumors lends strong support for context-dependent function. HOXA9 in particular appears to have tumor suppressor activity in ectodermally-derived tissue. The Hox 9 paralog group is required for mammary development during pregnancy in mice – lactation does not occur in knockout mice, and foster mothers are required for pup survival [97]. Interestingly, HOXA9 is frequently epigenetically silenced in human breast cancer [98]. In addition, HOXA9 CpG islands are frequent DNA methylation targets in stage I primary squamous carcinomas of the lung and there is evidence for HOXA9 dysregulation in human lung cancer cell lines compared to normal lung tissue [99, 100]. HOXA9 repression by promoter hypermethylation is also seen in testicular germ cell tumors [101]. Recently, the HOXA9 promoter was found to be hypermethylated in over 50% of ovarian cancer samples [102]; interestingly, MEIS1 (as well as MEIS2 and PBX3) show downregulation in a cisplatin-resistant ovarian tumor cell line when compared with chemosensitive parent cells [103]. Assuming that hypermethylation of HOX factors is not an epiphenomenon in solid tumors, the opposing phenotypes of HOXA9 in leukemia versus carcinoma emphasize the role of cellular context and argue against a general pro-proliferation or anti-apoptotic role for HOX proteins.
Current Approaches to Open Questions in HOX/TALE Biology
Recent advances in genome-wide technologies show promise for better defining the effector mechanisms employed by HOX proteins in leukemia. Multiple platforms are now available for unbiased identification of DNA binding sites on a genome-wide level, including ChIP-chip, ChIP-SAGE (serial analysis of gene expression), and ChIP-Sequencing. Comparing direct binding sites of Hoxa9 and Meis1 (when expressed individually and together) to the gene expression changes induced by each factor can help unravel the cascade of events leading to leukemic transformation. This approach will potentially resolve important molecular questions such as the mechanism of cooperation between Hoxa9 and Meis1, the transcriptional function of the two proteins, and the precise DNA motifs responsible for HOX recruitment.
Conclusion
Recent improvements in therapy for leukemia subsets (such as acute promyelocytic leukemia) rested upon advances in molecular pathogenesis. Characterizing downstream pathways involved in HOX-mediated leukemias, which are among the poorest prognosis AMLs and ALLs, will hopefully contribute to ameliorating the dismal outcomes seen in these groups. Whereas the topic of HOX and TALE factors in leukemia represents a newcomer to the decades-old field of HOX function in development, contemporary genomic approaches may further an understanding of HOX biology in embryogenesis and normal hematopoiesis as well as in neoplasia.
References
- 1.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 2.Owens BM, Hawley RG. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- 3.Harrison SC. Three-dimensional intricacies in protein-DNA recognition and transcriptional control. Nat Struct Mol Biol. 2007;14:1118–1119. doi: 10.1038/nsmb1207-1118. [DOI] [PubMed] [Google Scholar]
- 4.LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen WF, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen W-F, Rozenfeld S, Kwong A, Komuves LG, Lawrence HJ, Largman C. HOXA9 Forms Triple Complexes with PBX2 and MEIS1 in Myeloid Cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 8.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giampaolo A, Pelosi E, Valtieri M, Montesoro E, Sterpetti P, Samoggia P, Camagna A, Mastroberardino G, Gabbianelli M. Testa U, et al., editors. HOXB gene expression and function in differentiating purified hematopoietic progenitors. Stem Cells. 1995;13(Suppl 1):90–105. [PubMed] [Google Scholar]
