Abstract
Fish serum contains several specific binding proteins for insulin-like growth factors (IGFBPs). The structure and physiological function of these fish IGFBPs are unknown. Here we report the complete primary sequence of a zebrafish IGFBP deduced from cDNA clones isolated by library screening and rapid amplification of cDNA ends. The full-length 1,757-bp cDNA encodes a protein of 276 aa, which contains a putative 22-residue signal peptide and a 254-residue mature protein. The mature zebrafish IGFBP has a predicted molecular size of 28,440 Da and shows high sequence identity with human IGFBP-2 (52%). The sequence identities with other human IGFBPs are <37%. Chinese hamster ovary cells stably transfected with the zebrafish IGFBP-2 cDNA secreted a 31-kDa protein, which bound to IGF-I and IGF-II with high affinity, but did not bind to Des(1–3)IGF-I or insulin. Northern blot analyses revealed that the zebrafish IGFBP-2 transcript is a 1.8-kb band expressed in many embryonic and adult tissues. In adult zebrafish, IGFBP-2 mRNA levels were greatly reduced by growth hormone treatment but increased by prolonged fasting. When overexpressed or added to cultured zebrafish and mammalian cells, the zebrafish IGFBP-2 significantly inhibited IGF-I-stimulated cell proliferation and DNA synthesis. These results indicate that zebrafish IGFBP-2 is a negative growth regulator acting downstream in the growth hormone-IGF-I axis.
Keywords: growth regulation, evolution, fish, gene expression
Vertebrate animal growth, determined by the rate and duration of the growth process, is largely genetically controlled, but also strongly influenced by environmental factors (1). This is especially true for ectotherms like teleost fish, which rely on changes in water temperature, photoperiod, and food availability to trigger developmental processes such as hatching, metamorphosis (flatfishes, eels) or smoltification (salmonids), sexual maturation and spawning. Information from both external stimuli and the internal state is processed, integrated, and responded to by the brain for appropriate modification of growth through hormonally mediated pathways. A central step in this regulatory network is the growth hormone (GH)–insulin-like growth factor (IGF)-I axis. GH, synthesized in the pituitary gland and secreted into the bloodstream, is of critical importance for postnatal growth. The growth-promoting action of GH is primarily mediated by IGF-I, a 70-aa polypeptide structurally related to IGF-II and proinsulin. IGF-I is expressed in a wide variety of fish tissues with the highest level found in the liver (2–5). In teleosts, the hepatic IGF-I expression is under the regulation of GH and nutritional states. This dual regulation of IGF-I, which is an interface between nutrients and hormones acting in concert to control animal growth, is well conserved throughout vertebrates (5).
The IGF-I in fish serum and extracellular fluids is bound to specific binding proteins (IGFBPs) (2–4). IGFBPs are a family of proteins that specifically bind IGF-I and IGF-II with affinities that are equal to or greater than those of the IGF receptors. Six distinct IGFBPs, designated as IGFBP-1 to IGFBP-6, have been isolated and characterized in mammalian species to date (6, 7). These proteins act as carrier proteins in the bloodstream and control the efflux of IGFs from the vascular space. Locally expressed IGFBPs can alter IGF biological activity by modulating their interaction with the IGF receptors. In addition to the six high-affinity IGFBPs, a number of proteins, termed IGFBP-related proteins, have been reported to bind IGFs with low affinity (8). Using radioisotope-labeled human IGFs as ligands, several groups have demonstrated the presence of specific binding proteins for IGFs in fish sera (9–14). Kelley et al. (9) have characterized three distinct IGFBPs with the molecular sizes of 29 kDa, 31 kDa, and 45 kDa in four teleost fish species. The 45-kDa form is the most predominant and has been shown to be increased by GH treatment (9–13). Because its molecular size is similar to that of mammalian IGFBP-3 and because it is regulated by GH, this protein is likely to be a fish version of IGFBP-3. The 31-kDa and 29-kDa IGFBPs are similar to mammalian IGFBP-2 and IGFBP-1 in size, but their identities are not known. When fasted, the levels of the 29-kDa IGFBP increase significantly (14). Therefore, several IGFBPs are present in teleost fish and their levels are regulated by GH and nutritional state. The structures of these fish IGFBPs have not yet been determined and their functions are not clear.
In this study, we have determined the complete primary sequence of a zebrafish IGFBP. Sequence comparison revealed that this protein is the fish homologue of mammalian IGFBP-2. Zebrafish IGFBP-2 mRNA was expressed in many embryonic and adult tissues. Its tissue levels were decreased by GH treatment and increased by prolonged fasting. The zebrafish IGFBP-2 protein produced by Chinese hamster ovary (CHO) cells bound to IGFs with high specificity and affinity. When added exogenously or overexpressed in zebrafish and mammalian cells, the zebrafish IGFBP-2 inhibited IGF-I-stimulated cell growth and DNA synthesis. These results indicate that IGFBP-2 is a growth inhibitory protein acting downstream in the GH–IGF-I axis in zebrafish.
