Abstract
Phenotypic variation of Staphylococcus epidermidis involving the slime related ica operon results in heterogeneity in surface characteristics of individual bacteria in axenic cultures. Five clinical S. epidermidis isolates demonstrated phenotypic variation, i.e. both black and red colonies on Congo Red agar. Black colonies displayed bi-modal electrophoretic mobility distributions at pH 2, but such phenotypic variation was absent in red colonies of the same strain as well as in control strains without phenotypic variation. All red colonies had lost ica and the ability to form biofilms, in contrast to black colonies of the same strain. Real time PCR targeting icaA indicated a reduction in gene copy number within cultures exhibiting phenotypic variation, which correlated with phenotypic variations in biofilm formation and electrophoretic mobility distribution of cells within a culture. Loss of ica was irreversible and independent of the mobile element IS256. Instead, in high frequency switching strains, spontaneous mutations in lexA were found which resulted in deregulation of recA expression, as shown by real time PCR. RecA is involved in genetic deletions and rearrangements and we postulate a model representing a new mechanism of phenotypic variation in clinical isolates of S. epidermidis. This is the first report of S. epidermidis strains irreversibly switching from biofilm-positive to biofilm-negative phenotype by spontaneous deletion of icaADBC.
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-008-9249-8) contains supplementary material, which is available to authorized users.
Keywords: Biofilm, icaADBC, Phenotypic variation, Staphylococcus epidermidis
Introduction
Staphylococcus epidermidis is a normal constituent of the healthy human skin and mucosal microflora. In recent years, however, the bacterium has emerged as a frequent etiologic agent of infection associated with indwelling medical devices (Kozitskaya et al. 2004). A fundamental step in the pathogenesis of S. epidermidis-mediated foreign body infections is the ability of the organism to adhere and form a biofilm on the surface of a biomaterial. Adherence of S. epidermidis to prosthetic devices is thought to occur in two distinct steps: initial attachment to the biomaterial surface, and subsequent accumulation of bacteria through intercellular adhesion (Handke et al. 2004) mediated by polysaccharide intercellular adhesin (PIA). PIA mediates contact between bacterial cells, resulting in the accumulation of a multilayered biofilm and is an important virulence factor of S. epidermidis (Heilmann et al. 1996). The enzymes involved in PIA synthesis are encoded by the ica operon comprising icaA, icaD, icaB, and icaC.
S. epidermidis isolates exhibit phenotypic and genotypic flexibility, assumed to be an evolutionary advantage that helps staphylococci adapt to changing environmental conditions. Phenotypic variants can differ in terms of colony morphology, growth rate, haemolysis, biofilm formation, and antibiotic susceptibility (Christensen et al. 1990). In biofilm-producing S. epidermidis strains, expression of the intercellular adhesion genes (icaADBC) undergoes variation that is illustrated by a change of colony morphology on Congo Red agar (CRA) and altered biofilm formation (Ziebuhr et al. 1997). Currently, several mechanisms have been identified that regulate ica expression, including insertion of the transposon IS256 in the structural operon (Ziebuhr et al. 1999) and genes encoding proteins that regulate ica expression such as rsbU and sarA (Conlon et al. 2004). However, these mechanisms are reversible, meaning that after repeated sub-culturing, the biofilm-positive phenotype can be regained from biofilm-negative inocula. Recently, evidence was reported that in clinical S. epidermidis isolates, loss of ica might not only be due to IS256 insertions, and that irreversible switching occurs. Importantly, Arciola and coworkers (2004, 2005) have shown that a significant proportion of prosthesis derived clinical isolates are ica and IS256 negative.
The ability of S. epidermidis to adhere and form biofilm is associated with various physico-chemical properties of bacterial cell surfaces, such as hydrophobicity and electrophoretic mobility as a measure of the surface charge (Van der Mei et al. 1989). In the determination of a bacterial property, such as cell surface charge or hydrophobicity, pure cultures are generally considered as populations of identical organisms, although it is known that several strains display distinct subpopulations even in pure cultures. Subpopulations within one culture can differ in flagellation (Streger et al. 2002), natural competence (Dubnau 1991), autofluorescence (Kell et al. 1991), or electrophoretic mobility (Cowan et al. 1994; Van Merode et al. 2006b). Importantly, heterogeneity in electrophoretic mobility has been linked to bacterial adhesion to biotic (Cowan et al. 1994) and abiotic surfaces (Van Merode et al. 2006b) as well as to biofilm formation (Van Merode et al. 2006a). However, for none of the above described heterogeneities a mechanism has been forwarded.
The aim of the present study was to determine a relationship between phenotypic variation of clinical isolates of S. epidermidis, biofilm formation and electrophoretic mobility distribution. During the course of the study, genetic changes with respect to the ica operon were observed that were correlated to phenotypic variations in electrophoretic mobility and slime formation, enabling us to propose a new, possibly common, mechanism responsible for the observed phenotypic variation.