- 10.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 11.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 12.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 13.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 14.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 15.Amsellem S, Pflumio F, Bardinet D, Izac B, Charneau P, Romeo PH, Dubart-Kupperschmitt A, Fichelson S. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat Med. 2003;9:1423–1427. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- 16.Buske C, Feuring-Buske M, Abramovich C, Spiekermann K, Eaves CJ, Coulombel L, Sauvageau G, Hogge DE, Humphries RK. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood. 2002;100:862–868. doi: 10.1182/blood-2002-01-0220. [DOI] [PubMed] [Google Scholar]
- 17.Brun AC, Bjornsson JM, Magnusson M, Larsson N, Leveen P, Ehinger M, Nilsson E, Karlsson S. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004;103:4126–4133. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- 18.Ko KH, Lam QL, Zhang M, Wong CK, Lo CK, Kahmeyer-Gabbe M, Tsang WH, Tsang SL, Chan LC, Sham MH, Lu L. Hoxb3 deficiency impairs B lymphopoiesis in mouse bone marrow. Exp Hematol. 2007;35:465–475. doi: 10.1016/j.exphem.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Magnusson M, Brun AC, Miyake N, Larsson J, Ehinger M, Bjornsson JM, Wutz a, Sigvardsson M, Karlsson S. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood. 2007;109:3687–3896. doi: 10.1182/blood-2006-10-054676. [DOI] [PubMed] [Google Scholar]
- 20.Taghon T, Stolz F, De Smedt M, Cnockaert M, Verhasselt B, Plum J, Leclercq G. HOX-A10 regulates hematopoietic lineage commitment: evidence for a monocyte-specific transcription factor. Blood. 2002;99:1197–1204. doi: 10.1182/blood.v99.4.1197. [DOI] [PubMed] [Google Scholar]
- 21.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:12112–9. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- 23.Izon DJ, Rozenfeld S, Fong ST, Komuves L, Largman C, Lawrence HJ. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood. 1998;92:383–393. [PubMed] [Google Scholar]
- 24.Lawrence HJ, Christensen J, Fong S, Hu Y-L, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bijl J, Thompson A, Ramirez-Solis R, Krosl J, Grier DG, Lawrence HJ, Sauvageau G. Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood. 2006;108:116–122. doi: 10.1182/blood-2005-06-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, Jenkins NA, Copeland NG. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, Fulda S, Putz B, Eckstein G, Hauk S, Trenkwalder C, Zimprich A, Stiasny-Kolster K, Oertel W, Bachmann CG, Paulus W, Peglau I, Eisensehr I, Montplaisir J, Turecki G, Rouleau G, Gieger C, Illig T, Wichmann HE, Hosboer F, Muller-Myhsok B, Meitinger T. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 28.Eguchi M, Eguchi-Ishimae M, Greaves M. Molecular pathogenesis of MLL-associated leukemias. Int J Hematol. 2005;82:9–20. doi: 10.1532/IJH97.05042. [DOI] [PubMed] [Google Scholar]
- 29.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 30.Felix CA, Lange BJ, Chessells JM. Pediatric Acute Lymphoblastic Leukemia: Challenges and Controversies in 2000. Hematology Am Soc Hematol Educ Program. 2000:285–302. doi: 10.1182/asheducation-2000.1.285. [DOI] [PubMed] [Google Scholar]
- 31.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 32.De Braekeleer M, Morel F, Le Bris MJ, Herry A, Douet-Guilbert N. The MLL gene and translocations involving chromosomal band 11q23 in acute leukemia. Anticancer Res. 2005;25:1931–1944. [PubMed] [Google Scholar]