Materials and Methods
Materials.
All chemicals and reagents were purchased from Sigma unless noted otherwise. The rainbow molecular weight marker, [32P]dCTP, and [3H]thymidine were obtained from NEN. Human IGF-I, IGF-II, and Des(1–3)IGF-I were purchased from GroPep, Adelaide, Australia. FBS, DMEM, Ham's F12 medium, penicillin-streptomycin, and trypsin were purchased from GIBCO/BRL.
Experimental Animal and Cell Culture.
Wild-type zebrafish (Danio rerio) were maintained at 28°C in well water. The fish were fed twice daily unless indicated otherwise. The embryos were obtained by natural crosses. Zebrafish embryonic cells (ZF-4) and CHO-K1 cells were obtained from the American Type Culture Collection. The cells were grown in a 1:1 mixture of Ham's F12 and DMEM with penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% FBS.
Cloning and DNA Sequencing.
A 870-bp bovine IGFBP-2 cDNA, which was labeled with [32P]dCTP by a random priming kit (Boehringer Mannheim), was used to screen a 15- to 19-hr zebrafish embryonic cDNA library (a gift from Bruce Appel, Vanderbilt University, Nashville). Hybridization was carried out at 42°C. The blots were washed and exposed to x-ray film for autoradiography. The nucleotide sequence of resulting cDNA clones was determined by automated sequencing with the ABI Prism 377 by using a Big Dye Therminator cycle sequencing kit (Perkin–Elmer). A combination of primer walking and sequencing overlapping exonuclease III-digested clones, generated according to standard procedures, was performed on both strands. 5′ Rapid amplification of cDNA ends was performed by using a commercial kit (CLONTECH) following the manufacturer's instruction.
Plasmid Construction and Transfection.
A 1.75-kb zebrafish IGFBP-2 cDNA, constructed by restriction enzyme digestion and ligation, was subcloned into the pRc/cytomegalovirus (CMV) expression vector (Invitrogen) by using the HindIII and NotI sites. The plasmid was transfected into CHO-K1 cells by using Lipofecamine (GIBCO/BRL) as described (15). The pRc/CMV vector alone was used as control. The transfected cells were plated sparsely into 10-cm tissue culture dishes and cultured in selection medium (1.2 mg/ml G418). Selection medium was changed every 3–4 days. The discrete colonies were picked up after 2 weeks of selection and grown individually.
Western Ligand Blot, Affinity Crosslinking, and Immunodepletion.
Conditioned media (CM) were collected from confluent clones after 48 hr of incubation in serum-free medium, centrifuged, and stored at −80° C until use. Ligand blot analysis was performed as described (15). For affinity crosslinking, 100 μl of CM was incubated with 1 × 105 cpm [125I]human IGF-I or IGF-II with or without competitors in 1× PBS buffer containing 0.5% BSA and 10 mM PMSF overnight at 4°C. The proteins were crosslinked for 30 min by adding dissuccinimidyl suberate. After the reaction was stopped by adding Tris⋅HCl buffer, 25 μl of the samples was separated by SDS/PAGE. The gel was fixed, dried, and exposed to x-ray film for autoradiography. To deplete IGFBP-4 from CMs prepared from CHO cells, CMs were incubated with an anti-human IGFBP-4 antibody (a gift from David R. Clemmons, University of North Carolina, Chapel Hill) overnight at 4°C (1:500 dilution). Fifty microliters of protein A-Sepharose then was added and rocked for another 4 hr at 4°C. The samples were centrifuged, and the supernatants were collected and stored at −80.
DNA Synthesis and Growth Assays.
The effects of IGF-I and IGFBP-2 on cell growth and DNA synthesis were examined by direct cell counting and [3H]thymidine incorporation assay. Cell number was determined by using a hematocytometer. Growth curves were constructed by plotting cell number against time. The procedure for [3H]thymidine incorporation assay has been reported (16).
RNA Isolation and Northern Blot Analysis.
Total RNA was isolated from zebrafish embryos and adult tissues by using TriReagent (Molecular Research Center, Cincinnati) following the manufacturer's instruction. RNA samples were size-fractionated on a 1.2% agarose gel, blotted, and fixed onto a Hybond-N membrane (Amersham Pharmacia). The membrane was hybridized with 32P-dCTP-labeled zebrafish IGFBP-2 cDNA. A 300-bp salmon actin cDNA probe was used to determine levels of actin mRNA. Densitometry was performed by scanning the autoradiographs and then analyzing with scion image software.