Materials and methods
Strains and growth conditions
One hundred and five clinical isolates of S. epidermidis were obtained from blood, cerebrospinal fluid, pus and urine, in the Microbiology Department, Gadjah Mada University, Yogyakarta, Indonesia. None of the patients had a history of previous hospital admission. Identification of the isolates was done by Gram-staining, colony appearance on blood agar, coagulase and DNase testing and susceptibility to novobiocin and polymixin (Bannerman 2003). Clonal relatedness of strains was excluded using Pulsed Field Gel Electrophoresis (PFGE). The isolates were stored at −80°C in glycerol. S. epidermidis ATCC12228 and RP62A (ATCC35984) were used as reference strains where indicated.
Phenotypic characterization
The strains were cultured on CRA plates, prepared by adding 0.8 g of Congo Red (Sigma-Aldrich, Steinheim, Germany), 12 g bacto agar (Becton, Dickinson and Co, Sparks, MD, USA) and 36 g of saccharose (Merck, Darmstadt, Germany) to 1 l of brain heart infusion (Oxoid, Basingstoke, Hampshire, UK). The plates were subsequently incubated for 24 h at 37°C and additionally overnight at room temperature. The CRA plate assay was done in duplicate, and consistent results were obtained. To quantify the number of cells that had switched from black to red during culturing, single black colonies were used to inoculate tryptone soya broth (TSB, Oxoid, Basingstoke, Hampshire, UK). Following 24 h of incubation, the cultures were serially diluted on CRA. For each strain, this experiment was repeated five times with three individual black colonies each time and the results of these 15 individual colonies were averaged. Switching without prior broth culturing was assayed by emulsifying individual black colonies in phosphate-buffered saline (PBS, 10 mM potassium phosphate, 0.15 M NaCl pH 7.0) and serial dilution plating. Each experiment contained three individual black colonies and was repeated twice (i.e. results are averages of six individual colonies on two independent occasions).
Biofilm assay
A quantitative biofilm assay was performed, according to Christensen and co-workers (Christensen et al. 1987). Briefly, 1:100 dilutions of overnight cultures in TSB were used to inoculate wells in a microtiter polystyrene plate (Falcon, Becton Dickinson Labware, NJ, USA). After incubation for 24 h at 37°C, the plates were gently washed two times with phosphate-buffered saline (PBS, 10 mM potassium phosphate, 0.15 M NaCl pH 7.0), and stained with 1% (w/v) crystal violet solution; the excess stain was washed off with demineralised water. Subsequently the adherent cells were resuspended in acid–isopropanol (5% v/v 1 M HCl in isopropanol), and finally, the absorbance (A) was measured at 595 nm in a Fluostart Optima microplate reader (BMG Labtech).
In addition, biofilms were grown as described above in 12-well polystyrene tissue culture plates (Costar, Corning, NY, USA) for 24 h in 2 ml TSB medium and after washing stained with the bacterial Live/Dead stain BacLight (Molecular Probes, Leiden, the Netherlands) and incubated for 30 min in the dark. Confocal microscopic images were collected through the bottom of the plate using a Leica TCS SP2 confocal laser scanning microscope with a 40× objective (Van Merode et al. 2006a).
Electrophoretic mobility measurements
The electrophoretic mobility distribution of the isolates was measured as described previously (Van Merode et al. 2006b) using particulate microelectrophoresis. Briefly, each bacterial strain was resuspended in 30 ml of 10 mM potassium phosphate, set to a pH of 2, 3, 4, 5, 7 or 9 using either HCl or KOH, to a density of approximately 1 × 109 cells ml−1. The electrophoretic mobility at 150 V of the resuspended bacteria was then measured using a Lazer Zee Meter 501 (PenKem) which uses the scattering of incident laser light to detect the bacteria. The instrument was equipped with individual particle tracking software. Strains were designated to have heterogeneous electrophoretic mobility distributions if two or more populations could be distinguished that comprised at least 10% of the total number of cells measured. Isoelectric points (IEP) were estimated by extrapolating the graphs to the pH at which the electrophoretic mobility is 0 (the point where positive and negative charges on the bacterial cell surface counterbalance each other).
DNA isolation
Bacterial cells were pelleted, resuspended in 75 μl of demineralised water and treated with 25 μl of lysostaphin (Sigma-Aldrich, Steinheim, Germany) 1 mg ml−1 for 10 min at 37°C. Ten μl proteinase K (Sigma-Aldrich, Steinheim, Germany) solution of 1 mg ml−1 in demineralised water and 200 μl of 0.1 M Tris–HCl (pH 7.5) was added and incubation continued for 30 min at 37°C. Samples were heated to 100°C for 5 min after which cell debris and unlysed cells were removed by centrifugation (21,000g for 10 min). The DNA was ethanol precipitated, dissolved in 100 μl 10 mM Tris–HCl pH 7.5, 1 mM EDTA (TE) and the concentration was determined using a Nanodrop UV/VIS spectrophotometer (Isogen-Biosolutions Inc., Maarsen, the Netherlands). Samples were stored at −20°C until needed. For DNA colony isolation a procedure described elsewhere was used (Skow et al. 2005).
