Abstract
Widely used measures of the environment, especially the family environment of children, show genetic influence in dozens of twin and adoption studies. This phenomenon is known as gene-environment correlation in which genetically driven influences of individuals affect their environments. We conducted the first genome-wide association (GWA) analysis of an environmental measure. We used a measure called CHAOS which assesses ‘environmental confusion’ in the home, a measure that is more strongly associated with cognitive development in childhood than any other environmental measure. CHAOS was assessed by parental report when the children were 3 years and again when the children were 4 years; a composite CHAOS measure was constructed across the 2 years. We screened 490,041 autosomal single-nucleotide polymorphisms (SNPs) in a two-stage design in which children in low chaos families (N = 469) versus high chaos families (N = 369) from 3,000 families of 4-year-old twins were screened in Stage 1 using pooled DNA. In Stage 2, following SNP quality control procedures, 41 nominated SNPs were tested for association with family chaos by individual genotyping an independent representative sample of 3,529. Despite having 99% power to detect associations that account for more than 0.5% of the variance, none of the 41 nominated SNPs met conservative criteria for replication. Similar to GWA analyses of other complex traits, it is likely that most of the heritable variation in environmental measures such as family chaos is due to many genes of very small effect size.
Keywords: GE correlation, Family chaos, Genomewide association, Microarrays, Community sample
Introduction
Although genotype-environment (GE) interaction is the current focus of much behavioral genetic research, GE correlation appears to be a more widespread phenomenon (Plomin and Davis 2006). GE interaction refers to genetic sensitivity to environments in the sense that the effects of the environment can depend on genetics and the effects of genetics can depend on the environment (Kendler and Eaves 1986). In contrast, GE correlation refers to genetic exposure to the environment in that experiences can be correlated with genotype. Identifying GE correlation involves treating environmental measures as dependent measures in quantitative genetic analyses, and for this reason such research has been called the nature of nurture (Plomin and Bergeman 1991). Beginning with the pioneering work of Rowe (1981, 1983), dozens of twin and adoption studies have shown ubiquitous genetic influence on widely used measures of the environment (Plomin 1994). A recent review of 55 independent genetic studies using environmental measures found an average heritability of 0.27 across 35 different environmental measures (Kendler and Baker 2007).
In developmental psychology, the main long-term goal of GE correlation research is to explain the extent to which genetic factors mediate associations between ostensibly environmental measures and measures of children’s development (Plomin 1994). In terms of measures of parenting which are the mainstay of such studies, the key question is about the direction of effects, the extent to which parenting is the cause or the effect of children’s development (Bell 1968). For this reason, it is reasonable to investigate genetic influence of children on their parents’ parenting. For example, much GE correlation research in quantitative genetics uses twins who are children but analyzes parental reports of parenting towards the children (Plomin 1994). Finding evidence for genetic influence in such a design suggests that parenting reflects genetic differences in children, which is central to the direction-of-effects issue. However, the investigation of GE correlation is rich in other possibilities for understanding ways in which genetics impacts associations between parenting measures and children’s development (Plomin et al. 1977). It is also possible to use children’s self-reports of their perceptions of their parents’ parenting, which would assess genetic factors in the children themselves that contribute to heritability of such perceptions of parenting. In addition, it is possible to study genetic influences on parenting using a design in which the parents are the twins, which would assess genetic factors in the parents themselves that contribute to heritability regardless of whether the parenting measure is associated with children’s development.
Because of our interest in the direction-of-effects issue, we focused on a parenting measure known to relate to children’s development and conducted a genome-wide association study of this measure of parenting but using children. We used a parent-report measure called the Confusion, Hubbub, And Order Scale (CHAOS; Matheny et al. 1995) because it shows stronger (negative) associations with cognitive development in childhood than other environmental measures such as socioeconomic status (Petrill et al. 2004; Pike et al. 2006). Family chaos involves ‘environmental confusion’—the lack of organization and calm in the household. Although the CHAOS measure has not yet been used in published GE correlation research, the review mentioned above included results for three measures of family organization which yielded an average heritability of 0.25 (Kendler et al. 2007). We have found that a self-report version of the CHAOS measure produced a heritability estimate of 0.50 based on MZ and DZ twin correlations of 0.66 and 0.41 (unpublished) in a sample of 3,000 9-year-old twin pairs in our Twins Early Development Study (TEDS; Oliver and Plomin 2007), the sample used in the present study.
Research on GE correlation will be greatly advanced when genes are identified that are responsible for the heritability of environmental measures (Jaffee and Price 2007), just as research on GE interaction was advanced by identifying genes that interact with the environment in affecting behavioral development (Caspi et al. 2003, 2002). Candidate gene associations have been reported with marital status (Dick et al. 2006) and adults’ retrospective reports of how they were parented (Lucht et al. 2006). We used five SNPs reported to be associated with general cognitive ability in 7-year-olds (Butcher et al. 2005a, b) as a composite ‘SNP set’ and found that the SNP set correlated significantly with the CHAOS measure completed by parents when their children were 3 and 4 years old (Harlaar et al. 2005). Maternal education and paternal occupational class were not correlated with the SNP set. This evidence for GE correlation using measured genes and measured environments motivated us to conduct a genome-wide association scan for genes associated with family chaos.
Genomewide association scans are now possible using SNP microarrays (Hirschhorn and Daly 2005), although many issues remain to be resolved such as gene-centered versus genome-centered approaches (Neale and Sham 2004), common versus rare variants, sample size, and design (Carlson et al. 2004; Newton-Cheh and Hirschhorn 2005; Thomas et al. 2005; Wang et al. 2005). However, microarrays are expensive and can be used only once, which makes them impractical for genotyping the very large samples needed to detect gene associations of small effect size. One economical strategy for screening large samples for small effects is to pool DNA for groups such as cases versus controls for a disorder or low versus high groups for a quantitative trait (Darvasi and Soller 1994; Knight and Sham 2006; Norton et al. 2004). We have combined the strengths of microarrays and DNA pooling in a method we call SNP microarrays and pools (SNP-MaP). We and others have shown that pooled DNA can be genotyped reliably on microarrays (Butcher et al. 2004; Kirov et al. 2006; Meaburn et al. 2005; Meaburn et al. 2006; Pearson et al. 2007) and we have used the SNP-MaP method to identify genes associated with general cognitive ability (Butcher et al. 2005b, 2007) and with reading (Meaburn et al. 2007) using a multistage design that includes confirmation by individual genotyping of SNPs nominated in the SNP-MaP scan.