Statistical Analysis.
Values are means ± SE. Differences among groups were analyzed by one-way ANOVA followed by Fisher's protected least significance difference test using statview (Abacus Concept, Berkeley, CA).
Results
Isolation and Sequence Analysis of cDNA Clones Encoding Zebrafish IGFBP-2.
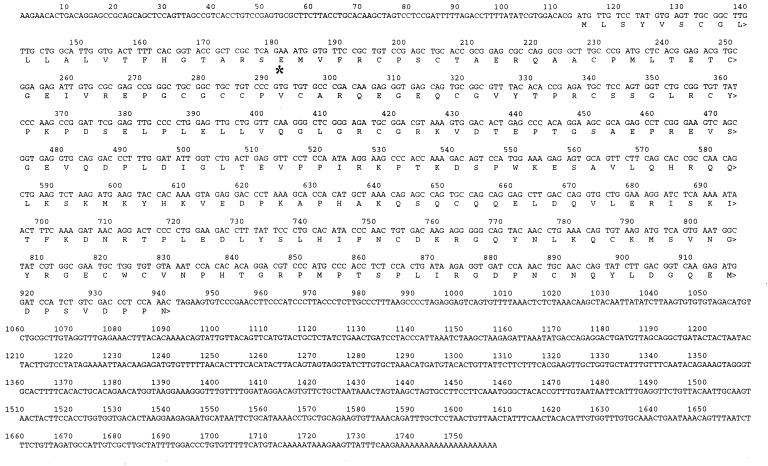
Screening of 50,000 recombinant clones from a zebrafish embryo cDNA library yielded a clone (≈1.6 kb) that exhibited high nucleotide sequence identity to human IGFBP-2 cDNA. This cDNA clone contained the 3′ untranslated region (UTR) but lacked the 5′ UTR region. Therefore, 5′ rapid amplification of cDNA ends was performed with poly(A)+ RNA obtained from zebrafish embryos. This process resulted in a unique cDNA, which represented the 5′ end of the zebrafish IGFBP-2 cDNA. This sequence has an initiation codon ATG that is flanked by sequences resembling the Kozak consensus sequence (17). The complete ORF of 828 bp encodes a protein of 276 aa (Fig. 1). The overall sequence identity of the entire protein with six known human IGFBPs is 32% (IGFBP-1), 52% (IGFBP-2), 32% (IGFBP-3), 37% (IGFBP-4), 34% (IGFBP-5), and 32% (IGFBP-6). The sequence identities between zebrafish IGFBP-2 and the IGFBP-related proteins are <17%. The zebrafish IGFBP-2 protein contains a 22-aa putative hydrophobic signal peptide (18). The mature 254-aa protein has a predicted molecular weight of 28,440 Da with no N-glycosylation site. The zebrafish IGFBP-2 cDNA contains a 3′ UTR of approximately 700 bp. A polyadenylation signal AATAAA is present at positions 1717–1722, and a poly(A) tail is at the 3′ end (Fig. 1).
Figure 1.
Nucleotide and deduced amino acid sequence of the zebrafish IGFBP-2. The first amino acid residue of the predicted mature protein is indicated by *. The GenBank database accession no. is AF198033.
The alignment of the zebrafish IGFBP-2 amino acid sequence with that of mammalian and chicken IGFBP-2 is shown in Fig. 2. The fish IGFBP-2 exhibits an overall sequence identity of 50–52% to chicken, rat, mouse, ovine, bovine, and human IGFBP-2. As shown in Fig. 2, zebrafish IGFBP-2 contains 18 cysteine residues, and their alignment is identical to those of mammalian and chicken IGFBP-2s. There are two highly conserved regions: the cysteine-rich N-terminal domain (residues 1–82) and C-terminal domain (residues 156–254). The central L domain (residues 83–155) shares little sequence identity (23%). The sequence identities in the N domain between the zebrafish IGFBP-2 and those of chicken, rat, and mouse are 65–69%. The identity with human IGFBP-2 in this domain is lower (57%) because of the presence of an extra 14 aa in the human molecule at positions 24–38. The identity in the entire C domain is approximately 53%. Because the last 17 residues of the C domain are highly variable, the sequence identity of the C domain without this region is considerably higher (62%). The Arg-Gly-Asp (RGD) sequence, present in the C-terminal domain of all mammalian and chicken IGFBP-2, is also present in the zebrafish at positions 232–234. The putative heparin binding motif (PKKXRP) located in the central domain in all of the known mammalian IGFBP-2 is absent in the zebrafish.
Figure 2.