PCR and real time PCR on genomic DNA
To determine the mechanism governing the appearance of heterogeneous cultures, the strains were screened for presence of the IS256 insertional element, as described previously (Kozitskaya et al. 2004). Standard PCR was performed on an ICycler machine (Biorad, Veenendaal, the Netherlands) using primers for icaA, icaC (Table 1a) and gyrB (Table 1b). Products were analyzed after standard agarose gel electrophoresis. Real time PCR was performed on a MyCycler real time PCR machine (Biorad, Veenendaal, the Netherlands) using SYBR green (Biorad, Veenendaal, the Netherlands) and the primer sets (Isogen-lifescience, Maarsen, the Netherlands) designed using Perlprimer (Marshall 2004) listed in Table 1b. For both types of PCR, total genomic DNA isolations were used as templates at 25 ng per sample. The following program was used for real time PCR: 95°C for 3 min, 40 cycles of 95°C for 15 s, primer specific annealing temperature (Table 1) for 15 s, elongation at 72°C for 15 s. CT values were computer-generated and the gene copy number was determined relative to gyrB gene copy number using the 2−ΔΔC T method (Livak and Schmittgen 2001).
Table 1.
Primer sequences for (a) PCR and (b) real time PCR used in this study
| Primer | Sequence (5′–3′) | Product size (bp) | |
|---|---|---|---|
| (a) PCR | |||
| icaA forwarda | CAGTATAACAACATTCTATTG | 1,425 | |
| icaA reversea | GAGAATTGATAAGAGTTCC | ||
| icaC forwarda | ATAAACTTGAATTAGTGTATT | 1,017 | |
| icaC reversea | CCATAGCTTGAATAAGGG | ||
| IS256 forwardb | TGAAAAGCGAAGAGATTCAAAGC | 1,102 | |
| IS256 reverseb | ATGTAGGTCCATAAGAACGGC | ||
| lexA forward | GAACTCACTAAGCGACAAA | 592 | |
| lexA reverse | ATTACTTTACCTACTACAATGACA | ||
| Primer | Sequence (5′–3′) | Product size (bp) | AT (°C) |
|---|---|---|---|
| (b) Real time PCR | |||
| icaA-1 forward | GGAAGTTCTGATAATACTGCTG | 124 | 56 |
| icaA-1 reverse | GATGCTTGTTTGATTCCCTC | ||
| gyrB-3 forward | GGAGGTAAATTCGGAGGT | 129 | 57.1 |
| gyrB-3 reverse | CTTGATGATAAATCGTGCCA | ||
| recA-1 forward | AAAGTTCAGGTAAGACGACAG | 277 | 54 |
| recA-1 reverse | TCCCATTTCACCTTCAATTTCAG | ||
Cloning and sequencing of lexA
lexA was amplified from genomic DNA isolated as described above using the primers listed in Table 1a. PCR products were cloned into pCR2.1 (TA-cloning kit, Invitrogen, Breda, the Netherlands) according to the manufacturer’s protocol. The presence of inserts was confirmed by restriction analysis using EcoRI. Positive clones were used for transformation of Escherichia coli DH5α and plates containing positive clones were sent for commercial sequencing (Baseclear, Leiden, the Netherlands).
Total RNA isolation and real time PCR analysis of recA expression
Total RNA was isolated from 24 h cultures grown in TSB at 37°C. Cells were harvested by centrifugation, and frozen at −80°C. Samples were thawed slowly on ice and resuspended in 100 μl water after which the bacterial suspension was frozen in liquid nitrogen. Frozen bacteria were ground using a mortar and pestle. Total mRNA was isolated using the Invisorb® Spin Cell RNA Mini Kit according to the manufacturer’s instructions. DNA was removed using the RNeasy mRNA clean-up protocol (Qiagen) and absence of genomic DNA was checked by PCR prior to reverse transcription. For all samples 35 cycles of PCR using the gyrB primer set (Table 1b) did not result in any detectable signal. One μg of total RNA was used for cDNA synthesis (iscript, Biorad) according to the manufacturer’s instructions. Real time PCR was performed as described above. Reactions were prepared using the CAS-1200™ pipetting robot (Corbett Life Science, Sydney, Australia). Expression levels of recA (see primers in Table 1b), were calculated using the 2−ΔΔC T method (Livak and Schmittgen 2001) using gyrB as a reference gene and S. epidermidis RP62A as the control strain.
Statistical analysis
Pearson product moment correlations between the presence of the icaA gene and phenotypic switching determined either by CRA plating or by electrophoretic mobility were calculated using free online software (http://www.wessa.net) (Wessa 2007).
Results
Colony appearance and ability to form biofilms
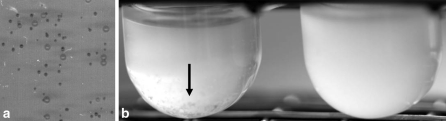
The phenotype of 105 clinical isolates, identified as S. epidermidis, on CRA was determined. Five isolates demonstrated phenotypic variation, indicated by the appearance of mixed black and red colonies on a single plate. To confirm phenotypic variation, a black colony was used to inoculate TSB, and the resulting culture was plated on CRA. A mixture of red and black colonies appeared. The percentage of black cells that had switched to red cells during overnight culturing was determined by serial dilution and colony counting and found to range from 15 to 93% (Table 2). When a red colony was used as an inoculum, only red colonies appeared on CRA plates. Both prolonged incubation (up to 5 days) and repeated sub-culturing for 5 days did not result in any black colonies re-appearing from a red colony inoculum. All strains investigated in the present study showed a clear difference in colony size on CRA plates between black and red colonies (Fig. 1). In addition, when grown in broth cultures inoculated with either red or black variants of the same strain, differences in growth were observed (Fig. 1).