In the present study, we apply the SNP-MaP method in a two-stage association scan of family chaos in a representative UK sample of 6,000 4-year-old children in 3,000 families. In the first stage, we used pooled DNA to screen for the largest SNP allele frequency differences from 490,041 autosomal SNPs comparing low chaos families (N = 463) and high chaos families (N = 402). In the second stage, we individually genotyped 48 SNPs nominated by SNP-MaP and tested them for association in an unselected representative sample of 3,529 children; genotyping an unselected sample allows us to test the quantitative trait locus (QTL) hypothesis by assessing the extent to which the SNPs are associated with CHAOS throughout the distribution. The goal of this two-stage design was to balance false positive and false negative results in the search for associations of small effect size.
Methods
Participants
The Twins Early Development Study (TEDS) is a large, longitudinal study set up to investigate the genetic and environmental bases of cognitive and behavioral development (Oliver et al. 2007; Trouton et al. 2002). TEDS recruited families of twins born in England and Wales in 1994, 1995 and 1996. Nearly 16,000 families were contacted, of whom over 11,000 agreed to participate. Parents completed questionnaire booklets in the year following the birth of the twins that assessed a range of background variables, with subsequent questionnaire booklets sent before the children’s birthdays. The sample is representative of the UK population (ascertained by comparison with the 1994 census data from the Office of National Statistics), although fewer mothers of twins are in full-time work outside the home. We excluded children with severe current medical problems, children who had suffered severe problems at birth or whose mothers had suffered severe problems during pregnancy. Unknown or uncertain zygosity was also grounds for exclusion. We also excluded twins whose first language was other than English. Finally, in order to avoid issues of population stratification, we included only twins whose parents reported their ethnicity as ‘white’, which is 94% of the sample (comparable to the UK population). The sample used in the present study included 4,650 families for whom DNA was available as well as environmental measures when the children were 4 years of age.
Measure
The degree of chaos in the home was assessed at both 3 and 4 years of age by parents (98% mothers) using the Confusion, Hubbub, and Order Scale (CHAOS; Matheny et al. 1995). The CHAOS questionnaire has been validated through comparison with direct observations in the home environment (Matheny et al. 1995). More than a dozen publications have used the CHAOS measure; a recent paper concludes that “the CHAOS scale provides an adequate and economical measure of home confusion and disorganization that should prove useful in clinical research with diverse populations” (Dumas et al. 2005).
The short version of the CHAOS measure that we used consisted of six items rated on a five-point scale (1 = definitely untrue, 5 = definitely true), including the following examples: “You can’t hear yourself think in our home” and “we are usually able to stay on top of things” (reverse scored). As mentioned earlier, in our research, the CHAOS measure assessed by parents when their children were 3 and 4 years of age correlated more highly with cognitive development than did other environmental measures such as socio-economic status; moreover, CHAOS correlated with cognitive development independently of socio-economic status (Pike et al. 2006), as has been found in other studies as well (Dumas et al. 2005). In our study when the children were 3 and 4 years old, CHAOS correlated 0.28 with low socio-economic status, 0.41 with maternal depression, 0.27 with negative maternal feelings towards the children, and 0.30 with harsh discipline towards the children (Pike et al. 2006).
A total chaos score was generated at 3 years and at 4 years by summing the items (following reverse scoring so that high values = high chaos). In our sample, the scale yielded acceptable internal consistency at both ages (Cronbach’s alpha = 0.63). The scale yielded a correlation of 0.69 from 3 to 4 years (N = 4650 families), indicating considerable stability across a year, which is a lower-limit estimate of test–retest reliability. Because reliability is increased by aggregating data at multiple measurement occasions, we averaged the CHAOS scores at 3 and 4 years.
Design and Procedures
The design and procedures are described briefly in this section; greater detail can be found in other publications (Butcher et al. 2007; Meaburn et al. 2007).
Stage 1: SNP microarrays and pooling (SNP-MaP) screen of low versus high groups
Low and high CHAOS families were selected from the TEDS sample of more than 4,000 families with twins for whom DNA on both twins and CHAOS data on the family were available. A 33% cut-off (i.e., the top and bottom third) was used to select families from the CHAOS score distribution. In addition, as part of an ongoing GE interaction study of general cognitive ability (‘g’), families were also required to score in either the top or bottom 15% of the general cognitive ability distribution. These criteria resulted in the selection of 469 low CHAOS families and 369 high CHAOS families. Allele frequencies for the low and high groups were indexed by the average of 10 independent DNA sub-pools (biological replicates) per group; each individual was randomly ascribed to one sub-pool.
Stage 2: Testing the QTL hypothesis by individually genotyping SNPs nominated by SNP-MaP in an unselected sample
In Stage 2 of the study, the QTL hypothesis was tested by individually genotyping an independent sample. Because the foundation sample contains twin pairs, only one twin per pair was selected (N = 4,655). We also excluded Stage 1 individuals and MZ co-twins of Stage 1 individuals. (Although it would be acceptable to include MZ co-twins of Stage 1 individuals in Stage 2, CHAOS is a family-wide measure, which means including an MZ co-twin is tantamount to including the pooled individual themselves because the genotype and the phenotype is the same.) After removing these individuals, 4,183 individuals remained; 3,529 had CHAOS data (z-score range of the sample was −2.3 to 4.0). The sample provides 100%, 99%, and 76% power to detect an additive single-locus genetic effect explaining 1%, 0.5% and 0.2% of the total variance of CHAOS scores, respectively, uncorrected for multiple testing (P < 0.05, one-tailed) (Purcell et al. 2003). These power estimates refer to the SNPs themselves; power is of course less to detect indirect associations with other polymorphisms in between the SNPs assessed on the Affymetrix GeneChip 500 K array. Nonetheless, with these SNPs, two-thirds of the SNPs are in high LD (r 2 > 0.8) with a SNP genotyped in HapMap (Pe’er et al. 2006); with this in mind, power to capture a truly contributory variant by indirect association is equivalent to genotyping a sample 2,823 individuals. Such a scenario has 100%, 96% and 66% to detect QTLs with the same parameters as previously mentioned.
DNA pool construction
Each individual selected for the low or high CHAOS groups was randomly assigned to one of ten sub-pools for each group. Genomic DNA for each individual, extracted from buccal swabs (Freeman et al. 2003) and suspended in EDTA TE buffer (0.01 M Tris–HCl, 0.001 M EDTA, pH 8.0), was quantified in triplicate using PicoGreen™ dsDNA quantitation reagent (Invitrogen). Upon obtaining reliable triplicate readings, each individual contributed the same amount of DNA to their respective sub-pool. Because individual samples differed in their concentrations, a range of volumes of individual DNAs was added to permit equimolar DNA contributions to the sub-pools. We deemed 1 μl the minimum volume that could be added to a sub-pool without compromising pipette error. Therefore, the amount of DNA contributed to the sub-pools was determined by the mass of DNA contained in 1 μl of the most concentrated individual, in this case 98.6 ng/μl. Each individual therefore contributed 98.6 ng to the DNA pool. The range of concentrations for the 20 sub-pools was: 14.7–17.2 ng/μl (low CHAOS), and 15.7–17.2 ng/μl (high CHAOS).