Comparison of the zebrafish IGFBP-2 primary sequence with that of human (27), bovine (37), ovine (38), mouse (39), rat (29), and chicken (40). Sequence alignment was obtained with macvector by the Clustal method. Gaps were introduced to maximize sequence homologies. The first amino acid residue of the predicted mature protein is indicated by *. Residues that are identical or conserved are shaded. The consensus sequence is shown on the bottom line.
Expression and Functional Characterization of the Zebrafish IGFBP-2 Protein.
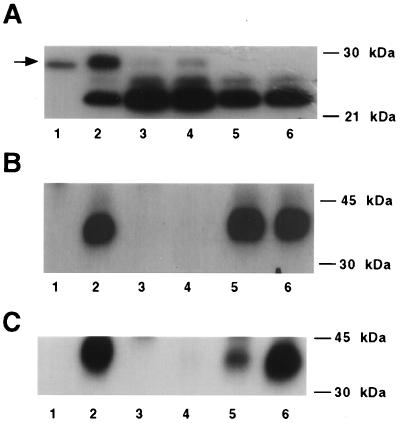
To determine that this zebrafish cDNA encodes a functional IGFBP, we constructed a zebrafish IGFBP-2 expression plasmid and introduced it into CHO cells by stable transfection. Of the numerous clones obtained, three IGFBP-2/pRc/MCV-transfected clones and two empty vector-transfected (mock) control clones were randomly selected for further analysis. Wild-type CHO cells secreted a 24-kDa IGFBP. This IGFBP has been identified as IGFBP-4 by immunoblot. The three IGFBP-2/pRc/CMV-transfected clones, but not the mock-transfected or wild-type cells, secreted variable levels of a 31-kDa IGFBP in addition to the endogenous IGFBP-4 (Fig. 3A). This IGFBP comigrated with pure bovine IGFBP-2. To determine the relative IGF binding specificity of this fish IGFBP, crosslinking experiments were performed by using [125I]IGF-I or [125I]IGF-II. When [125I]IGF-I was used as the ligand, a 38-kDa band was detected (Fig. 3B, lane 2). [125I]IGF-I was competed effectively by unlabeled IGF-I and IGF-II but not by insulin. Des(1–3)IGF-I, which has significantly reduced affinity for mammalian IGFBPs (19), was unable to compete at a very high concentration. Similar results were obtained when [125I]IGF-II was used as radioligand (Fig. 3C). The 38-kDa IGF-II/IGFBP-2 band was competed most effectively by IGF-II, to a less extent by IGF-I, but not Des(1–3)IGF-I or insulin. Therefore, the cDNA cloned from zebrafish encodes a functional IGFBP that binds to IGF-I and IGF-II with high affinity and specificity.
Figure 3.
The zebrafish IGFBP-2 cDNA encodes a protein that binds to IGFs but not to insulin and Des(1–3)IGF-I. (A) Ligand blot analysis of CM prepared from the pRc/CMV-IGFBP-2-transfected clones 2, 5, and 6 (lanes 2–4), the pRC/CMV vector-transfected clones 1 and 2 (lanes 5 and 6), and pure bovine IGFBP-2 (lane 1). (B) Affinity crosslinking of [125I] IGF-I to zebrafish IGFBP-2 produced by CHO cells (IGFBP-2-transfected clone 2). Binding was competed by the addition of no competitor (lane 2), 500 ng of IGF-I (lane 3), IGF-II (lane 4), Des(1–3)IGF-I (lane 5), and insulin (lane 6). Lane 1 is the negative control that did not have any zebrafish IGFBP-2. The competition of [125I] IGF-I was similar in all other IGFBP-2-transfected clones examined (clones 5 and 6). (C) Affinity crosslinking of [125I] IGF-II to zebrafish IGFBP-2 produced by CHO cells (IGFBP-2-transfected clone 2). Binding was competed by the addition of no competitor (lane 2), 500 ng of IGF-I (lane 3), IGF-II (lane 4), Des(1–3)IGF-I (lane 5), and insulin (lane 6). Lane 1 is the negative control that did not have any zebrafish IGFBP-2. The competition of [125I] human IGF-II was similar in other IGFBP-2-transfected clones examined (clones 5 and 6).
Zebrafish IGFBP-2 Is Expressed in Multiple Tissues Under the Regulation of GH and Nutritional State.