Table 2.
Six of the clinical S. epidermidis isolates with their site of isolation, showing phenotypic variation, i.e. black and red colonies on CRA plates and percentage of switched cells observed on these plates
| Strain | Origin of isolation | Switched cells (%) | |
|---|---|---|---|
| Broth culture | Emulsified colony | ||
| 46 | Cerebrospinal fluid | 0 ± 0 | 0 ± 0 |
| 45 | Pus | 39 ± 5 | 0 ± 0 |
| 196 | Pus | 93 ± 5 | 2 ± 0 |
| 493 | Cerebrospinal fluid | 15 ± 1 | 0 ± 0 |
| 906 | Blood | 79 ± 2 | 12 ± 4 |
| 1098 | Cerebrospinal fluid | 93 ± 11 | 1.5 ± 0 |
Strain 46 is a black, non-switching strain used as the control strain
Fig. 1.
Growth effects of ica presence. Panel a shows images of CRA plate of strain 196 as a representative of all switching strains. Note that black (dark in picture) colonies are consistently smaller than red (light grey in picture) colonies. Panel b shows photographs of overnight broth culture of strain 45 inoculated with either black (left tube) or red (right tube) colonies from the same CRA plate. Note the extensive flocculation (black arrow) in the black colony inoculated cultures and the reduced turbidity
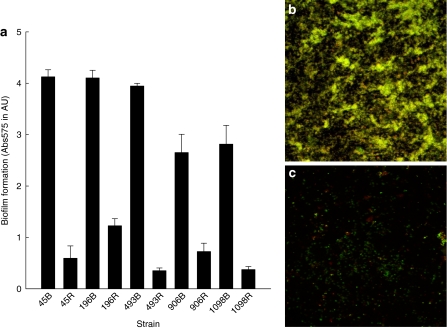
The ability of black and red variants to form biofilm was quantitatively assayed in microtiter plates. Black colonies grew thick biofilms, in contrast to red colonies that were poor biofilm formers (Fig. 2a). To exclude any effects of crystal violet bound by PIA (present in black, absent in red colonies), the biofilms were also studied by confocal laser scanning microscopy (CLSM) following Live/Dead staining (Fig. 2b, c). These results confirmed that the quantitative differences in biofilm formation after crystal violet staining were not associated with the presence or absence of PIA.
Fig. 2.
Panel a: Quantitative biofilm formation of S. epidermidis strains, showing phenotypic variation and grown either from black colonies (indicated with B) or red colonies (indicated with R). Absorbances are means over triplicate wells (±standard deviations). Panels b and c show representative CLSM images of biofilms grown from a black colony (panel b) and a red colony (panel c) of strain 45
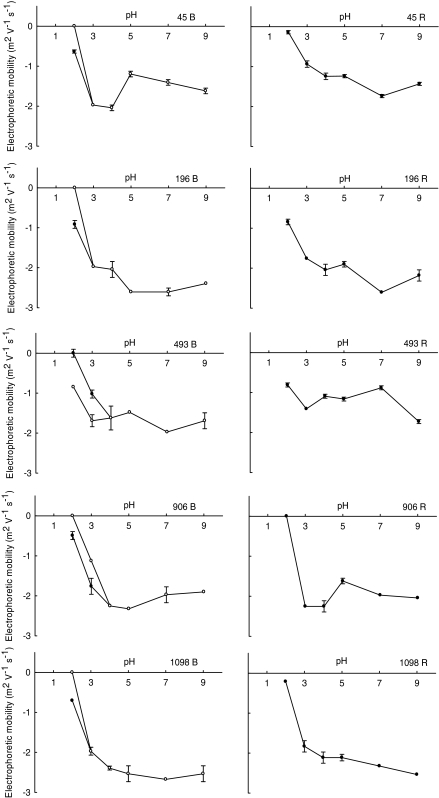
Electrophoretic mobility distributions
For the five strains that showed phenotypic variation, the pH-dependent electrophoretic mobility distributions of black colonies and the corresponding red colonies were determined (Fig. 3). At pH 2 all black inocula yielded cultures with a heterogeneous electrophoretic mobility distribution, which was absent in the red derivatives of the same strain. As controls, several black, non-switching, strains were used as inocula, invariably yielding cultures with homogeneous electrophoretic mobility distributions (data not shown). The absence of heterogeneous electrophoretic mobility distributions in red colonies as well as in the black control strain without phenotypic variation indicates that development of a heterogeneous population after overnight culturing is related to phenotypic variation.
Fig. 3.