SNP microarray allelotyping of pooled DNA
Each of the 20 DNA pools was allelotyped using the GeneChip® Mapping 500 K Array set in accordance with the standard protocol for individual DNA samples (see the GeneChip® Mapping 500 K Assay Manual for full protocol). Each microarray was scanned using the GeneChip® Scanner 3000 with High-Resolution Scanning Upgrade, which was controlled using GeneChip® Operating software (GCOS) v1.4. Cell intensity (.cel) files were analyzed using GTYPE. Each of the twenty DNA sub-pools was assayed on a separate microarray set; for quality control checks, a reference DNA individual provided by the manufacturer (sample number 100103) was also assayed on a separate microarray set.
Generation of SNP-MaP allele frequency estimates
Relative Allele Signal (RAS) scores, calculated using the 10 K MPAM Mapping algorithm, have been shown to be reliable and valid indices of allele frequency in pooled DNA (Brohede et al. 2006; Butcher et al. 2004; Craig et al. 2005; Kirov et al. 2006; Liu et al. 2005; Meaburn et al. 2005, 2006; Simpson et al. 2005). Details of how probesets on Affymetrix Mapping GeneChip® microarrays are used to calculate allele frequency estimates as described elsewhere (Butcher et al. 2007). Allele frequency estimates for the 500 K microarray set were calculated manually from the raw probe intensity data exported as a .txt file.
Selection of SNPs from Stage 1
To select SNPs for individual genotyping, we derived a rank-based composite score based on five criteria from the Stage 1 dataset. The derivation of this composite score is presented elsewhere (Butcher et al. 2007). Briefly, the five criteria were: (1) greater average allele frequency difference between low and high CHAOS groups, (2) smaller average variance of the low and high CHAOS groups (i.e., variance across the DNA pooled allele frequency estimates for each group), (3) smaller average variance within each microarray (i.e., variance across the multiple probesets that form the microarray’s allele frequency estimate), (4) greater number of successful replicate pools, and (5) greater minor allele frequency, as indexed by the average of the low and high CHAOS groups. Because we expect many more putatively significant associations from Stage 1 than could be realistically individually genotyped (>5,000, P < 0.01), we used this composite to choose the top 48 SNPs with the highest composite scores. The SNP screen was restricted to the autosomes because the DNA pools included both boys and girls, which complicates analyses of SNPs on the X chromosome.
Individual genotyping
After excluding Stage 1 individuals and selecting just one twin per pair as described earlier, the 3,529 individuals were genotyped using the Applied Biosystems’ SNPlex™ genotyping system and analyzed using GeneMapper v4.0 software (Applied Biosystems). SNPlex is a capillary electrophoresis-based multiplex genotyping system capable of genotyping up to 48 SNPs per sample per well (Tobler et al. 2005). In addition to the 3,529 TEDS individuals, 88 CEPH individuals who have been genotyped as part of the HapMap Project (The International HapMap Consortium 2003; The International HapMap Consortium 2005) were obtained from the Coriell Institute to assess genotyping quality and error rate. Reference genotypes of CEPH individuals for the selected SNPs were downloaded from HapMart, the data mining tool for downloading HapMap data (http://hapmart.hapmap.org/BioMart/martview).
Because quantitative genetic research strongly suggests that the majority of genetic effects are additive, we were primarily interested in testing SNPs for their additive effect. Therefore, genotypes of SNPs passing quality control (see below) were tested for additive genetic effects using a Pearson correlation (r). In addition, we followed a procedure recommended by Balding (2006) to test whether a non-additive model (ANOVA) predicted significantly better than an additive model (linear regression).
Genotyping quality control for individual genotyping
The following sequential criteria were applied: SNPs were omitted from analysis if poor genotype clusters prevented GeneMapper software from making calls or if a SNP showed >1 genotype mismatch between CEPH genotypes deposited in HapMap and those derived using in-house genotyping methods. Individuals were omitted if their SNP call rate was <65%. Finally, for each SNP, low peak height genotypes (<25% of the average peak height) were removed; we apply this procedure because poor quality samples often exhibit high background noise that SNPlex can mistake as heterozygotes. It is important to control for this as an excess of heterozygotes will artificially inflate the type-I error rate of Hardy–Weinberg equilibrium tests.
Results
Stage 1: SNP microarrays and pooling (SNP-MaP) screen of low versus high groups
SNP-MaP allele frequencies for the 20 DNA pools were calculated. In order to increase the reliability of SNP-MaP allele frequency estimates, we required allele frequency estimates from a minimum of 6 (out of 10) replicates for both high and low groups. We also excluded SNPs with minor allele frequencies lower than 0.05 as power to detect association in this range is greatly reduced. After these exclusion criteria, the autosomal genomewide screen consisted of 448,944 SNPs from the 500 K microarray set.
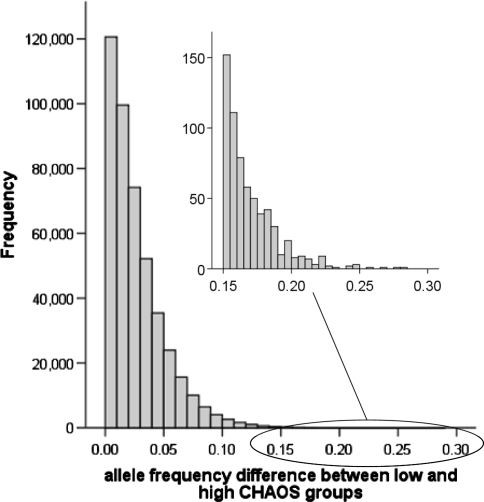
The average allele frequency for the low and high CHAOS groups was calculated for each SNP. The correlation between the low and high CHAOS groups was 0.992, indicating that the rank order of allele frequencies was highly reliable overall—a test analogous to genome control. Accordingly, between-group differences were small: Fig. 1 illustrates that 90% of the SNPs exhibited between-group differences smaller than 0.05, with a mean between-group absolute difference of 0.027 for the whole dataset (range: 0.00–0.28).
Fig. 1.