- 33.McCormack E, Bruserud O, Gjertsen BT. Review: genetic models of acute myeloid leukaemia. Oncogene. 2008 doi: 10.1038/onc.2008.16. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 35.Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D, Brock HW, Slany R, Hess JL. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 36.Zeisig BB, Milne T, Garcia-Cuellar M-P, Schreiner S, Martin M-E, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, Hess JL, Slany RK. Hoxa9 and Meis1 Are Key Targets for MLL-ENL-Mediated Cellular Immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 39.Rozovskaia T, Ravid-Amir O, Tillib S, Getz G, Feinstein E, Agrawal H, Nagler A, Rappaport EF, Issaeva I, Matsuo Y, Kees UR, Lapidot T, Lo Coco F, Foa R, Mazo A, Nakamura T, Croce CM, Cimino G, Domany E, Canaani E. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc Natl Acad Sci USA. 2003;100:7853–7858. doi: 10.1073/pnas.1132115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 41.Casas S, Nagy B, Elonen E, Aventin A, Larramendy ML, Sierra J, Ruutu T, Knuutila S. Aberrant expression of HOXA9, DEK, CBL and CSF1R in acute myeloid leukemia. Leuk Lymphoma. 2003;44:1935–1941. doi: 10.1080/1042819031000119299. [DOI] [PubMed] [Google Scholar]
- 42.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, Downing JR. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 43.Casorelli I, Tenedini E, Tagliafico E, Blasi MF, Giuliani A, Crescenzi M, Pelosi E, Testa U, Peschle C, Mele L, Diverio D, Breccia M, Lo-Coco F, Ferrari S, Bignami M. Identification of a molecular signature for leukemic promyelocytes and their normal counterparts: Focus on DNA repair genes. Leukemia. 2006;20:1978–1988. doi: 10.1038/sj.leu.2404376. [DOI] [PubMed] [Google Scholar]
- 44.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, Zeng C, Baron A, Hunger SP, m Varella-Garcia M, Gemmill R, Brizard F, Brizard A, Roche J. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 45.Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedeu J, Montserrat E, Campo E. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–6954. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 46.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 47.Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, Baruchel A, toribio ML, Sigaux F. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 48.Speleman F, Cauwelier B, Dastugue N, Cools J, Verhasselt B, Poppe B, Van Roy N, Vandesompele J, Graux C, Uyttebroeck A, Boogaerts M, De Moerloose B, Benoit Y, Selleslag D, Billiet J, Robert A, Huguet F, Vandenberghe P, De Paepe A, Marynen P, Hagemeijer A. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia. 2005;19:358–366. doi: 10.1038/sj.leu.2403657. [DOI] [PubMed] [Google Scholar]
- 49.Baleydier F, Decouvelaere AV, Bergeron J, Gaulard P, Canioni D, Bertrand Y, Lepretre S, Petit B, Dombret H, Beldjord K, Molina T, Asnafi V, Macintyre E. T Cell Receptor Genotyping and HOXA/TLX1 Expression Define Three T Lymphoblastic Lymphoma Subsets which Might Affect Clinical Outcome. Clin Cancer Res. 2008;14:692–700. doi: 10.1158/1078-0432.CCR-07-1927. [DOI] [PubMed] [Google Scholar]
- 50.Morgan R, Begum R, Theti D, Chansa M, Pettengell R, Sohal J. HOXA9 expression increases with age in human haemopoietic cells. Leukemia Res. 2005;29:1221–1222. doi: 10.1016/j.leukres.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Curr Opin Hematol. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- 52.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 53.Serrano E, Lasa A, Perea G, Carnicer MJ, Brunet S, Aventin A, Sierra J, Nomdedeu JF. Acute myeloid leukemia subgroups identified by pathway-restricted gene expression signatures. Acta Haematol. 2006;116:77–89. doi: 10.1159/000093636. [DOI] [PubMed] [Google Scholar]
- 54.Afonja O, Smith JE, Cheng DM, Goldenberg AS, Amorosi E, Shimamoto T, Nakamura S, Ohyashiki K, Ohyashiki J, Toyama K, Takeshita K. MEIS1 and HOXA7 genes in human acute myeloid leukemia. Leukemia Res. 2000;24:849–855. doi: 10.1016/s0145-2126(00)00059-x. [DOI] [PubMed] [Google Scholar]
