Abstract
This study sought to determine whether the presence of hypermethylated genes in the surgical margins can predict local recurrences in head and neck squamous cell carcinomas (HNSCCs). We prospectively collected tumour and surgical margin specimens from patients with HNSCCs who had undergone surgical resections. Quantitative methylation-specific PCR (QMSP) of CDKN2A, CCNA1 and DCC were performed in these specimens and correlated with clinical data. Of the 42 patients eligible for the study, 27 were hypermethylation informative for the above three genes. This latter group was associated with longer disease-free survivals (P=0.007) and longer time to disease-specific deaths (P=0.004). Multivariate analyses confirmed hypermethylation non-informative tumours as an independent prognosticating factor for disease-specific deaths (risk ratio 3.8, P=0.026). Quantitative MSP of the margins of 24 hypermethylation informative tumours revealed that 11 patients had molecularly positive margins, of which, five developed disease-specific events (DSEs, three local recurrences and two metastases), compared to none in patients with molecularly negative margins, after a median follow-up of 48 months. Log-rank analyses showed that molecularly positive margins were associated with shorter time to local recurrences and disease-specific deaths (P=0.03 and 0.01, respectively). This study demonstrated that QMSP of hypermethylated promoters in surgical margins predicted all the local recurrences in our series of HNSCC patients. We have also identified hypermethylation non-informative tumours as an independent predictor for the development of DSEs.
Keywords: hypermethylation, margins, HNSCC, recurrence, QMSP, prognostic
Head and neck squamous cell carcinoma (HNSCC) afflicts more than 500 000 patients per year (Parkin et al, 2005). Despite recent advances in therapeutic options, the mortality remains about 50% in 5 years. This is in part due to the high incidence of local recurrences (up to 50%) after surgery despite achieving histopathologically clear resection margins (Jesse and Sugarbaker, 1976; Davidson et al, 1981; Kowalski et al, 1993). Two possible mechanisms could account for these observations (Hockel and Dornhofer, 2005). First, minimal residual cancer cells might remain undetectable by standard histological margin assessment (Brennan et al, 1995; van Houten et al, 2004) and second, local recurrences may have evolved from a field of genetically altered mucosal cells (Slaughter et al, 1953; Califano et al, 1996; van Houten et al, 2004; Braakhuis et al, 2005; Martone et al, 2007). The detection of these ‘high risk’ lesions will allow clinicians to tailor adjuvant therapeutic options that can diminish the incidence of recurrences.
The improved understanding of tumour biology has uncovered many novel tumour markers that can distinguish between malignant and non-malignant cells (Sidransky, 2002). Brennan et al, (1995) first reported the use of TP53 mutation for the detection of the residual tumour cells in surgical resection margins to predict locoregional recurrences. Various groups have also investigated surgical margins with other molecular markers such as eIF4E, PSA, tyrosinase and microsatellite instability to predict locoregional recurrences (Proebstle et al, 1996; Theodorescu et al, 1999; Sardi et al, 2000; Nathan et al, 2002; Temam et al, 2004).
More recently, silencing of tumour suppressor genes by hypermethylation of selected promoter regions has been identified as an important mechanism of carcinogenesis and implicated in a variety of solid and haematological malignancies (Laird and Jaenisch, 1994; Esteller et al, 2002; Momparler, 2003). As hypermethylated promoters often occur from 100- to 1000-folds more frequently in tumour cells compared to normal cells, they are therefore good candidate molecular markers for detection of low numbers of tumour cells in the milieu of normal cells (Laird, 2003). Methylation-specific PCR (MSP) and quantitative MSP (QMSP) have been used to detect methylated DNA in the body fluids of patients with cancers of the breast (Silva et al, 1999, 2002), lung (Esteller et al, 1999), prostate (Goessl et al, 2000), bladder (Dominguez et al, 2002), gastrointestinal tract (Kawakami et al, 2000; Eads et al, 2001; Zou et al, 2002) and head and neck region (Sanchez-Cespedes et al, 2000; Righini et al, 2007). There are however few reports of methylation analyses of resection margins. Goldenberg et al, (2004) demonstrated in a pilot study with six patients that intra-operative assessments of surgical margins methylation status were feasible using QMSP. No recurrence was observed in any of the three patients with positive molecular resection margin but the follow-up period was short. Shaw et al, (2007b) performed quantitative methylation analyses of resection margins in 20 patients using pyrosequencing methylation analyses (PMA) . However, 6 out of 13 positive margins subsequently developed locoregional recurrences, there were however two recurrences in seven negative margins. The authors suggested that the small sample size and the use of fixed tissue DNAs may have contributed to the inconclusive findings. Furthermore, the use of pyrosequencing with a sensitivity of 1 out of 20 to 1 out of 50 may not be appropriate for detection of small numbers of tumour cells in surgical margins. Most recently, Martone et al, (2007) identified similar methylation profiles between tumours and margins in 9 out of 11 patients with HNSCC-specific deaths. As this study consisted only of retrospectively selected patients who had relapses and HNSCC-specific deaths but no disease-free controls, it did not identify significant association between methylation statuses in surgical margins with disease-free survivals.