A histogram illustrating the distribution of absolute allele frequency differences between low and high CHAOS groups derived through pooled DNA on microarrays. The y-axis indicates the number of SNPs and x-axis shows absolute allele frequency differences between low and high CHAOS groups. The figure shows that the vast majority of allele frequency differences are small and that the mean allele frequency between low and high CHAOS groups is about 0.027. The x-axis is elongated to accommodate outliers, which are a logical source of candidate SNPs to follow up. The total number of SNPs is 448,944 because SNPs represented by fewer than 6 out of 10 replicates were removed
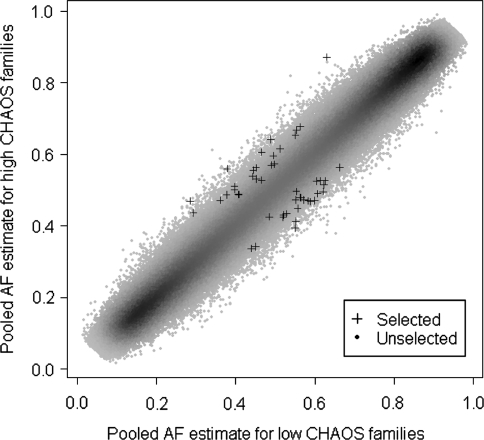
As explained in Methods, SNPs selected for individual genotyping in Stage 2 were chosen on the basis of a ranked composite score which took into account the between-group allele frequency difference, variance between- and within-biological replicate microarrays, number of successfully assayed arrays and minor allele frequency. Due to financial restrictions, we were limited to individually genotyping in SNPlex a single probeset of 48 SNPs with the highest composite scores. The mean absolute difference between low and high SNP-MaP allele frequency estimates for these 48 SNPs was 0.11 (ranging from 0.05 to 0.24). The seven SNPs with the largest between-group allele frequency differences were not selected as they exhibited high levels of variance, which counted unfavorably in the composite selection score. Figure 2 places the 48 selected SNPs in the context of the full dataset by plotting the average allele frequency of the low CHAOS group against that of the high CHAOS group. Details about the 48 selected SNPs can be found in Table 1.
Fig. 2.
A scatterplot showing the 48 top-ranked SNPs (crosses) against the background of 448,994 unselected SNPs comparing allele frequencies for the low CHAOS group (x-axis) and the high CHAOS group (y-axis). The figure also displays the density of SNPs as a function of low CHAOS versus high CHAOS allele frequency differences; density of SNP clusters increases as the heat map changes from light grey (sparse clusters) though to dark grey (dense clusters). Allele frequency differences are small with the majority of small differences occurring for SNPs with minor allele frequencies of 0.10–0.25, which reflects the representation of SNPs with these allele frequencies on the Affymetrix microarray. The correlation between low and high CHAOS allele frequencies was 0.992 indicating high reliability of the rank order of allele frequencies across the low and high CHAOS groups
Table 1.
Summary of Stage 1 and Stage 2 genotyping for the 48 SNPs selected by ranked composite score using pooled DNA to screen the genome
| SNP information | Stage 1 (SNP-MaP) | Stage 2 (individual genotyping) | Correct direction? | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AffyID | dbSNP RS ID | Chr | Genomic relation | Strand | Alleles | Low | High | L−H diff | Strand | QTL r | QTL p | N | H−W | |
| SNP_A-4295547 | rs10001415 | 4p15.1 | Intergenic | – | A/G | 0.63 | 0.872 | –0.242 | + | –0.003 | 0.436 | 2,794 | 0.000 | |
| SNP_A-1865002 | rs1030303 | 9q21.33 | AGTPBP1: intron | + | A/T | 0.397 | 0.502 | –0.105 | + | 0.018 | 0.172 | 2,905 | 0.005 | No |
| SNP_A-1964916 | rs10496711 | 2q21.2 | Intergenic | + | C/G | 0.604 | 0.526 | 0.078 | + | –0.016 | 0.189 | 2,976 | 0.647 | No |
| SNP_A-1813456 | rs10783746 | 12q13.2 | mRNA evidence | + | A/G | 0.62 | 0.496 | 0.124 | + | 0.019 | 0.152 | 2,879 | 0.562 | Yes |
| SNP_A-2005014 | rs10884390 | 10q25.1 | SORCS1: intron | + | A/G | 0.571 | 0.472 | 0.099 | + | 0.028 | 0.068 | 2,951 | 0.914 | Yes |
| SNP_A-1893780 | rs10915207 | 1p35.3 | EPB41: intron | – | A/G | 0.489 | 0.57 | –0.081 | + | 0.006 | 0.378 | 2,739 | 0.978 | |
| SNP_A-1887744 | rs11084785 | 19q13.11 | Intergenic | + | C/T | 0.551 | 0.654 | –0.103 | + | 0.007 | 0.364 | 2,703 | 0.929 | |
| SNP_A-2305187a | rs11263591 | 11q13.3 | FGF3: intron | – | C/G | 0.406 | 0.488 | –0.082 | + | – | – | – | – | |
| SNP_A-2049663 | rs11264825 | 1q23.1 | FCRL1: intron | – | C/T | 0.465 | 0.528 | –0.063 | + | −0.007 | 0.356 | 2,587 | 0.971 | |
| SNP_A-1822015 | rs11950448 | 5p12 | Intergenic | – | C/G | 0.52 | 0.425 | 0.095 | + | −0.001 | 0.483 | 2,880 | 0.021 | |
| SNP_A-1921659 | rs1214451 | 10q23.31 | Intergenic | + | C/G | 0.495 | 0.596 | –0.101 | + | −0.027 | 0.072 | 2,920 | 0.969 | Yes |
| SNP_A-2112236 | rs1275051 | 11q24.1 | Intergenic | + | A/G | 0.564 | 0.479 | 0.085 | + | −0.007 | 0.351 | 2,959 | 0.595 | |
| SNP_A-1869107 | rs12820468 | 12q23.1 | TMEM16D: intron | – | A/G | 0.528 | 0.434 | 0.094 | + | −0.031 | 0.048 | 2,966 | 0.810 | Yes |
| SNP_A-1957323 | rs13262547 | 8q13.2 | mRNA evidence | + | G/T | 0.556 | 0.449 | 0.107 | + | 0.005 | 0.402 | 2,880 | 0.997 | |
| SNP_A-2161686 | rs1414314 | 1p32.1 | Intergenic | – | A/G | 0.45 | 0.342 | 0.108 | + | 0.038 | 0.023 | 2,712 | 0.416 | No |
| SNP_A-2072154 | rs1501630 | 3q24 | mRNA evidence | + | C/T | 0.451 | 0.531 | –0.08 | – | −0.015 | 0.207 | 2,916 | 0.998 | No |
| SNP_A-2234256 | rs1667301 | 2q37.1 | mRNA evidence | – | C/T | 0.488 | 0.642 | –0.154 | – | 0.004 | 0.406 | 2,960 | 1.000 | |