- 55.Sauvageau G, Thorsteinsdottir U, Hough MR, Hugo P, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myelo-proliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 56.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischbach NA, Rozenfeld S, Shen W, Fong S, Chrobak D, Ginzinger D, Kogan SC, Radhakrishnan A, Le Beau MM, Largman C, Lawrence HJ. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105:1456–1466. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- 58.Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 Immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced Meis expression. Mol Cell Biol. 2000;20:3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pineault N, Buske C, Feuring-Buske M, Abramovich C, Rosten P, Hogge DE, Aplan PD, Humphries RK. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003;101:4529–4538. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 62.Pineault N, Abramovich C, Humphries RK. Transplantable cell lines generated with NUP98-Hox fusion genes undergo leukemic progression by Meis1 independent of its binding to DNA. Leukemia. 2005;19:636–643. doi: 10.1038/sj.leu.2403696. [DOI] [PubMed] [Google Scholar]
- 63.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 66.Wermuth PJ, Buchberg AM. Meis1-mediated apoptosis is caspase dependent and can be suppressed by coexpression of HoxA9 in murine and human cell lines. Blood. 2005;105:1222–1230. doi: 10.1182/blood-2004-03-0802. [DOI] [PubMed] [Google Scholar]
- 67.Wang GG, Pasillas MP, Kamps MP. Persistent Transactivation by Meis1 Replaces Hox Function in Myeloid Leukemogenesis Models: Evidence for Co-Occupancy of Meis1-Pbx and Hox-Pbx Complexes on Promoters of Leukemia-Associated Genes. Mol Cell Biol. 2006;26:3902–3916. doi: 10.1128/MCB.26.10.3902-3916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mamo A, Krosl J, Kroon E, Bijl J, Thompson A, Mayotte N, Girard S, Bisaillon R, Beslu N, Featherstone M, Sauvageau G. Molecular dissection of Meis1 reveals 2 domains required for leukemia induction and a key role for Hoxa gene activation. Blood. 2006;108:622–629. doi: 10.1182/blood-2005-06-2244. [DOI] [PubMed] [Google Scholar]
- 69.Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias [see comment] Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 70.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 72.Eklund EA. The role of HOX genes in malignant myeloid disease. Curr Opin Hematol. 2007;14:85–89. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- 73.Faber J, Armstrong SA. Mixed lineage leukemia translocations and a leukemia stem cell program. Cancer Res. 2007;67:8425–8428. doi: 10.1158/0008-5472.CAN-07-0972. [DOI] [PubMed] [Google Scholar]
- 74.Faber J, Krivtsov AV, Stubbs MC, Wright R, van den Heuvel-Eibrink M, Kung AL, et al. HoxA9 Knockdown Inhibits Proliferation and Induces Cell Death in Human MLL-Rearranged Leukemias. [Abstract #734] Blood. 2006:108. [Google Scholar]
- 75.Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, Moulding DA, Kioussis D, Lappin TR, Brady HJ, Williams O. Continuous MLL-ENL expression is necessary to establish a “Hox Code” and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen IM, Feinberg AP, Jenkins NA, Copeland NG, Shaughnessy JD. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11) (p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 77.Slape C, Aplan PD. The role of NUP98 gene fusions in hematologic malignancy. Leuk Lymphoma. 2004;45:1341–1350. doi: 10.1080/10428190310001659325. [DOI] [PubMed] [Google Scholar]
- 78.Dorsam ST, Ferrell CM, Dorsam GP, Derynck MK, Vijapurkar U, Khodabakhsh D, Pau B, Bernstein H, Haqq CM, Largman C, Lawrence HJ. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103:1676–1684. doi: 10.1182/blood-2003-07-2202. [DOI] [PubMed] [Google Scholar]
- 79.Ferrell CM, Dorsam ST, Ohta H, Humphries RK, Derynck MK, Haqq C, Largman C, Lawrence HJ. Activation of stem-cell specific genes by HOXA9 and HOXA10 homeodomain proteins in CD34+ human cord blood cells. Stem Cells. 2005;23:644–655. doi: 10.1634/stemcells.2004-0198. [DOI] [PubMed] [Google Scholar]
- 80.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- 81.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 82.Morgado E, Albouhair S, Lavau C. Flt3 is dispensable to the Hoxa9/Meis1 leukemogenic cooperation. Blood. 2007;109:4020–4022. doi: 10.1182/blood-2006-01-039586. [DOI] [PubMed] [Google Scholar]