The pH dependence of the electrophoretic mobilities of cultures started with black (left panel) and red colonies (right panel) of the same strain in 10 mM potassium phosphate. The error bars denote standard deviations over three experiments with separately cultured bacteria, with each experiment comprising at least 100 bacteria
In the heterogeneous populations, one subpopulation had an estimated isoelectric point (IEP) at pH 2.0, while the second subpopulation showed a slightly lower IEP at pH ≤ 1.5 (Fig. 3, Table 3). By virtue of the use of individual particle tracking software in particulate microelectrophoresis, the percentage distribution of both sub-populations could be measured as well and was found to range between 15 and 85% (see also Table 3).
Table 3.
Percentage distribution of subpopulations of the heterogeneous S. epidermidis strains in electrophoretic mobility at pH 2 and their isoelectric points (IEP)
| S. epidermidis strains | Subpopulation #1 | Subpopulation #2 | ||
|---|---|---|---|---|
| IEP | Size (%) | IEP | Size (%) | |
| 45B | 1.5 | 44 ± 10 | 2 | 56 ± 10 |
| 196B | <1.5 | 78 ± 5 | 2 | 22 ± 5 |
| 493B | <1.5 | 26 ± 4 | 2 | 74 ± 4 |
| 906B | 1.5 | 85 ± 4 | 2 | 15 ± 4 |
| 1098B | 1.5 | 82 ± 4 | 2 | 18 ± 4 |
Subpopulations #1 represents the more negatively charged population, while subpopulation #2 represents the less negatively charged population. The population sizes are mean ± SD over three independent experiments
Presence of icaA and icaC
Phenotypic variation in S. epidermidis can occur by insertion of the mobile element IS256 into icaA and icaC (Ziebuhr et al. 1999) or into rsbU and sarA, encoding proteins involved in regulating expression of the ica operon (Conlon et al. 2004). We therefore assayed the presence of IS256 in these strains as well as the presence of icaA and icaC in black and red colonies of the same strains. IS256 was not found in any of the black or red strains (data not shown). Conventional PCR revealed the presence of icaA and icaC in all cultures started from black colonies, while both genes were absent in all cultures started from red colonies. To verify that this loss of icaA and icaC was the major reason for the phenotypic variation observed in biofilm formation and electrophoretic mobility, DNA was isolated from 10 individual red colonies selected on CRA plates for each switching strain. All of these colonies were negative for icaA and icaC, while all were positive for gyrB. Apparently, in these strains expression of the icaA and icaC genes is not altered through reversible insertion of IS256, but the genes are completely deleted.
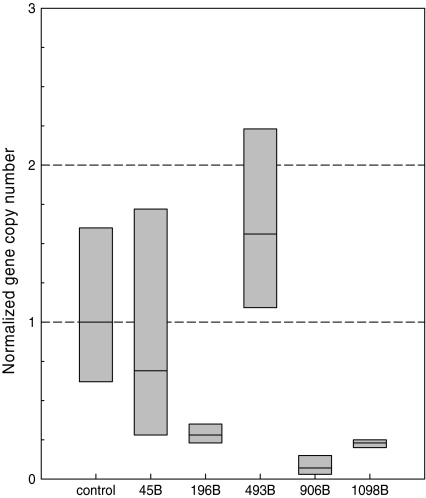
Gene copy number analysis within cultures
Because icaA and icaC were deleted during growth of these strains, it is possible to quantify the occurrence of phenotypic variation by determining the gene copy number of icaA in a culture. To this end, real time PCR was performed on the genomic DNA isolations of cultures inoculated with a black, switching colony. The gene copy number of the target gene (icaA) was quantified relative to the reference gene gyrB, using the DNA of strain 46 (a black non-switching strain, Table 2) as a control. For strains 493 and 45 no significant change in icaA copy number could be observed (normalized gene copy number >0.5 relative to gyrB), but for strains 196, 906 and 1098 a significant decrease (normalized gene copy number <0.5 relative to gyrB) in icaA presence was found (Fig. 4).
Fig. 4.
Determination of the relative gene copy number for icaA within a culture started with a black colony using real time PCR. Amounts were calculated from three replicate wells for gyrB (reference gene) and five for icaA (target gene). As a control, the DNA of a black strain that does not show phenotypic variation was used. The mean and variation (grey bars) are depicted for each strain. The 2−ΔΔC T was calculated from the average CT values and standard deviations
Cloning and sequencing of lexA
In Yersinia pestis high frequency mutations have been observed that also involve black–red switching on CRA plates (Hare and McDonough 1999). One of the proposed mechanisms responsible for these mutations involves recombinase A (RecA) that catalyses recombination between homologous regions of DNA. One component involved in regulation of expression of recA is LexA. In the genomic database of S. epidermidis ATCC12228 (Zhang et al. 2003), lexA appeared to contain a frame shift mutation resulting in a truncated protein. However, upon sequencing of lexA of S. epidermidis ATCC12228 in our laboratory, no frame shift mutation was found. Instead, multiple mutations resulting in two amino acid substitutions were found (Fig. S1), both of which were within the essential N-terminal part of the protein (Knegtel et al. 1995). From the five switching clinical isolates in the present study, one was found to contain amino acid substitutions at different positions within LexA (Fig. S1). The other four strains contained mutations in lexA, but none resulted in apparent amino acid substitutions. Sequencing of the black control strain (46) showed a conserved amino acid change (lysine for arginine) which probably does not affect LexA activity.