The objective of our study was to measure the amount of hypermethylated promoters of selected genes, in a series of prospectively collected surgical margins of HNSCC patients, to predict local recurrences and cancer-specific survivals. We utilised QMSP to interrogate the methylation profiles of CDKN2A, CCNA1 and DCC in the resection margins in a cohort of patients who underwent curative resections of HNSCCs. We selected these genes on the basis of reports that CDKN2A (Shaw et al, 2006), CCNA1 (Shaw et al, 2006) and DCC (Carvalho et al, 2006) are hypermethylated in 28, 53 and 75% of HNSCCs respectively. Significantly, all three genes have very low level of methylation in normal mucosa and are thus good candidate genes for the purpose of our study. Functionally, CDKN2A (Kim and Sharpless, 2006) and DCC (Mehlen and Fearon, 2004; Carvalho et al, 2006) are known tumour suppressor genes, whereas CCNA1 (Ji et al, 2005) has been shown to play a role in cell cycle regulation. The study was conducted in two phases: (1) to determine the hypermethylation informative statuses of the primary tumours and (2) to detect the presence of promoter hypermethylation in the surgical margins of those tumours that were hypermethylation informative; all findings were then correlated with clinical data.
Materials and methods
Sample collection and histopathological examination
We prospectively collected fresh frozen tumour biopsies and surgical margin specimens from patients treated for HNSCCs at Institut Gustave-Roussy (Villejuif, France) between January and December 2000. The Institutional Review Board approved the study, and informed consent was obtained from all patients. Tumour staging (TNM and pTNM) was performed according to the Union International Contre Cancer 1997 criteria. The study design, samples collection and histopathological examinations of surgical specimens were performed as previously described (Temam et al, 2004).
Patients with close resection margins (⩽5 mm) on surgical specimens or moderate-to-severe dysplasia on surgical margins were excluded. To ensure adequate representation of the margins, patients with less than three margins available were excluded from the second phase of the study.
During the study period, 42 patients met the inclusion criteria and were recruited for the study. All patients underwent as first treatment surgical resection of the primary tumours and unilateral or bilateral neck dissections depending on tumour sites and nodal status. The indication for postoperative radiotherapy depended on tumour stage, tumour site and nodal status.
DNA extraction and bisulphite treatment
All tumour biopsy specimens included in the analysis were diagnosed as invasive HNSCCs. Serial 40-μm thick frozen sections of each tumour or surgical margin specimen were performed with H&E staining for histopathologic control of the first and the last 5-μm thick section. A pathologist (PF), blinded to the clinical data, reviewed all the slides to confirm (1) the percentage of tumour cells present in the tumour biopsies or (2) the absence of carcinomas or moderate-to-severe dysplasias in each surgical margin.
DNA from tumour and surgical margin specimens was extracted using the QIAamp Tissue kit (Qiagen, Courtaboeuf, France). DNA quality was verified using GeneQuant II (Amersham Pharmacia Biotech, Cambridge, UK).
The extracted DNA was modified by sodium bisulphite in accordance to the manufacturer's protocol (Chemicon no. 7280) and resuspended in 100 μl of TE buffer (EDTA 2.5 mmol l−1 and Tris-HCl 10 mmol l−1) and stored at −20°C.
Methylation analysis
The bisulphite-modified DNA was used as a template for QMSP, as previously described (Harden et al, 2003) with some modifications. In brief, the primers and probes were designed to specifically amplify the bisulphite-converted promoter of the gene of interest. The primers and probe sequences for CDKN2A and β-actin have been previously described (Lo et al, 1999; Harden et al, 2003). The sequences of CCNA1 and DCC are listed below in the order of forward primer, reverse primer and probe: CCNA1; 5′-GCGGTTTCGGAGAGCGTAC-3′, 5′-GACGCCCCCGAACCTAAC-3′, 6FAM5′-TTTGTCGCGGTCGGTATGGAAACG-3′TAMRA, DCC; 5′-TGTTCGCGATTTTTGGTTTC-3′, 5′-ACCGATTACTTAAAAATACGCG-3′, 6FAM5′-TTTTCGGAGTTTTTTTGTTTAGCGC-3′TAMRA.