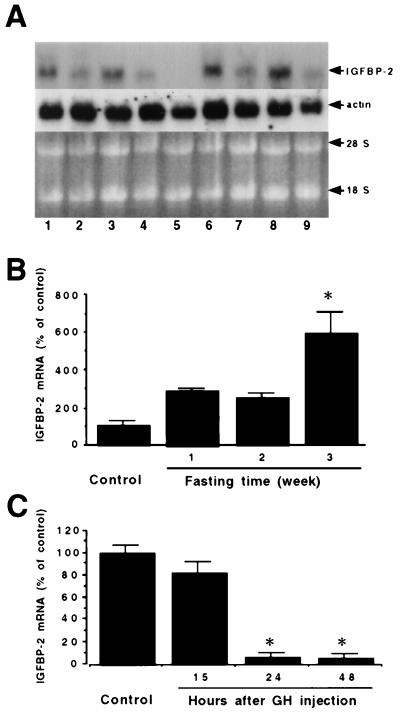
Northern blot analysis of adult fish and embryos revealed a single transcript of approximately 1.8 kb (Fig. 4A). IGFBP-2 mRNA is relatively abundant in brain, liver, muscle, eye, and intestine of adult zebrafish. To determine the nutritional and hormonal regulation of IGFBP-2 expression, we fasted adult zebrafish for up to 3 weeks. The levels of IGFBP-2 mRNA in normal, well-fed adult animals were low. When the animals were fasted, the steady-state levels of IGFBP-2 increased significantly. Three-week fasting increased the IGFBP-2 mRNA level by 5-fold (Fig. 4B). No significant change was seen in actin mRNA levels. We next determined the effect of GH on IGFBP-2 gene expression. As shown in Fig. 4C, a single i.p. injection of fish GH (1 μg/g body weight) resulted in a significant decrease in the steady-state levels of IGFBP-2 mRNA in adult fish. At 24 and 48 hr after the GH injection, the levels of IGFBP-2 mRNA were reduced to 6% and 5% of the control levels, respectively (Fig. 4C). This effect was specific because no significant change was found with the levels of actin mRNA. These data suggest that the IGFBP-2 expression is increased by prolonged fasting but negatively regulated by GH in zebrafish.
Figure 4.
The expression of IGFBP-2 gene is increased by fasting and decreased by GH treatment in zebrafish. (A) Tissue distribution pattern of IGFBP-2 mRNA. Northern blot analysis of RNA samples isolated from 20-hr embryos (lane 1), whole body of adult zebrafish (lane 2), and brain (lane 3), eye (lane 4), ovary (lane 5), liver (lane 6), intestine (lane 7), muscle (lane 8) and fin (lane 9) collected from adult fish. (B) Effect of fasting on IGFBP-2 mRNA expression. RNA was isolated from fed (control) or fasted adult zebrafish. The values are means ± SE of three animals. *, P < 0.05 compared with the control. (C) Effect of GH on IGFBP-2 mRNA expression. Adult zebrafish were injected with pure seabass GH (1 μg/ml) or saline and sacrificed 15, 24, or 48 hr after the injection. The values are means ± SE of three animals. *, P < 0.05 compared with the control.
Zebrafish IGFBP-2 Inhibits IGF-I-Stimulated DNA Synthesis and Cell Proliferation.
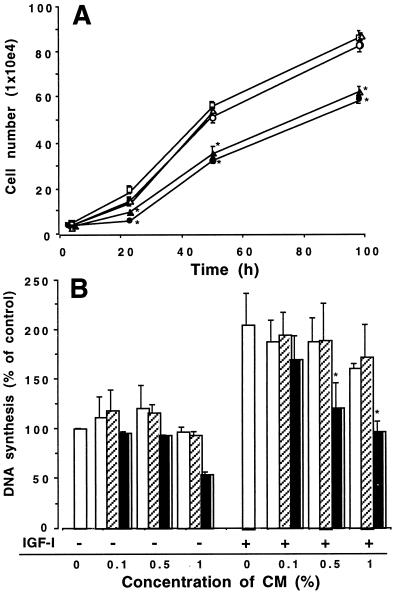
To determine the biological effect of IGFBP-2 on cell growth, the growth rates of selected CHO clones were determined in the presence of 1% FBS. Mock-transfected cells had similar growth curves to those of the wild-type CHO cells (Fig. 5A). In contrast, IGFBP-2-transfected clones 6 and 2, which highly expressed zebrafish IGFBP-2, had significantly reduced cell numbers compared with those in the wild-type and mock-transfected cells at 24, 48, and 98 hr (P < 0.01). These data indicate that overexpression of zebrafish IGFBP-2 inhibits CHO cell growth. We next sought to determine the effect of IGFBP-2 in modulating IGF actions in zebrafish cells by [3H]thymidine incorporation assay. Exposure of confluent ZF-4 cells to IGF-I (100 ng/ml) resulted in a 205 ± 32% increase in thymidine incorporation (P < 0.05, n = 3). To eliminate the potential interference of the endogenous CHO IGFBP-4 present in the CMs, immunodepletion was performed by using an antibody against IGFBP-4. This procedure depleted the endogenous IGFBP-4 without changing the levels of zebrafish IGFBP-2 (data not shown). When added to the cultures at concentrations up to 5%, immunodepleted CMs prepared from either wild-type or mock-transfected cells did not cause any notable change in basal DNA synthesis levels. Addition of the CM containing zebrafish IGFBP-2 resulted in a moderate, but statistically insignificant, decrease. When coincubated with IGF-I, the zebrafish IGFBP-2 containing CM inhibited IGF-I-stimulated DNA synthesis in zebrafish cells in a concentration-dependent manner (Fig. 5B). At the concentration of 1%, it completely abolished the IGF-I effect. Wild-type and mock-transfected CMs had no such inhibitory effect. Similar results also were obtained by immunostaining of 5-bromo-2-deoxyuridine incorporation in the nuclei of dividing ZF-4 cells (data not shown). These results suggest that zebrafish IGFBP-2 inhibits cell proliferation in cultured mammalian and zebrafish cells.