- 83.Palmqvist L, Argiropoulos B, Pineault N, Abramovich C, Sly LM, Krystal G, Wan A, Humphries RK. The Flt3 receptor tyrosine kinase collaborates with NUP98-HOX fusions in acute myeloid leukemia. Blood. 2006;108:1030–1036. doi: 10.1182/blood-2005-12-007005. [DOI] [PubMed] [Google Scholar]
- 84.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stubbs MC, Faber J, Kung AL, Cameron S, Armstrong SA. HOXA9 Represses Bim Expression in MLL Rearranged Leukemia: Implications for Drug Therapy. [Abstract #57] Blood. 2007;110:26a. [Google Scholar]
- 86.Faber J, Krivtsov AV, Stubbs MC, Van den Heuvel-Eibrink M, Kung AL, Zwaan CM, et al. The Myocyte Enhancer Factor 2C (MEF2C) Is a Direct Transcriptional Target of HOXA9 and Mediates Leukemia Survival in Human Mixed-Lineage Leukemias. Blood. 2007;110:26a. [Abstract #58] [Google Scholar]
- 87.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 88.Whelan JT, Ludwig DL, Bertrand FE. HoxA9 induces insulin-like growth factor-1 receptor expression in B-lineage acute lymphoblastic leukemia. Leukemia. 2008 doi: 10.1038/leu.2008.57. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 89.Dintilhac A, Bihan R, Guerrier D, Deschamps S, Pellerin I. A conserved non-homeodomain Hoxa9 isoform interacting with CBP is co-expressed with the ‘typical’ Hoxa9 protein during embryogenesis. Gene Expr Patterns. 2004;4:215–222. doi: 10.1016/j.modgep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 90.Shen WF, Krishnan K, Lawrence HJ, Largman C. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol Cell Biol. 2001;21:7509–7522. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. J Biol Chem. 2005;280:12359–12370. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 92.Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. PBX proteins: much more than Hox cofactors. Int J Dev Biol. 2008;52:9–20. doi: 10.1387/ijdb.072304al. [DOI] [PubMed] [Google Scholar]
- 93.Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KLB. Eukaryotic Translation Initiation Factor 4E Activity Is Modulated by HOXA9 at Multiple Levels. Mol Cell Biol. 2005;25:1100–1112. doi: 10.1128/MCB.25.3.1100-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–751. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 95.Bandyopadhyay S, Ashraf MZ, Daher P, Howe PH, DiCorleto PE. HOXA9 Participates in the Transcriptional Activation of E-Selectin in Endothelial Cells. Mol Cell Biol. 2007;27:4207–4216. doi: 10.1128/MCB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diehl F, Rossig L, Zeiher AM, Dimmeler S, Urbich C. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood. 2007;109:1472–1478. doi: 10.1182/blood-2006-08-039651. [DOI] [PubMed] [Google Scholar]
- 97.Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyard J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 99.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, Rosell R, Gemmill RM, Drabkin HA. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci USA. 2000;97:12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lind GE, Skotheim RI, Fraga MF, Abeler VM, Esteller M, Lothe RA. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and SCGB3A1 (HIN-1) J Pathol. 2006;210:441–449. doi: 10.1002/path.2064. [DOI] [PubMed] [Google Scholar]
- 102.Wu Q, Lothe RA, Ahlquist T, Silins I, Trope CG, Micci F, Nesland JM, Suo Z, Lind GE. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crijns AP, de Graeff P, Geerts D, Ten Hoor KA, Hollema H, van der Sluis T, Hofstra RM, de Bock GH, de Jong S, vander Zee AG, de Vries EG. MEIS and PBX homeobox proteins in ovarian cancer. Eur J Cancer. 2007;43:2495–2505. doi: 10.1016/j.ejca.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 104.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhourse MJ, van der Spek PJ, Lowenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 105.Kasper LH, Brindle PK, Schnabel CA, Pritchard CEJ, Cleary ML, van Deursen JMA. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]