Fluorogenic PCRs were carried out in a reaction volume of 50 μl consisting of 300 nM of each primer; 150 nM of probe. 10 μl of treated DNA solution were used in each real-time MSP reaction. Amplifications were carried out in 96-well plates in a 7900 Sequence Detector System (Perkin-Elmer Applied Biosystems, Norwalk, CT, USA). Commercially available methylated DNA (CpGenome Universal Methylated DNA AbCys S.A. no. S7821) were used as positive controls and serial dilutions of this DNA were used for constructing the calibration curves to detect between five genomic equivalents and 15 000 genomic equivalents of DNA on each plate. Identical laboratory procedures and intermixing were performed in the same laboratory for each batch tested. All tumours and margins DNA for each test batch were bisulphite modified at the same time with their positive and negative controls to minimise variations in experimental conditions. All plates contained multiple water blanks and bisulphite modified Human Genomic DNA (CpGenome Universal Unmethylated DNA no. S7822) as negative controls. Every QMSP was performed in duplicates or triplicates.
Calculation of methylation index and weighted tumour methylation index
Methylation index (MI) was computed as the ratios between the methylation values of the gene of interest and the internal reference gene, β-actin, which were obtained by Taqman analysis (Harden et al, 2003). These ratios were used as measures to represent the relative levels of methylation in any given sample:
MI=gene of interest methylation value/β-actin methylation value × 100%.
Given that tumour specimens comprised between 50 and 100% of malignant cells, the MI obtained from tumour specimens therefore represented the average methylation levels of a mixture of the normal and malignant cells. As methylation of CDKN2A, CCNA1 and DCC exist only in very low levels in normal cells (Carvalho et al, 2006; Shaw et al, 2006), we calculated a separate index, which we termed as the weighted tumour methylation index (WTMI) to better reflect methylation levels in the tumour cells:
Weighted tumour methylation index (WTMI)=MI of the tumour specimens divided by the percentage of malignant cells present in the tumour specimens (MI/percentage of tumour × 100%).
Stratification of tumours and margins
In contrast to the semiquantitative nature of MSP, QMSP generates precise MI in continuous variables and thus necessitates that a threshold be set to dichotomise the data. We employed two thresholds to stratify (a) tumours into hypermethylation informative or non-informative using WTMI of the tumour specimens and (b) margins into molecularly positive or negative using MI of the margin specimens:
Stratification of tumours. Tumours were assigned to be hypermethylation informative only if WTMI are ⩾5%. The cutoff at 5% was chosen to select only tumours that have very high levels of hypermethylation to allow discriminative detection of very small numbers of informative tumour cells mixed in a large numbers of normal cells in the surgical margins.
Stratification of margins. Quantitative MSP for the margins were performed only on the tumours that were hypermethylation informative and only on the genes that the tumours were informative for. We then ranked the margins in ascending order according to the MI obtained for each of the gene tested. The margins with MI above the 90th percentile of all the margins tested for that gene were deemed molecularly positive for that corresponding gene. This threshold was set to allow exclusions of margins with low levels of MI of unknown significance.
Statistical analysis
This prospective study was powered based on a previous study conducted by one of our co-authors (Temam et al, 2004), which demonstrated an disease-free survival (DFS) at 30 months of 85% among patients with negative margins and 30% among patients with positive margins (using microsatellite instability as markers). To achieve a study with 80% power, we calculated prospectively that 22 informative patients were required to demonstrate a similar difference in DFS at 30 months with an α-risk of 5%, working on the basis that half of the informative patients have positive margins for hypermethylation in our study. Univariate analyses using Fisher's exact test and Student's t-test were performed for categorical and continuous data, respectively. Clinical and biological characteristics were analysed for their association with time to local recurrence using Cox proportional hazards models. Estimates of survival curves were calculated according to the Kaplan–Meier product-limit method and were calculated from the time of surgery to the time of death or the last follow-up visit. Times to local recurrence for various prognostic groups were compared using the log-rank test. The Cox proportional hazards regression model was used to assess the prognostic effect of patient characteristics and molecular markers to estimate DFS and time to disease-specific death. Predictive variables with P-values of ⩽0.10 for the univariate Cox proportional hazards model were included in a multicovariate model. All of the computations carried out were performed using NCSS and SPSS software.
Result
Methylation profiles of tumours
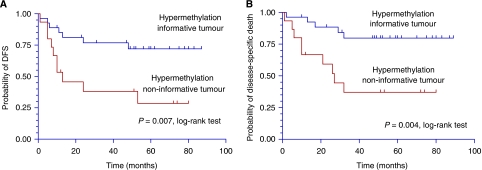
Quantitative MSP of the three selected genes CDKN2A, CCNA1 and DCC was performed on 42 tumours. High levels of hypermethylation (WTMI ⩾5%, range 5.1–95%) were detected in one or more of the three genes in 64.3% (n=27/42) of tumours. These were designated as hypermethylation informative as described (Materials and Methods). Baseline clinical characteristics such as age, sex, site, stage, histopathological gravity signs and adjuvant therapy did not differ significantly according to the tumour methylation profiles (Table 1). The median clinical follow-up period was 48 months (range, 1–89). Only three patients were lost to follow-up within 24 months of diagnosis. Mean follow-up period for patients who remained events free were 70 months. Sixteen patients developed one or more disease-specific events (DSEs, defined as local and/or regional recurrences and/or distant metastasis) with median DFS of 12.25 months (range, 1–53). The hypermethylation informative group fared better, compared to the non-informative group; with longer DFSs and longer time to disease-specific deaths (P=0.007 and 0.004, respectively, Kaplan–Meier analysis with log-rank test) as shown in Figure 1. Multi-covariate analyses with possible confounding factors such as age, sex, site, stage, histopathological gravity signs and adjuvant therapy identified hypermethylation non-informative tumours as an independent prognosticating factor for DFS with a risk ratio of 3.82 (Table 2).