Expression analysis of recA
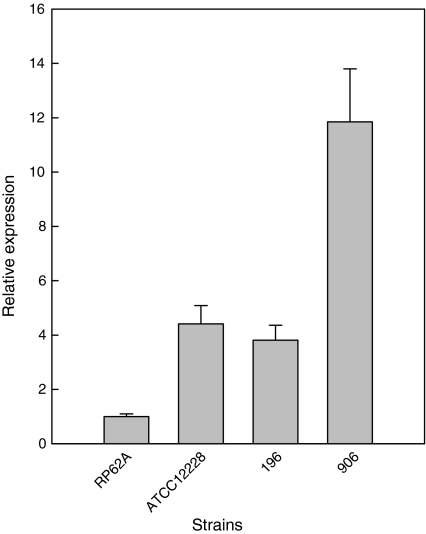
The effect of the mutations in lexA, as observed in S. epidermidis ATCC12228 and two of the clinical isolates, on recA expression was analyzed using real time PCR. Relative to the control strain, S. epidermidis RP62A, recA was over-expressed four times in S. epidermidis ATCC12228 (Fig. 5). Upon comparison of the other strains in this study, 196 and 906 showed significant over-expression of recA (4 and 12 times, respectively) compared to S. epidermidis RP62A.
Fig. 5.
Relative expression of recA. The relative expression of recA was plotted using RP62A as a control strain and gyrB as the reference gene. The 2−ΔΔC T was calculated from the average CT values of two reactions and standard deviations are given
Discussion
Phenotypic variation in biofilm formation is common in clinical isolates of S. epidermidis. In the current study, nearly 5% of all clinical isolates analyzed showed phenotypic variation in biofilm forming ability and electrophoretic mobility. None of the isolates analyzed originated from hospital related infection, but all were derived from patients that came into the clinic with infections thus limiting the chances of these isolates to be clonally related. This was confirmed by PFGE (data not shown). In line with previous publications, black colony variants on CRA were good biofilm formers, in contrast to their red derivatives (Ziebuhr et al. 1999; Conlon et al. 2004).
Previous work with Enterococcus faecalis has shown that the electrophoretic mobility distribution of a culture is an important determinant for biofilm formation (Van Merode et al. 2006a). Axenic heterogeneous cultures were shown to be excellent biofilm formers, while cultures with homogeneous electrophoretic mobility distributions formed poor biofilms. In line with this, the electrophoretic mobility distributions of black, switching strains were found to be heterogeneous, while black non-switching strains were homogeneous. Isoelectric points of ica-positive and ica-negative strains have been determined previously (Ruzicka et al. 2007), but this study did not include any strains with phenotypic variation. The IEP for ica-negative strains was slightly lower at pH 2.3 than the IEP of ica-positive strains (IEP at pH 2.6). Based on this small difference in IEP, we can associate subpopulation #2 of our switching S. epidermidis strains, possessing the highest IEP (see Table 3), with the biofilm-positive (ica-positive) phenotype, although our IEP values are somewhat lower than those reported by Ruzicka et al. (2007). This is probably due to the different techniques used, capillary isoelectric focusing (Ruzicka et al. 2007) versus microelectrophoresis (this study).
Previous reports have shown that phenotypic variation in biofilm formation is governed by the on/off switching of expression of the ica operon by at least two mechanisms; directly, through insertion of the mobile element IS256 into one or more structural genes (Ziebuhr et al. 1999), or indirectly, through insertion of IS256 into the regulatory genes rsbU or sarA that govern ica expression (Conlon et al. 2004). Here however, we rule out insertional inactivation of the icaADBC genes by IS256 for several reasons: (i) screening for the sequence of IS256 in the DNA of the strains used in this study by PCR (Kozitskaya et al. 2004) demonstrated that all strains were IS256 negative. (ii) Screening using PCR and real time PCR for the presence of the icaA and icaC genes and gene fragments showed that these were absent in the red, but present in the black cultures. (iii) The inability of red colonies to form black colonies again, even after prolonged incubation for up to 5 days or subsequent culturing, showed that the switch from the biofilm-positive to the biofilm-negative phenotype is irreversible, in contrast to insertional inactivation by IS256 which results in reversible switching (Ziebuhr et al. 1999). Therefore it is concluded that the red variants from the five strains studied had permanently lost icaA and icaC.