| SNP_A-2183115 | rs17203935 | 18q11.2 | mRNA evidence | – | G/T | 0.625 | 0.526 | 0.099 | + | 0.015 | 0.215 | 2,742 | 0.288 | No |
| SNP_A-1804934 | rs17221652 | 10p15.3 | ADARB2: intron | + | C/G | 0.551 | 0.473 | 0.078 | + | 0.022 | 0.124 | 2,701 | 0.162 | Yes |
| SNP_A-1915452 | rs1861594 | 12p12.3 | PTPRO: intron | + | A/G | 0.439 | 0.337 | 0.102 | + | 0.022 | 0.118 | 2,949 | 0.125 | Yes |
| SNP_A-2269742 | rs2043478 | 5q31.2 | SPOCK1: intron | + | A/G | 0.598 | 0.471 | 0.127 | + | –0.005 | 0.397 | 2,716 | 0.739 | |
| SNP_A-2229263 | rs289647 | 10p11.21 | Intergenic | + | A/G | 0.55 | 0.394 | 0.156 | + | 0.004 | 0.414 | 2,854 | 0.471 | |
| SNP_A-2252700 | rs297255 | 10q26.13 | mRNA evidence | + | A/G | 0.511 | 0.616 | –0.105 | – | −0.019 | 0.150 | 2,948 | 1.000 | No |
| SNP_A-1955113 | rs316191 | 5q15 | RIOK2: intron | – | G/T | 0.408 | 0.488 | –0.08 | – | 0.001 | 0.468 | 2,941 | 0.441 | |
| SNP_A-1828397 | rs318931 | 10q26.3 | Intergenic | – | A/T | 0.613 | 0.526 | 0.087 | + | −0.014 | 0.220 | 2,897 | 0.043 | Yes |
| SNP_A-2025774 | rs333785 | 11p12 | Intergenic | + | G/T | 0.397 | 0.51 | –0.113 | + | 0.001 | 0.482 | 2,720 | 0.208 | |
| SNP_A-1800569a | rs3843872 | 3q13.13 | MYH15: intron | + | C/G | 0.451 | 0.562 | –0.111 | – | – | – | – | – | |
| SNP_A-2239462 | rs392670 | 3p26.3 | CHL1: intron | – | A/T | 0.521 | 0.431 | 0.09 | – | −0.014 | 0.227 | 2,958 | 0.556 | No |
| SNP_A-1809780 | rs4637723 | 7p12.3 | Intergenic | – | A/C | 0.497 | 0.573 | –0.076 | + | −0.026 | 0.090 | 2,734 | 0.848 | No |
| SNP_A-2161472 | rs4761289 | 12q15 | Intergenic | – | A/C | 0.293 | 0.437 | –0.144 | + | −0.032 | 0.046 | 2,792 | 0.162 | No |
| SNP_A-2086969 | rs4766691 | 12q24.13 | Intergenic | – | C/T | 0.285 | 0.47 | –0.185 | + | 0.001 | 0.478 | 2,697 | 0.072 | |
| SNP_A-2196642 | rs4787125 | 16p13.2 | Intergenic | + | A/G | 0.622 | 0.515 | 0.107 | + | 0.010 | 0.285 | 2,967 | 0.602 | Yes |
| SNP_A-1974629a | rs4839628 | 3q24 | SLC9A9: intron | – | A/G | 0.662 | 0.563 | 0.099 | + | – | – | – | – | |
| SNP_A-2106553 | rs4853578 | 2q32.3 | MYO1B: intron | + | A/G | 0.378 | 0.56 | –0.182 | + | 0.014 | 0.228 | 2,699 | 0.986 | No |
| SNP_A-4241706 | rs538385 | 13q12.3 | Intergenic | + | C/T | 0.442 | 0.538 | –0.096 | + | 0.026 | 0.087 | 2,716 | 0.965 | No |
| SNP_A-2281229 | rs6048344 | 20p11.21 | Intergenic | + | C/G | 0.562 | 0.48 | 0.082 | + | 0.001 | 0.483 | 2,911 | 0.535 | |
| SNP_A-2092290 | rs6130222 | 20q12 | PTPRT: intron | – | C/G | 0.444 | 0.556 | –0.112 | + | 0.011 | 0.275 | 2,950 | 0.605 | Yes |
| SNP_A-2020710 | rs619689 | 11q23.2 | Intergenic | + | C/G | 0.553 | 0.496 | 0.057 | – | 0.010 | 0.290 | 2,964 | 0.907 | No |
| SNP_A-2283323 | rs6583654 | 8q24.3 | Intergenic | + | C/T | 0.583 | 0.47 | 0.113 | + | 0.009 | 0.311 | 2,738 | 0.266 | |
| SNP_A-2133000 | rs6751663 | 2p14 | Intergenic | – | C/G | 0.377 | 0.487 | –0.11 | + | −0.006 | 0.368 | 2,967 | 0.087 | |
| SNP_A-2035401 | rs6776618 | 3q28 | Intergenic | + | C/G | 0.589 | 0.468 | 0.121 | + | −0.008 | 0.334 | 2,911 | 0.514 | |
| SNP_A-2277540 | rs7122693 | 11q23.2 | CADM1: intron | + | A/G | 0.465 | 0.606 | –0.141 | + | −0.005 | 0.387 | 2,734 | 0.751 | |
| SNP_A-2265151 | rs7233747 | 18q23 | Intergenic | + | C/T | 0.562 | 0.677 | –0.115 | + | 0.028 | 0.062 | 2,941 | 0.999 | No |
| SNP_A-2178721 | rs7873355 | 9p23 | Intergenic | + | C/G | 0.551 | 0.414 | 0.137 | + | −0.003 | 0.432 | 2,952 | 0.277 | |
| SNP_A-2270656 | rs7970012 | 12q24.32 | mRNA evidence | – | A/G | 0.606 | 0.49 | 0.116 | + | −0.015 | 0.206 | 2,987 | 0.002 | Yes |
| SNP_A-2042483 | rs9533962 | 13q14.11 | Intergenic | – | A/G | 0.36 | 0.472 | –0.112 | + | −0.026 | 0.078 | 2,955 | 0.940 | No |
| SNP_A-1959377 | rs9606213 | 22q11.21 | C22orf25: intron | + | A/G | 0.485 | 0.425 | 0.06 | + | 0.005 | 0.402 | 2,918 | 0.115 | |
| SNP_A-1785409 | rs9934231 | 16p13.13 | CLEC16A: intron | – | C/G | 0.553 | 0.668 | –0.115 | + | 0.010 | 0.286 | 2,942 | 0.690 | Yes |
These SNPs were individually genotyped in an unselected sample of 3,529 individuals in order to test the QTL hypothesis. The allele whose frequency is shown in the ‘Low’ and ‘High’ columns is the allele that is alphabetically first in the ‘Alleles’ column (e.g., A rather than G in the first row). Note that the alleles measured here (using Affymetrix microarrays) may not be the same as the alleles measured using individual genotyping (using the SNPlex™ assay) because the Affymetrix and SNPlex platforms may be based on different DNA strands (e.g., for the SNP in the first row, alleles on different strands were assessed in the Affymetrix microarray and in the SNPlex—which was of course taken into account in determining the direction of the association in Stage 1 and Stage 2. The Low–High allele frequency difference (‘l-h’) shown in the 9th column was calculated by subtracting the allele frequency of the High CHAOS group from the Low CHAOS group. ‘QTL r’ (column 11) is the correlation between the composite CHAOS scores and additive genotypic values (0, 1, 2) as scored in the direction of high CHAOS as predicted by Stage 1. For example, for the SNP in row 1, because the frequency of the A allele was higher in the high CHAOS group in Stage 1, the genotypic values in Stage 2 were scored as follows: GG = 0, AG = 1, AA = 2). QTL p (column 12) is the p value of the QTL r for the unselected sample of individuals (N shown in column 13)