Table 1. Patient characteristics according to tumour methylation status.
|
Tumour hypermethylation
|
|||
|---|---|---|---|
| Informative (n=27)a | Non-informative (n=15) | P-valueb | |
| Age, years | |||
| Mean±s.d. | 59.67±9.7 | 57.07±10.46 | |
| Median (maximum, minimum) | 61 (32, 77) | 61 (39, 73) | 0.72 |
| Sex, n (%) | |||
| Male | 22 (81.5) | 12 (80) | |
| Female | 5 (18.5) | 3 (20) | 1 |
| Site, n (%) | |||
| Oral cavity | 10 (37.0) | 6 (40.0) | |
| Oropharynx | 10 (37.0) | 2 (13.3) | |
| Hypopharynx | 5 (18.5) | 3 (20.0) | |
| Larynx | 2 (7.4) | 4 (26.7) | 0.218 |
| Tumour stage, n (%) | |||
| I–II | 8 (29.6) | 2 (13.3) | |
| III–IV | 19 (70.4) | 13 (86.7) | 0.286 |
| Histopathological gravity signsc, n (%) | |||
| No | 24 (88.9) | 10 (66.7) | |
| Yes | 3 (11.1) | 5 (33.3) | 0.11 |
| Adjuvant therapyd, n (%) | |||
| No | 14 (51.9) | 12 (80.0) | |
| Yes | 13 (48.1) | 3 (20.0) | 0.102 |
| Disease-specific events,n (%) | |||
| No | 20 (74.1) | 6 (40.0) | |
| Yes | 7 (25.9) | 9 (60.0) | 0.047 |
| Disease-specific death,n (%) | |||
| No | 22 (81.5) | 6 (40.0) | |
| Yes | 5 (18.5) | 9 (60.0) | 0.015 |
Informative: high level of gene promotor hypermethylation (WTMI ⩾ 5%).
Fisher's exact test.
Histopathological gravity signs: perineural invasion, angiolymphatic invasion or both.
Concurrent chemoradiotherapy or radiotherapy alone.
Figure 1.
Kaplan–Meier curve of hypermethylation informative tumour vs hypermethylation non-informative tumour in (A) disease-free survival and (B) time to disease-specific death.
Table 2. Univariate and multivariate Cox proportional hazards models in estimating time to disease-specific death.
| Covariates | Regression coefficient | s.e. | Risk ratio | Probability |
|---|---|---|---|---|
| Age, years | −0.014 | 0.025 | 0.986 | 0.567 |
| Sex (male vs female) | 0.813 | 0.758 | 2.254 | 0.284 |
| Stage (I–II vs III–IV) | 0.012 | 0.667 | 1.012 | 0.985 |
| Adjuvant therapya (yes vs no) | 0.306 | 0.548 | 1.358 | 0.577 |
| Histopathological gravity sign (yes vs no) | 0.377 | 0.602 | 1.458 | 0.531 |
| Hypermethylation informative (− vs +) | 1.341 | 0.602 | 3.823 | 0.026b |
Concurrent chemoradiotherapy or radiotherapy alone.
No covariate other than tumour methylation status was entered in the multivariate analyses, because P>0.10 for all other covariates.
Molecular analyses of surgical margins
In the second phase of this study, we performed QMSP on the resection margins of the hypermethylation informative tumours. Three patients with informative tumours were excluded from this phase because there were less than three margins available for analysis (Materials and Methods). One hundred-thirteen margins from the remaining 24 patients (3–7 margins per patient) underwent QMSP analyses for the genes that were informative in the primary tumours. The majority of the margins had very low methylation levels for CDKN2A, CCNA1 and DCC, with median MI at 0.08% (range 0–0.39%), 0.06% (range 0–31.9%) and 0.08% (range 0–2.28%), respectively. The threshold MI levels for defining margins molecular positivity, set at 90th percentile for each of the gene tested, were 0.36% (CDKN2A), 0.87% (CCNA1) and 0.91% (DCC). Twelve margins from 11 patients were deemed molecularly positive; 10 patients had one margin that were positive for one or more gene tested, whereas one other patient had two margins that were positive for the same gene.
Baseline clinical characteristics did not differ according to margins methylation status (Table 3). Five of the 11 patients with molecularly positive surgical margins developed DSE (three local recurrence, two distant metastases) compared to none in the 13 patients with molecularly negative surgical margins (P=0.011, Fisher's exact test).