Figure 5.
Zebrafish IGFBP-2 inhibits cell growth and DNA synthesis. (A) Overexpression of zebrafish IGFBP-2 inhibits CHO cell growth. Growth curves of wild-type CHO cells (□), the pRC/CMV vector-transfected clone 1 (○) and clone 2 (▵), and the pRc/CMV-IGFBP-2-transfected clone 2 (●) and clone 6 (▴) are shown. *, P < 0.01 (n = 8) compared with the wild-type controls. (B) Effect of zebrafish IGFBP-2 on IGF-I-stimulated DNA synthesis in cultured zebrafish cells. Confluent cells were exposed to serum-free medium or IGF-I (100 ng/ml) in the presence of various concentrations of CM prepared from wild-type (open bars), mock-transfected (hatched bars), and zebrafish IGFBP-2-transfected cells (filled bars). The values are means ± SE of three separate experiments, each of which was performed in triplicate. *, P < 0.05 compared with the IGF-I alone group.
Discussion
In this study we report the cloning, characterization, and expression of a zebrafish cDNA encoding a protein homologous to mammalian IGFBP-2. The zebrafish IGFBP-2 has an apparent molecular size of 31 kDa and binds to IGF-I and IGF-II with high affinity and specificity. When overexpressed and added exogenously to zebrafish and mammalian cells, it inhibited IGF-I and serum-stimulated cell proliferation and DNA synthesis.
The six high-affinity mammalian IGFBPs share a common domain organization: a highly cysteine-rich N-terminal domain, a cysteine-rich C-terminal domain, and a central domain with no cysteine residues except in IGFBP-4. This domain arrangement is well conserved in the zebrafish IGFBP-2. Sequence alignment reveals that the N and C domains are highly conserved across species. In contrast, the central L domain shows little sequence identity except for two short stretches of amino acids (R116KPXK120 and Q131HRGXXK137, where X represents any residue). Twelve of the 18 conserved cysteines are located in the N domain and the remaining six in the C-terminal domain. The cysteine residues in the N-terminal domain are arranged into two Zn-finger motifs (CX2CX7C). The high degree of conservation of N and C domains is probably not surprising. Recent studies have indicated that both N- and C-terminal domains are required for IGF binding. It has been shown in bovine IGFBP-2 in particular that residue Tyr-60 in the N domain and a stretch of amino acids (K222HGLYNLKQCKMSLN236) in the C domain are important for IGF binding (20, 21). These residues are well conserved in the zebrafish IGFBP-2 (Tyr-61 and K194RGQYNLKQCKMSVN208). When the zebrafish IGFBP-2 protein was produced and analyzed, it bound to human IGF-I and IGF-II with high affinity but did not bind to human Des(1–3)IGF-I or insulin. These results suggest that IGF binding determinant(s) of IGFBP-2 have been highly conserved during the several hundred million years of vertebrate evolution.