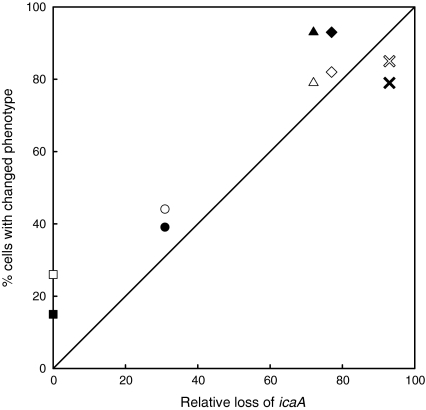
The percentage of bacteria in a culture with the lowest IEP (Table 3), associated with the biofilm-negative phenotype (Ruzicka et al. 2007), corresponds quantitatively with the percentage of bacteria that had switched from black to red colonies as determined on CRA (compare Table 2), i.e. the percentage of icaA-negative organisms. Moreover, the development of the icaA-negative genotype as quantitatively determined with real time PCR correlated with the phenotypic switching observed on CRA (Pearson r = 0.93, two-side P = 0.02) as well as with the variation in electrophoretic mobility (Pearson r = 0.99, two-sided P = 0.001) (Fig. 6). Previous studies have shown that absence of ica is common among clinical S. epidermidis isolates (Arciola et al. 2005) and that in most of these ica-negative strains, IS256 is not present (Arciola et al. 2004), as also found here. Possibly our observation that clinical isolates rapidly lose ica might explain that strains isolated from biofilms are ica-negative once in the laboratory environment.
Fig. 6.
Relation between genotypic and phenotypic analysis. Relative loss of icaA was plotted against relative phenotypic changes (percentage of red colonies) determined on CRA (closed symbols) and the percentage of bacteria with the lowest IEP (open symbols). A 1:1 correlation is indicated by the line of identity drawn in the graph (● strain 45, ▲ strain196, ■ strain 493, × strain 906 and ♦ strain 1098)
In Y. pestis, RecA dependent high frequency mutations have been observed that involve black–red switching on CRA plates (Hare and McDonough 1999). RecA catalyses recombination between homologous regions of DNA. Its activity is tightly regulated since inappropriate recombination can have serious consequences, such as deletion of genomic regions between repeats (reviewed in Cox 2007). In line with observations in Y. pestis, a high frequency of mutations is observed in several clinical S. epidermidis isolates. S. epidermidis ATCC12228, commonly used as an ica-negative reference strain, contains many inversions and rearrangements in its genome (Zhang et al. 2003). Although the frame shift mutations in lexA, as present in the published genome sequence, could not be confirmed for the S. epidermidis ATCC12228 strain in our laboratory, two other mutations were found, both resulting in an amino acid change. In addition, two of the clinical isolates also contained mutations resulting in amino acid substitutions (Fig. S1). In addition, all other sequenced lexA clones contained mutations, that although not resulting in any amino acid changes, could influence expression of lexA. We hypothesized that the high frequency of deletion events is a consequence of mutations within lexA resulting in deregulation of recA expression and therewith a high frequency of recombination, insertion and genetic rearrangement events, causing a deletion of the icaADBC operon. Therefore recA expression was analyzed in strains that had lost the icaADBC operon and appeared indeed upregulated in the strains ATCC12228, 196 and 906, as compared to RP62A (Fig. 5) whereas the other three strains had no detectable expression of recA (not shown). Remarkably, strains 196 and 906 are the clinical isolates with the highest % of switched colonies on CRA plates (compare Table 2 and Fig. 5).
For Y. pestis it was proposed that the high frequency of mutations could occur because of the lack of selection for Congo Red binding (Hare and McDonough 1999). Because of the observed growth advantages of ica-negative S. epidermidis strains, it is possible that there is in fact a positive selection for such deletion strains to develop due to increased fitness. Therefore we propose the following model for the observed switching (Fig. 7). Due to growth limitation of strains showing high expression of the ica operon, selective pressure exists for mutants that lack this operon. These mutants develop due to deregulation of recA as a consequence of (probably spontaneous) mutations in lexA. As a result of this mechanism, high frequency deletion of the ica operon occurs. In the current study only 5% of the strains tested showed this mechanism of switching, but this likely constitutes a clear underestimate. Arciola and coworkers surveyed the presence of ica in 400 clinical S. epidermidis isolates from prosthesis-associated infections using a PCR method and found that 42% of all isolates were ica-negative (Arciola et al. 2005). The new switching mechanism described in the current study could explain the high occurrence of ica negative clinical isolates as observed previously.
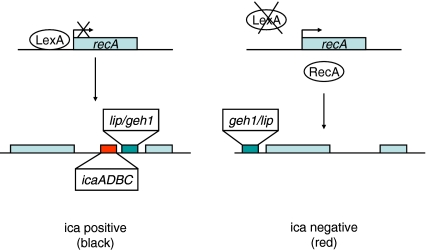
Fig. 7.