Table 3. Patient characteristics according to margin methylation status.
|
Margin hypermethylation
|
|||
|---|---|---|---|
| + | − | P-valuea | |
| Age, years | |||
| Mean±s.d. | 61.6±11.1 | 58.3±9.7 | |
| Median (maximum, minimum) | 64 (40, 77) | 61 (32, 68) | 0.72 |
| Sex | |||
| Male | 8 (72.7) | 12 (92.3) | |
| Female | 3 (27.3) | 1 (7.7) | 0.3 |
| Site, n (%) | |||
| Oral cavity | 6 (54.5) | 4 (30.8) | |
| Oropharynx | 5 (45.5) | 4 (30.8) | |
| Hypopharynx | 0 (0) | 4 (30.8) | |
| Larynx | 0 (0) | 1 (7.7) | 0.15 |
| Tumour stage, n (%) | |||
| I–II | 4 (36.4) | 3 (23.1) | |
| III–IV | 7 (63.4) | 10 (76.9) | 0.67 |
| Histopathological gravity signsb, n (%) | |||
| No | 9 (81.8) | 12 (92.3) | |
| Yes | 2 (18.2) | 1 (7.7) | 0.58 |
| Adjuvant therapyc, n (%) | |||
| No | 4 (36.4) | 7 (53.8) | |
| Yes | 7 (63.4) | 6 (46.2) | 0.17 |
| Disease-specific event, n (%) | |||
| No | 5 (45.5) | 13 (100) | |
| Yes | 6 (54.5) | 0 (0) | 0.01 |
| Locoregional recurrence, n (%) | |||
| No | 8 (72.7) | 13 (100) | |
| Yes | 3 (27.3) | 0 (0) | 0.03d |
| Disease-specific death, n (%) | |||
| No | 7 (63.4) | 13 (100) | |
| Yes | 4 (36.4) | 0 (0) | 0.01d |
Fisher's exact test.
Histopathological gravity signs: perineural invasion, angiolymphatic invasion or both.
Concurrent chemoradiotherapy or radiotherapy alone.
Log-rank test.
The local recurrences occurred in two oropharyngeal cancers (stage IV) and one oral cavity cancer (stage II). The distant metastases were both lung metastases and had occurred in one oral cavity cancer (stage II) and one oropharyngeal cancer (stage IV).
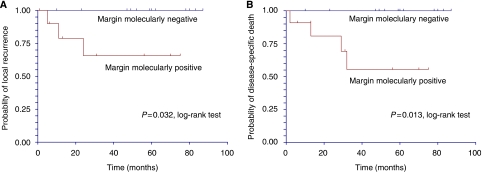
Molecular positivity in surgical margins were significantly associated with decreased time to local recurrences (P=0.03, Kaplan–Meier analysis with log-rank test, Figure 2A). The latter was not significantly associated with other covariates (age, sex, site, stage, histopathological gravity signs and adjuvant therapy) by Cox regression analysis (not shown). Furthermore, molecularly positive surgical margins were associated with decreased time to disease-specific deaths (P=0.013, Kaplan–Meier analysis with log-rank test, Figure 2B).
Figure 2.
Kaplan–Meier curves of margin molecularly positive vs margin molecularly negative in (A) time to local recurrence and (B) time to disease-specific death.
Discussion
The objective of our study was to measure the relative amount of hypermethylated promoters of selected genes in the surgical margins of HNSCC patients and to determine whether these could predict local recurrences and cancer-specific survivals. The most important finding in our study is that the molecularly positive margins identified by QMSP correctly predicted all five DSE (three local recurrences and two distant metastases), whereas none of the patients with molecularly negative margins developed any DSE at a median follow-up period of 48 months.
WTMI and prognosis
Sixty-four per cent of the HNSCCs in our cohort of patients were informative for hypermethylation. This compares favourably with the ratio of informative tumour using other molecular markers such as TP53 mutation (50% informative) (Boyle et al, 1993; Brennan et al, 1995) and MSI (48% informative) (Temam et al, 2004). We noted that tumour stage was not associated with recurrence in this series, probably because we selected patients after exhaustive pathological assessments or because the sample size was small for subset analyses.