Analysis of mRNA size and levels in zebrafish revealed that the IGFBP-2 gene is expressed at high levels in various tissues and developmental stages. The zebrafish IGFBP-2 transcript is 1.8 kb, larger than its counterparts in human, bovine, rat, and mouse (all of which are around 1.5 kb) but smaller than that of the chicken (2.3 kb). IGFBP-2 mRNA is easily detected in zebrafish embryos and adult tissues. In adult zebrafish, high levels of IGFBP-2 mRNA are found in brain, liver, muscle, eye, and intestine (Fig. 4). The high levels of IGFBP-2 expression in various tissues suggest that IGFBP-2 is being synthesized locally in zebrafish. This finding is in good agreement with previous observations that various fish tissues in culture produce a 31-kDa IGFBP (13, 23, 24). Therefore, IGFBP-2 is produced locally and may play a role in controlling IGF availability and activity in defined tissues in fish. Previous studies have shown that mammalian IGFBP-2 expression is significantly increased in diabetic or fasted animals, or with protein or energy restriction (25, 26). Elevation in IGFBP-2 mRNA levels also is observed in hypophysectomized rats whereas GH therapy partially reduced it (25). A recent study indicated that hypophysectomy induced the presence of an IGFBP in the serum of tilapia (41). These features of IGFBP-2 gene expression are well conserved in zebrafish. As shown in this study, prolonged fasting greatly increased whereas GH injection decreased the tissue levels of IGFBP-2 in zebrafish.
The discovery that the zebrafish IGFBP-2 is a growth inhibitory protein acting downstream in the GH–IGF-I axis is of particular interest. This finding is supported by (i) the retarded growth phenotype of CHO cells that overexpressed zebrafish IGFBP-2, and (ii) that addition of zebrafish IGFBP-2 abolished IGF-I-stimulated DNA synthesis and cell proliferation in cultured zebrafish cells. This conclusion also is supported by the gene expression data. To date, several mammalian IGFBP-2 proteins have been isolated and characterized functionally. Mammalian IGFBP-2 has been shown to both inhibit and potentiate IGF actions depending on cellular context (28, 30–34). Because the IGF-I receptor mediates most of the biological actions of IGFs, and because the soluble form of IGFBP-2 binds to IGFs with higher affinity than do the IGF-I receptors, the inhibitory effects can be rationally explained by its competition for the ligand with the IGF receptors. The mechanism of potentiation of IGF activity by IGFBP-2, however, is not completely understood. Accumulating evidence indicates that cell surface/extracellular matrix (ECM) association of IGFBP-2 is a contributing factor (35, 36). The cell surface/ECM association of IGFBP-2 has a lower affinity for IGFs compared with IGFBP-2 in solution. It has been postulated that this switch of binding affinity may result in the equilibrium favoring releasing of IGF-I to its receptor and may account for its potentiating effect. The RGD motif embedded in a conserved pentapeptide (IRGDP) is present in mammalian, chicken, and zebrafish IGFBP-2 (between residues 221 and 226). A similar RGD motif in human IGFBP-1 has been shown to be involved in the integrin binding of IGFBP-1, and this interaction mediates IGF action of IGFBP-1 on cell migration (22). Previous studies, however, have failed to demonstrate a similar functional role of the RGD motif in IGFBP-2 (36). Recent studies have suggested that the association of IGFBP-2 with cell surface or ECM is caused by its ability to bind to proteoglycans (35, 36). Although it has been proposed that the putative heparin-binding motif (PKKXRP) located in the C domain of all mammalian IGFBP-2 is responsible for the cell surface/proteoglycan binding property of this molecule, direct functional test of this motif has not been reported. Intriguingly, the PKKXRP heparin-binding motif is absent in the zebrafish IGFBP-2: the sequence in the corresponding region in the zebrafish is PK—AP (where — represents a gap). Consistent with the finding in structure, our functional analysis results indicate the zebrafish IGFBP-2 acts predominantly as an IGF-I inhibitor in both zebrafish and mammalian cells. Furthermore, our preliminary results suggested that unlike mammalian IGFBP-2 zebrafish IGFBP-2 did not localize to the cell surface (data not shown). It is possible that the predominantly inhibitory action of zebrafish IGFBP-2 is caused by its lack of the heparin-binding motif and consequently its inability to bind cell surface/ECM. Future mutagenesis studies are needed to directly test this hypothesis by mutating the PKKXRP motif in the human IGFBP-2 and introducing this sequence into fish IGFBP-2.
In summary, we have isolated and characterized cDNA clones encoding zebrafish IGFBP-2. Structural, functional, and gene expression analyses indicate that zebrafish IGFBP-2 is an evolutionarily conserved growth inhibitory protein acting downstream in the GH-IGF axis. This finding further supports the concept that IGFBP-2 is an important growth regulator in vertebrates. The acquisition of the zebrafish IGFBP-2 sequence and the availability of its cDNA probes and protein will now enable us to elucidate the developmental role of IGFBP-2 in a well-established teleost model organism.
Acknowledgments
We thank Dr. B. Appel at Vanderbilt University for providing the zebrafish cDNA library, Dr. D. R. Clemmons at the University of North Carolina for providing the IGFBP-4 antibody, and C. Rosario for critical reading of this manuscript. This study was supported in part by National Science Foundation Grant IBN-9728911 and National Institutes of Health Grant 5P60DK-20572 through a Pilot/Feasibility grant from Michigan Diabetes Research and Training Center.