The last column (‘correct direction’) column indicates whether the direction of allele frequency differences is the same in Stage 2 as it was in Stage 1. We did not compare directions of effect between Stage 1 and Stage 2 for SNPs yielding correlations in the range –0.01 and 0.01 in Stage 2.
aAssay failure: no data available
Note. Value in bold for SNP rs12820468 is the only SNP yielding a nominally significant association in the unselected independent sample in Stage 2
Individual genotyping quality control
In our SNPlex analysis, three out of 48 SNPs (rs11263591, rs3843872 and rs4839628) exhibited poor call rates across plates due to poor genotype clustering and were omitted from further analyses. The remaining SNPs showed acceptable genotyping error rates as measured by the concordance between our in-house derived genotypes for 88 CEPH individuals and the genotypes of the same CEPH individuals available from HapMap: We observed 3 mismatches out of 3,954 genotypes (error rate < 0.1%). Of these errors, homozygotes were erroneously called as heterozygotes.
393 individuals (11%) showing low call rates across SNPs were omitted; fragmented DNA is a pre-requisite to running SNPlex and sub-optimal fragmentation is the likely cause of these low call rates. We also excluded an additional 9% of genotypes per SNP whose peak heights were <25% of the average peak height for that SNP across the study. Finally, with 45 SNPs, none would be expected to depart from Hardy–Weinberg equilibrium at P < 0.01; however, 4 SNPs (rs10001415, rs1030303, rs11950448 and rs7970012) did show significant departures and these SNPs were omitted from subsequent analysis. At the cost of reduced sample size, these conservative criteria improved observed genotypic distributions under Hardy–Weinberg equilibrium, tightened genotype clusters in SNPlex, and left the distribution of CHAOS unchanged. After excluding the 7 aforementioned SNPs, we observe 128,299 (88.7%) of a possible 144,689 genotypes. After excluding samples with poor call rates and low peak heights, we used 117,062 genotypes to perform association analysis. The distribution of the CHAOS measure was unchanged after genotype exclusionary criteria.
Stage 2: Testing the QTL hypothesis by individually genotyping SNPs nominated by SNP-MaP in an unselected sample
The 41 successfully genotyped SNPs nominated by Stage 1 were individually genotyped across the unselected sample of 3,529 children in order to test the QTL hypothesis directly by assessing the extent to which the SNPs are associated with CHAOS throughout the distribution. Each individual’s genotypes for the 41 SNPs were tested for additive genotypic effects. With 41 tests and an alpha of 0.05, 2 significant results would be expected on the basis of chance alone using a nominal one-tailed alpha level of 0.05. (We used a one-tailed test because the difference observed in Stage 2 was required to be in the same direction as that seen in Stage 1 screening.). Only one SNP (rs12820468) was significantly associated in the predicted direction with individual differences in CHAOS throughout the distribution. A summary of Stage 1 and Stage 2 results for the 48 SNPs selected (including SNP locations) is provided in Table 1.
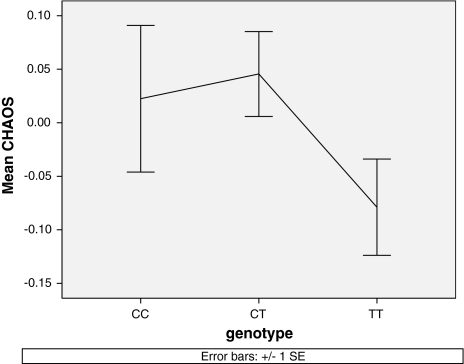
Figure 3 presents the results for rs12820468 in terms of standardized mean quantitative trait CHAOS scores for the three SNP genotypes. As can be seen from Fig. 3, the SNP appears to show dominance for the rarer C allele with homozygotes and heterozygote carriers appearing to be susceptible for selecting more disordered environments. However, following the procedure suggested by Balding (2006), we compared additive and non-additive models and found that the non-additive model did not fit significantly better than the additive model. We also examined the associations separately for boys and girls, but no significant differences were found; because our Stage 1 design included boys and girls it would favor SNPs that show effects in both sexes.
Fig. 3.
Genotype-by-phenotype plot for SNP rs12820468 illustrating the effect of genotype (x-axis) on standardized CHAOS scores (y-axis). The best-fitting genetic model was additive despite the apparent effect of dominance
Discussion
In the first genomewide association scan of an environmental measure, we chose to study family chaos using the parent-report CHAOS measure with DNA of the children because we are interested in the role of GE correlation in the mediation of associations between parenting and children’s development. CHAOS is an especially interesting parenting measure because it correlates more highly with children’s cognitive development than do other environmental measures, including socio-economic status. Like other measures of the family environment, there is evidence from quantitative genetic studies for genetic influence. The present study attempted to bring the power of genome-wide association (GWA) to bear on identifying some of the DNA variation in children responsible for genetic influence on parent-reported CHAOS.