Our findings that patients with hypermethylated tumours have better prognosis corroborated with recent reports of others. A recent study reported promoter methylation profiles of hMLH1, MGMT and CDKN2A in 51 cases of HNSCCs using MSP and demonstrated that tumours with two or more methylated genes had improved DFS at 2 years (Puri et al, 2005). Koscielny et al, (2007) reported in his series of 67 patients that hypermethylation and LOH were the two main mechanisms responsible for inactivating CDKN2A but only the hypermethylated group were associated with a lower incidence of locoregional recurrence. Shaw et al, (2007a) observed in a series of 76 patients that tumours with CpG island methylation phenotype (CIMP) were associated with marked inflammatory response and less aggressive tumour biology. These reports and our findings in this study are emerging evidence that hypermethylation of certain genes in HNSCCs may be associated with longer survival. Unfortunately, available literature does not provide clear insight to the mechanism behind this counter-intuitive observation that hypermethylation of tumour suppressor genes in HNSCCs can lead to better prognosis. Nevertheless, a possible parallel mechanism can be drawn from studies in colorectal cancers. There is convincing evidence that CIMP and chromosomal instability represent two independent and inversely related mechanisms of genetic and epigenetic instability in sporadic colorectal carcinomas (Goel et al, 2007). Furthermore, CIMP underlies a subtype of colorectal cancer with microsatellite instability with distinctive clinical features (Ribic et al, 2003; Weisenberger et al, 2006). We postulate that a similar mechanism may be at work in HNSCCs, where hypermethylated tumours constitute a subgroup of HNSCCs with a distinctive clinical profile.
The possible presence of LOH in CDKN2A (Koscielny et al, 2007) and DCC (Papadimitrakopoulou et al, 1998) may increase the likelihood of tumours being non-informative for these two markers. However, this is unlikely to confound our findings in the margin analyses of informative tumours. Furthermore, the absence of data that clearly outline the interaction between LOH and hypermethylation has made it difficult to adjust our scoring system to cater for the presence of LOH.
MI of surgical margins and recurrence
All three local recurrences and two distant metastases occurred in the 11 (out of 24) patients with molecularly positive margins. We noted that 7 out of the 11 patients with molecularly positive margins underwent adjuvant radiotherapy and, of these, only one patient developed local recurrence. In contrast, of the remaining four patients who did not have adjuvant radiotherapy therapy, two had developed local recurrences. It is likely that adjuvant treatments have prevented local recurrences that might have otherwise occurred in some of these patients with molecularly positive margins and thus may partly account for the ‘false-positive’ patients.
In contrast, all 13 patients with molecularly negative margins turned out to be true negative. We noted that four of these were hypopharyngeal tumours (often associated with the poorest prognosis), while there was none in the group with positive margins. Although this disparity in tumour sites did not reach statistical significance (P=0.067, Fisher's exact test), probably due to the small sample size, we were nonetheless reassured that the specificity of the molecularly negative margins could not be attributed to the selection of tumour sites with favourable prognosis.
Implication
Our findings have both clinical and scientific implications to HNSCCs. Clinically, the poorer prognosis of patients with non-informative tumours or molecularly positive surgical margins may warrant either closer follow-up or recommendation of adjuvant therapy, even in the absence of other risk factors such as advanced stage, perineural invasion and perivascular invasion. Conversely, in cases where the benefit of adjuvant therapy is equivocal, methylation profiles of the tumours or the margins can serve as additional indicators to tailor sensible treatment strategies so as to minimise unnecessary treatment morbidities. However, these results must be verified in an independent prospective study before it can be used to formulate clinical guidelines. We are in the process of incorporating such a validation study into an ongoing international, multicentric, prospective controlled trial led by Institute Gustave-Roussy.
For any tumour molecular marker to be clinically relevant, it should ideally be informative in the majority of that specific tumour. In previous reports of similarly designed studies, molecular markers such as TP53 and MSI were both informative in about 50% of HNSCCs. In comparison, hypermethylation was informative in almost two-thirds of the HNSCCs using our panel of three genes, even though we only designated tumours with very high level of hypermethylation as informative. The percentage of informative tumours can conceivably be further increased by screening other suitable candidate genes. Hypermethylated genes are therefore good molecular markers for HNSCCs in the clinical settings.
Scientifically, our findings together with other reports would suggest that hypermethylation of certain gene loci may confer a tumour phenotype with distinctive clinical characteristics. Further research is warranted to delineate the exact roles of hypermethylation and other alternative mechanisms such as genetic deletion and point mutation in HNSCC carcinogenesis.
This study also illustrates that QMSP is ideally suited for detection of hypermethylated markers in surgical margins. It detects promoter hypermethylation with great sensitivity and specificity. In addition, it is technically robust and easily reproducible. Most importantly, it offers significant advantage to conventional MSP because of its ability to generate accurate quantitative data as a continuous variable. The latter allows the dichotomy of data at a threshold to facilitate stratification, a feature that we leverage on to determine the tumour methylation profile (hypermethylation informative vs non-informative) and the margin methylation status (molecularly positive vs negative).
Summary
This is to our knowledge the first study to report that presence of hypermethylated promoters in surgical margins of HNSCCs can predict local recurrences and disease-specific deaths. Performing QMSP with a panel of three genes (CDKN2A, CCNA1 and DCC), we found 64% of HNSCC tumours were hypermethylation informative. Methylation analyses of their resection margins correctly predicted all the recurrences in this cohort of patients. Furthermore, we have showed that HNSCCs with hypermethylation of these three gene promoters have a better prognosis.