Schematic overview of the proposed series of events, leading to the deletion of the ica operon. In the presence of LexA, transcription of recA is prevented, there are no genomic rearrangements and the icaADBC region is present thus resulting in black colonies on CRA plates. Due to mutations, LexA can no longer bind to the promoter region of recA resulting in RecA transcription and activity and the occurrence of genetic rearrangements and deletions. As a result the geh1/lip genes are inverted and the icaADBC region disappears thus resulting in red colonies on CRA plates
In conclusion, while investigating the role of culture heterogeneity towards IEP in biofilm formation of clinical isolates of S. epidermidis, we observed a new mechanism of phenotypic variation. This phenotypic variation, as observed using CRA plates, correlates with a genotype characterized by deletion of the ica operon. The percentage of genotypic variation is highly correlated with IEP. Deletion of ica appears to be caused by spontaneous mutations in lexA, which results in deregulation of recA expression leading to genetic rearrangements and deletions.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Willy de Haan for her help with the real time PCR and Prof. Dr Ir Willem Norde for helpful discussion. We also thank Dr G. A. Kampinga and co-workers from the Medical Microbiology department at the UMCG for PFGE analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Arciola CR, Campoccia D, Gamberini S, Rizzi S, Donati ME, Baldassarri L, Montanaro L. Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials. 2004;25:4117–4125. doi: 10.1016/j.biomaterials.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Gamberini S, Campoccia D, Visai L, Speziale P, Baldassarri L, Montanaro L. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J Biomed Mater Res A. 2005;75:408–413. doi: 10.1002/jbm.a.30445. [DOI] [PubMed] [Google Scholar]
- Bannerman TL. Gram-Positive Cocci. In: Murray PR, editor. Manual of clinical microbiology. Washington, DC: ASM Press; 2003. pp. 394–404. [Google Scholar]
- Christensen GD, Baddour LM, Simpson WA. Phenotypic variation of Staphylococcus epidermidis slime production in vitro and in vivo. Infect Immun. 1987;55:2870–2877. doi: 10.1128/iai.55.12.2870-2877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GD, Baddour LM, Madison BM, Parisi JT, Abraham SN, Hasty DL, Lowrance JH, Josephs JA, Simpson WA. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1169. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- Conlon KM, Humphreys H, O’Gara JP. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol. 2004;186:6208–6219. doi: 10.1128/JB.186.18.6208-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan MM, Mikx FHM, Busscher HJ. Electrophoretic mobility and hemagglutination of Treponema denticola ATCC 33520. Colloids Surf B Biointerfaces. 1994;2:407–410. doi: 10.1016/0927-7765(94)80004-9. [DOI] [Google Scholar]
- Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke LD, Conlon KM, Slater SR, Elbaruni S, Fitzpatrick F, Humphreys H, Giles WP, Rupp ME, Fey PD, O’Gara JP. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J Med Microbiol. 2004;53:367–374. doi: 10.1099/jmm.0.05372-0. [DOI] [PubMed] [Google Scholar]
- Hare JM, McDonough KA. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J Bacteriol. 1999;181:4896–4904. doi: 10.1128/jb.181.16.4896-4904.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- Kell DB, Ryder HM, Kaprelyants AS, Westerhoff HV. Quantifying heterogeneity: flow cytometry of bacterial cultures. Antonie Van Leeuwenhoek. 1991;60:145–158. doi: 10.1007/BF00430362. [DOI] [PubMed] [Google Scholar]
- Knegtel RM, Fogh RH, Ottleben G, Rüterjans H, Dumoulin P, Schnarr M, Boelens R, Kaptein R. A model for the LexA repressor DNA complex. Proteins. 1995;21:226–236. doi: 10.1002/prot.340210305. [DOI] [PubMed] [Google Scholar]
- Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72:1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- Ruzicka F, Horka M, Hola V, Votava M. Capillary isoelectric focusing—useful tool for detection of the biofilm formation in Staphylococcus epidermidis. J Microbiol Methods. 2007;68:530–535. doi: 10.1016/j.mimet.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Skow A, Mangold KA, Tajuddin M, Huntington A, Fritz B, Thomson RB, Jr, Kaul KL. Species-level identification of staphylococcal isolates by real-time PCR and melt curve analysis. J Clin Microbiol. 2005;43:2876–2880. doi: 10.1128/JCM.43.6.2876-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streger SH, Vainberg S, Dong H, Hatzinger PB. Enhancing transport of Hydrogenophaga flava ENV735 for bioaugmentation of aquifers contaminated with methyl tert-butyl ether. Appl Environ Microbiol. 2002;68:5571–5579. doi: 10.1128/AEM.68.11.5571-5579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Mei HC, Brokke P, Dankert J, Feijen J, Rouxhet PG, Busscher HJ. Physicochemical surface properties of nonencapsulated and encapsulated coagulase-negative staphylococci. Appl Environ Microbiol. 1989;55:2806–2814. doi: 10.1128/aem.55.11.2806-2814.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Merode AE, Van der Mei HC, Busscher HJ, Krom BP. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. J Bacteriol. 2006;188:2421–2426. doi: 10.1128/JB.188.7.2421-2426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Merode AE, Van der Mei HC, Busscher HJ, Waar K, Krom BP. Enterococcus faecalis strains show culture heterogeneity in cell surface charge. Microbiology. 2006;152:807–814. doi: 10.1099/mic.0.28460-0. [DOI] [PubMed] [Google Scholar]
- Wessa P (2007) Free statistics software. Office for research development and education, version 1.1.21-r4, URL http://www.wessa.net/
- Zhang YQ, Ren SX, Li HL, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Ziebuhr W, Heilmann C, Gotz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.