We found one SNP associated with family chaos that reached a nominal significance level of P < 0.05. However, with 41 SNPs nominated in a first SNP-MaP stage using pooled DNA for low and high CHAOS groups we would expect 2 SNPs to remain significant on the basis of chance alone. For this reason, we conclude that despite having 99% power to detect SNP associations that account for more than 0.5% of the variance, we were unable to detect any SNP associations that met conservative criteria for significance. That is, because two significant associations in the second stage of the design would be expected on the basis of chance alone, we assume that the single significant association that emerged (rs12820468) is a chance result. Despite this, it is worth noting that rs12820468 is located in intron 7 of transmembrane protein 16D (TMEM16D) and although the likelihood of the SNP showing functionality is low, it lies in an LD block containing 4 indels (including a 9 bp deletion) and numerous repeat elements. TMEM16D is a large (∼334 Kb) protein coding gene of unknown function located on chromosome 12q23.1-q23.2 and exists in 3 known isoforms. Overall, TMEM16D shows some conservation of features with primates but little with placental mammals or vertebrates. Because functional plausibility is unclear, more work on the TMEM16D gene might be warranted in future molecular genetic research on family chaos and its correlates.
The power of the present design leads us to draw a more far-reaching conclusion: We conclude that it is unlikely that any SNPs of large effect contribute to heritable influence on family chaos as assessed by parents of young children using DNA of the children. As mentioned in the Introduction, we would only find SNP associations if parental reports of CHAOS are correlated with SNPs in their children—this is the key test of GE correlation mediation of the relationship between parenting and children’s development. However, if this were not one’s goal, it may be easier to find SNP associations using children’s own reports of CHAOS or to find SNP associations between parents’ reports of CHAOS and the parents’ DNA. Alternatively, power to detect associations underscoring such evocative GE correlations may be increased by directly studying the behaviors of twins that evoke the parenting. If these behaviors of twins are more heritable then these studies might also yield more associations.
Another possibility is that SNPs not represented on the Affymetrix 500 K array, as well as other polymorphisms (e.g., copy number variation, indels, microsatellites etc.), may have passed through our screen unnoticed. In this regard, it is noteworthy that the same design and sample used in this study have been successful in identifying six SNPs associated with general cognitive ability even though the average effect size of the six SNP associations was only 0.2% (Butcher et al. 2007). This finding suggests that the two-stage SNP-MaP design followed by individual genotyping of an independent unselected sample can identify SNP associations of small effect size; the present finding is important in demonstrating that the design does not always yield positive findings. One important difference between the two studies is that general cognitive ability is nearly twice as heritable as measures of family environment which could indicate that it will be more difficult to find genes associated with family environment, although it is not necessarily the case that it is easier to find genes for more heritable traits. Another difference between the two studies is that in the SNP-MaP stage, we used top and bottom thirds of the CHAOS distribution to select our low and high CHAOS groups, whereas the low and high general cognitive ability groups were selected from the top and bottom sixths. Given roughly equal sample sizes in the two studies, the less severe selection for CHAOS results in less power than in the study of general cognitive ability.
Nonetheless, the evidence for the heritability of measures of the family environment such as family chaos is persuasive (e.g., Kendler and Baker 2007), which implies that differences in DNA sequence are ultimately responsible for the heritability. It is likely that the DNA differences responsible for this heritability have such small or subtle effects that even more powerful strategies will be needed to detect them. Identifying genes associated with environmental measures will be worth the effort because they will foster research on an active model of experience in which individuals select, modify and create environments on the basis of their genetic proclivities (Plomin 1994). In other words, genetic effects on behavior do not stop at the skin—genetic effects need to be considered in relation to an ‘extended phenotype’ that includes effects on individuals’ environments (Dawkins 1982, 2004).
Acknowledgements
Our genomewide association scan of CHAOS was supported by grants from the Wellcome Trust (GR75492) and the U.S. National Institute of Child Health and Human Development (HD44454, HD46167, HD49861). The Twins Early Development Study (TEDS) has been funded since 1995 by a program grant from the U.K. Medical Research Council (G9424799, now G0500079). We are grateful to the TEDS families for their participation and support for more than a decade.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Edited by Danielle Dick.
References
- Balding DJ. A tutorial on statistical methods for population association studies. Nat Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Bell RQ. A reiteration of the direction of effects in socialization. Psycholog Rev. 1968;75:81–95. doi: 10.1037/h0025583. [DOI] [PubMed] [Google Scholar]
- Brohede J, Dunne R, McKay JD, Hannan GN. PPC: an algorithm for accurate estimation of SNP allele frequencies in small equimolar pools of DNA using data from high density microarrays. Nucleic Acids Res. 2006;33:e142. doi: 10.1093/nar/gni142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Liu L, Hill L, Al-Chalabi A, Plomin R, et al. Genotyping pooled DNA on microarrays: A systematic genome screen of thousands of SNPs in large samples to detect QTLs for complex traits. Behav Genet. 2004;34:549–555. doi: 10.1023/B:BEGE.0000038493.26202.d3. [DOI] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Dale PS, Sham P, Schalkwyk L, Craig IW, et al. Association analysis of mild mental impairment using DNA pooling to screen 432 brain-expressed SNPs. Mol Psychiatry. 2005;10:384–392. doi: 10.1038/sj.mp.4001589. [DOI] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, Craig IW, et al. SNPs, microarrays, and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6,000 children. Hum Mol Genet. 2005;14:1315–1325. doi: 10.1093/hmg/ddi142. [DOI] [PubMed] [Google Scholar]
- Butcher LM, Davis OSP, Craig IW, Plomin R (2007) Genomewide QTL association scan of general cognitive ability using pooled DNA and 500 K SNP microarrays. Genes, Brain and Behavior, (Epub ahead of print; doi: 10.1111/j.1601-183X.2007.00368.x) [DOI] [PMC free article] [PubMed]
- Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Craig DW, Huentelman MJ, Hu-Lince D, Zismann VL, Kruer MC, Lee AM, et al. Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics. 2005;6:138. doi: 10.1186/1471-2164-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics. 1994;138:1365–1373. doi: 10.1093/genetics/138.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R. The extended phenotype. Oxford: WH Freeman; 1982. [Google Scholar]
- Dawkins R. Extended phenotype—but not too extended. A reply to Laland, Turner and Jjablonka. Biol Philos. 2004;19:377–396. doi: 10.1023/B:BIPH.0000036180.14904.96. [DOI] [Google Scholar]
- Dick DM, Agrawal A, Schuckit MA, Bierut L, Hinrichs A, Fox L, et al. Marital status, alcohol dependence, and GABRA2: evidence for gene-environment correlation and interaction. J Stud Alcohol. 2006;67:185–194. doi: 10.15288/jsa.2006.67.185. [DOI] [PubMed] [Google Scholar]
- Dumas JE, Nissley J, Nordstrom A, Smith EP, Prinz RJ, Levine DW. Home chaos: sociodemographic, parenting, interactional, and child correlates. J Clin Child Adolesc Psychol. 2005;34:93–104. doi: 10.1207/s15374424jccp3401_9. [DOI] [PubMed] [Google Scholar]
- Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig I. DNA from buccal swabs recruited by mail: evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet. 2003;33:67–72. doi: 10.1023/A:1021055617738. [DOI] [PubMed] [Google Scholar]
- Harlaar N, Butcher LM, Meaburn E, Craig IW, Plomin R. A behavioural genomic analysis of DNA markers associated with general cognitive ability in 7-year-olds. J Child Psychol Psychiatry. 2005;46:1097–1107. doi: 10.1111/j.1469-7610.2005.01515.x. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Jaffee SR, Price TS. Gene-environment correlations: a review of the evidence and implications for prevention of mental illness. Mol Psychiatry. 2007;12:432–442. doi: 10.1038/sj.mp.4001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychol Med. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Eaves LJ. Models for the joint effects of genotype and environment on liability to psychiatric illness. Am J Psychiatry. 1986;143:279–289. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O’Donovan MC. Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics. 2006;7:27. doi: 10.1186/1471-2164-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J, Sham P. Design and analysis of association studies using pooled DNA from large twin samples. Behav Genet. 2006;36:665–677. doi: 10.1007/s10519-005-9016-9. [DOI] [PubMed] [Google Scholar]
- Liu Q-R, Drgon T, Walther D, Johnson C, Poleskaya O, Hess J, et al. Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proc Nat Acad Sci USA. 2005;102:11864–11869. doi: 10.1073/pnas.0500329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht M, Barnow S, Schroeder W, Grabe HJ, Finckh U, John U, et al. Negative perceived paternal parenting is associated with dopamine D2 receptor exon 8 and GABA(A) alpha 6 receptor variants: an explorative study. Am J Med Genet Part B Neuropsychiatr Genet. 2006;141:167–172. doi: 10.1002/ajmg.b.30255. [DOI] [PubMed] [Google Scholar]
- Matheny AP, Wachs TD, Ludwig JL, Phillips K. Bringing order out of chaos: psychometric characteristics of the Confusion, Hubbub and Order Scale. J Appl Dev Psychol. 1995;16:429–444. doi: 10.1016/0193-3973(95)90028-4. [DOI] [Google Scholar]
- Meaburn E, Butcher LM, Liu L, Fernandes C, Hansen V, Al-Chalabi A, et al. Genotyping DNA pools on microarrays: tackling the QTL problem of large samples and large numbers of SNPs. BMC Genomics. 2005;6:52. doi: 10.1186/1471-2164-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn E, Butcher LM, Schalkwyk LC, Craig IW, Plomin R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res. 2006;34:e27. doi: 10.1093/nar/gnj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn EL, Harlaar N, Craig IW, Schalkwyk LC, Plomin R (2007) Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100 K SNP microarrays in a sample of 5760 children. Molecular Psychiatry, Aug 7, (Epub ahead of print; doi:10.1038/sj.mp.4002063) [DOI] [PubMed]
- Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005;573:54–59. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Norton N, Williams NM, O’Donovan MC, Owen MJ. DNA pooling as a tool for large-scale association studies in complex traits. Ann Med. 2004;36:146–152. doi: 10.1080/07853890310021724. [DOI] [PubMed] [Google Scholar]
- Oliver B, Plomin R. Twins’ Early Development Study (TEDS): a multivariate, longitudinal genetic investigation of language, cognition and behavior problems from childhood through adolescence. Twin Res Hum Genet. 2007;10(1):96–105. doi: 10.1375/twin.10.1.96. [DOI] [PubMed] [Google Scholar]
- Pe’er I, de Bakker PIW, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, et al. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrill SA, Pike A, Price TS, Plomin R. Chaos in the home and socioeconomic status are associated with cognitive development in early childhood: environmental risks identified in a genetic design. Intelligence. 2004;32:445–460. doi: 10.1016/j.intell.2004.06.010. [DOI] [Google Scholar]
- Pike A, Iervolino A, Eley T, Price TS, Plomin R. Environmental risk and young children’s cognitive and behavioral development. Int J Behav Dev. 2006;30:55–66. doi: 10.1177/0165025406062124. [DOI] [Google Scholar]
- Plomin R. Genetics and experience: the interplay between nature and nurture. Thousand Oaks, California: Sage Publications Inc; 1994. [Google Scholar]
- Plomin R, Bergeman CS. The nature of nurture: genetic influences on “environmental” measures. Behav Brain Sci. 1991;14:373–427. [Google Scholar]
- Plomin R, Davis OSP. Gene-environment interactions and correlations in the development of cognitive abilities and disabilities. In: MacCabe J, O’Daly O, Murray RM, McGuffin P, Wright P, editors. Beyond nature and nurture: genes, environment and their interplay in psychiatry. Andover, UK: Thomson Publishing Services; 2006. pp. 35–45. [Google Scholar]
- Plomin R, DeFries JC, Loehlin JC. Genotype-environment interaction and correlation in the analysis of human behaviour. Psycholog Bull. 1977;85:309–322. doi: 10.1037/0033-2909.84.2.309. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Rowe DC. Environmental and genetic influences on dimensions of perceived parenting: a twin study. Dev Psychol. 1981;17:203–208. doi: 10.1037/0012-1649.17.2.203. [DOI] [Google Scholar]
- Rowe DC. Biometrical genetic models of self-reported delinquent behavior: a twin study. Behav Genet. 1983;13:473–489. doi: 10.1007/BF01065923. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Knight J, Butcher LM, Hansen VK, Meaburn E, Schalkwyk LC, et al. A central resource for accurate allele frequency estimation from pooled DNA genotyped on DNA microarrays. Nucleic Acids Res. 2005;33:e25. doi: 10.1093/nar/gni028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nat Genet. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Haile RW, Duggan D. Recent developments in genomewide association scans: a workshop summary and review. Am J Hum Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- Trouton A, Spinath FM, Plomin R. Twins early development study (TEDS): a multivariate, longitudinal genetic investigation of language, cognition and behaviour problems in childhood. Twin Res. 2002;5:444–448. doi: 10.1375/136905202320906255. [DOI] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]