Acknowledgments
We are greatly indebted to the head and neck surgeons who helped to collect the samples (Drs Patrick Marandas, Anne-Marie Leridant, Morbize Julieron and Gerard Mamelle). We thank the Health Manpower Development Programme (Singhealth, Singapore) who provided funding support for Dr HK Tan. We also thank the Diplôme Universitaire d'Etudes et de Recherche en Cancérologie Clinique for its continuous support of this research initiative.
References
- Boyle JO, Hakim J, Koch W, van der Riet P, Hruban RH, Roa RA, Correo R, Eby YJ, Ruppert JM, Sidransky D (1993) The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res 53: 4477–4480 [PubMed] [Google Scholar]
- Braakhuis BJ, Leemans CR, Brakenhoff RH (2005) Expanding fields of genetically altered cells in head and neck squamous carcinogenesis. Semin Cancer Biol 15: 113–120 [DOI] [PubMed] [Google Scholar]
- Brennan JA, Mao L, Hruban RH, Boyle JO, Eby YJ, Koch WM, Goodman SN, Sidransky D (1995) Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med 332: 429–435 [DOI] [PubMed] [Google Scholar]
- Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D (1996) Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 56: 2488–2492 [PubMed] [Google Scholar]
- Carvalho AL, Chuang A, Jiang WW, Lee J, Begum S, Poeta L, Zhao M, Jeronimo C, Henrique R, Nayak CS, Park HL, Brait MR, Liu C, Zhou S, Koch W, Fazio VM, Ratovitski E, Trink B, Westra W, Sidransky D, Moon CS, Califano JA (2006) Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res 66: 9401–9407 [DOI] [PubMed] [Google Scholar]
- Davidson TM, Nahum AM, Astarita RW (1981) Microscopic controlled excisions for epidermoid carcinoma of the head and neck. Otolaryngol Head Neck Surg 89: 244–251 [DOI] [PubMed] [Google Scholar]
- Dominguez G, Carballido J, Silva J, Silva JM, Garcia JM, Menendez J, Provencio M, Espana P, Bonilla F (2002) p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res 8: 980–985 [PubMed] [Google Scholar]
- Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, Laird PW (2001) Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 61: 3410–3418 [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Paz MF, Campo E, Colomer D, Novo FJ, Calasanz MJ, Galm O, Guo M, Benitez J, Herman JG (2002) Cancer epigenetics and methylation. Science 297: 1807–1808; discussion 1807–1808 [DOI] [PubMed] [Google Scholar]
- Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG (1999) Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 59: 67–70 [PubMed] [Google Scholar]
- Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR (2007) The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology 132: 127–138 [DOI] [PubMed] [Google Scholar]
- Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, Miller K (2000) Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res 60: 5941–5945 [PubMed] [Google Scholar]
- Goldenberg D, Harden S, Masayesva BG, Ha P, Benoit N, Westra WH, Koch WM, Sidransky D, Califano JA (2004) Intraoperative molecular margin analysis in head and neck cancer. Arch Otolaryngol Head Neck Surg 130: 39–44 [DOI] [PubMed] [Google Scholar]
- Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D (2003) Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res 9: 1370–1375 [PubMed] [Google Scholar]
- Hockel M, Dornhofer N (2005) The hydra phenomenon of cancer: why tumors recur locally after microscopically complete resection. Cancer Res 65: 2997–3002 [DOI] [PubMed] [Google Scholar]
- Jesse RH, Sugarbaker EV (1976) Squamous cell carcinoma of the oropharynx: why we fail. Am J Surg 132: 435–438 [DOI] [PubMed] [Google Scholar]
- Ji P, Agrawal S, Diederichs S, Baumer N, Becker A, Cauvet T, Kowski S, Beger C, Welte K, Berdel WE, Serve H, Muller-Tidow C (2005) Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell cycle progression in somatic cells. Oncogene 24: 2739–2744 [DOI] [PubMed] [Google Scholar]
- Kawakami K, Brabender J, Lord RV, Groshen S, Greenwald BD, Krasna MJ, Yin J, Fleisher AS, Abraham JM, Beer DG, Sidransky D, Huss HT, Demeester TR, Eads C, Laird PW, Ilson DH, Kelsen DP, Harpole D, Moore MB, Danenberg KD, Danenberg PV, Meltzer SJ (2000) Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst 92: 1805–1811 [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE (2006) The regulation of INK4/ARF in cancer and aging. Cell 127: 265–275 [DOI] [PubMed] [Google Scholar]
- Koscielny S, Dahse R, Ernst G, von Eggeling F (2007) The prognostic relevance of p16 inactivation in head and neck cancer. ORL J Otorhinolaryngol Relat Spec 69: 30–36 [DOI] [PubMed] [Google Scholar]
- Kowalski LP, Magrin J, Waksman G, Santo GF, Lopes ME, de Paula RP, Pereira RN, Torloni H (1993) Supraomohyoid neck dissection in the treatment of head and neck tumors. Survival results in 212 cases. Arch Otolaryngol Head Neck Surg 119: 958–963 [DOI] [PubMed] [Google Scholar]
- Laird PW (2003) The power and the promise of DNA methylation markers. Nat Rev Cancer 3: 253–266 [DOI] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R (1994) DNA methylation and cancer. Hum Mol Genet 3Spec No: 1487–1495 [DOI] [PubMed] [Google Scholar]
- Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM (1999) Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res 59: 3899–3903 [PubMed] [Google Scholar]
- Martone T, Gillio-Tos A, De Marco L, Fiano V, Maule M, Cavalot A, Garzaro M, Merletti F, Cortesina G (2007) Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res 13: 5089–5094 [DOI] [PubMed] [Google Scholar]
- Mehlen P, Fearon ER (2004) Role of the dependence receptor DCC in colorectal cancer pathogenesis. J Clin Oncol 22: 3420–3428 [DOI] [PubMed] [Google Scholar]
- Momparler RL (2003) Cancer epigenetics. Oncogene 22: 6479–6483 [DOI] [PubMed] [Google Scholar]
- Nathan CO, Amirghahri N, Rice C, Abreo FW, Shi R, Stucker FJ (2002) Molecular analysis of surgical margins in head and neck squamous cell carcinoma patients. Laryngoscope 112: 2129–2140 [DOI] [PubMed] [Google Scholar]
- Papadimitrakopoulou VA, Oh Y, El-Naggar A, Izzo J, Clayman G, Mao L (1998) Presence of multiple incontiguous deleted regions at the long arm of chromosome 18 in head and neck cancer. Clin Cancer Res 4: 539–544 [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108 [DOI] [PubMed] [Google Scholar]
- Proebstle TM, Huber R, Sterry W (1996) Detection of early micrometastases in subcutaneous fat of primary malignant melanoma patients by identification of tyrosinase-mRNA. Eur J Cancer 32A: 1664–1667 [DOI] [PubMed] [Google Scholar]
- Puri SK, Si L, Fan CY, Hanna E (2005) Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinoma. Am J Otolaryngol 26: 12–17 [DOI] [PubMed] [Google Scholar]
- Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S (2003) Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, Favrot MC (2007) Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res 13: 1179–1185 [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D (2000) Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res 60: 892–895 [PubMed] [Google Scholar]
- Sardi I, Franchi A, Ferriero G, Frittelli A, Bruschini L, Montali E, Gallo O (2000) Prediction of recurrence by microsatellite analysis in head and neck cancer. Genes Chromosomes Cancer 29: 201–206 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Lowe D, Bowers NL, Liloglou T, Field JK, Woolgar JA, Risk JM (2007a) CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol 43: 878–886 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Woolgar JA, Lowe D, Rogers SN, Field JK, Liloglou T, Risk JM (2007b) Quantitative methylation analysis of resection margins and lymph nodes in oral squamous cell carcinoma. Br J Oral Maxillofac Surg 45: 617–622 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM (2006) Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer 94: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D (2002) Emerging molecular markers of cancer. Nat Rev Cancer 2: 210–219 [DOI] [PubMed] [Google Scholar]
- Silva JM, Dominguez G, Garcia JM, Gonzalez R, Villanueva MJ, Navarro F, Provencio M, San Martin S, Espana P, Bonilla F (1999) Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res 59: 3251–3256 [PubMed] [Google Scholar]
- Silva JM, Garcia JM, Dominguez G, Silva J, Miralles C, Cantos B, Coca S, Provencio M, Espana P, Bonilla F (2002) Persistence of tumor DNA in plasma of breast cancer patients after mastectomy. Ann Surg Oncol 9: 71–76 [DOI] [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, Smejkal W (1953) Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6: 963–968 [DOI] [PubMed] [Google Scholar]
- Temam S, Casiraghi O, Lahaye JB, Bosq J, Zhou X, Julieron M, Mamelle G, Lee JJ, Mao L, Luboinski B, Benard J, Janot F (2004) Tetranucleotide microsatellite instability in surgical margins for prediction of local recurrence of head and neck squamous cell carcinoma. Clin Cancer Res 10: 4022–4028 [DOI] [PubMed] [Google Scholar]
- Theodorescu D, Frierson Jr HF, Sikes RA (1999) Molecular determination of surgical margins using fossa biopsies at radical prostatectomy. J Urol 161: 1442–1448 [PubMed] [Google Scholar]
- van Houten VM, Leemans CR, Kummer JA, Dijkstra J, Kuik DJ, van den Brekel MW, Snow GB, Brakenhoff RH (2004) Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: a prospective study. Clin Cancer Res 10: 3614–3620 [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793 [DOI] [PubMed] [Google Scholar]
- Zou HZ, Yu BM, Wang ZW, Sun JY, Cang H, Gao F, Li DH, Zhao R, Feng GG, Yi J (2002) Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin Cancer Res 8: 188–191 [PubMed] [Google Scholar]