Abbreviations
- CM
conditioned medium
- GH
growth hormone
- IGF
insulin-like growth factor
- IGFBP
IGF binding protein
- CHO
Chinese hamster ovary
- CMV
cytomegalovirus
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF198033).
References
- 1.Cock A G. Q Rev Biol. 1966;41:131–190. doi: 10.1086/404940. [DOI] [PubMed] [Google Scholar]
- 2.Siharath K, Bern H A. Proc Zool Soc (Calcutta) 1993;3:113–124. [Google Scholar]
- 3.Dickhoff W W, Beckman B R, Larsen D A, Duan C, Moriyama S. Fish Physiol Biochem. 1997;17:231–236. [Google Scholar]
- 4.Plisetskaya E M. Comp Biochem Physiol B. 1998;121:3–11. doi: 10.1016/s0305-0491(98)10107-4. [DOI] [PubMed] [Google Scholar]
- 5.Duan C. J Nutr. 1998;128:306S–314S. doi: 10.1093/jn/128.2.306S. [DOI] [PubMed] [Google Scholar]
- 6.Jones I J, Clemmons D R. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 7.Kelley K M, Oh Y, Gargosky S E, Gucev Z, Matsumoto T, Hwa V, Ng L, Simpson D M, Rosenfeld R G. Int J Biochem Cell Biol. 1995;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 8.Oh Y, Yamanaka H-S, Kim P, Vorwerk P, Wilson E, Hwa V, Yand D-H, Spagnoli A, Wanek D, Rosenfeld R G. In: Molecular Mechanisms to Regulate the Activities of Insulin-Like Growth Factors. Tanaka K, Hizuka N, Takahashi S-I, editors. Amsterdam: Elsevier; 1998. pp. 125–133. [Google Scholar]
- 9.Kelley K M, Siharath K, Bern H A. J Exp Zool. 1992;263:220–224. doi: 10.1002/jez.1402630213. [DOI] [PubMed] [Google Scholar]
- 10.Niu P-D, Le Bail P-Y. J Exp Zool. 1993;265:627–636. [Google Scholar]
- 11.Anderson T A, Bonnet L R, Conlon M A, Owens P C. J Endocrinol. 1993;136:191–198. doi: 10.1677/joe.0.1360191. [DOI] [PubMed] [Google Scholar]
- 12.Siharath K, Nishioka R S, Madsen S S, Bern H A. Mol Mar Biol Biotech. 1995;4:171–178. [Google Scholar]
- 13.Siharath K, Nishioka R S, Bern H A. Aquaculture. 1995;135:195–202. [Google Scholar]
- 14.Siharath K, Kelley K M, Bern H A. Gen Comp Endocrinol. 1996;102:307–316. doi: 10.1006/gcen.1996.0074. [DOI] [PubMed] [Google Scholar]
- 15.Duan C, Hawes S B, Prevette T, Clemmons D R. J Biol Chem. 1996;271:4280–4288. doi: 10.1074/jbc.271.8.4280. [DOI] [PubMed] [Google Scholar]
- 16.Duan C, Clemmons D R. J Biol Chem. 1998;273:16836–16842. doi: 10.1074/jbc.273.27.16836. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Nucleic Acids Res. 1987;15:8125–8133. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson M E. Nucleic Acids Res. 1984;12:5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis G L, Ross M, Ballard F J, Milner S J, Senn C, McNeil K A, Wallace J C, King R, Wells J R E. J Mol Endocrinol. 1992;8:213–223. doi: 10.1677/jme.0.0080213. [DOI] [PubMed] [Google Scholar]
- 20.Hobba G D, Lothgren A, Holmberg E, Forbes B, Francis G L, Wallace J C. J Biol Chem. 1998;273:19691–19698. doi: 10.1074/jbc.273.31.19691. [DOI] [PubMed] [Google Scholar]
- 21.Forbes E F, Turner D, Hodge S J, McNeil K A, Forsberg G, Wallace J C. J Biol Chem. 1998;273:4647–4652. doi: 10.1074/jbc.273.8.4647. [DOI] [PubMed] [Google Scholar]
- 22.Jones J I, Gockerman A, Busby W H, Wright G, Clemmons D R. Proc Natl Acad Sci USA. 1993;90:10553–10557. doi: 10.1073/pnas.90.22.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukazawa Y, Siharath K, Iguchi T, Bern H A. Gen Comp Endocrinol. 1995;99:239–247. doi: 10.1006/gcen.1995.1107. [DOI] [PubMed] [Google Scholar]
- 24.Moriyama S, Kagawa H, Duan C, Dickhoff W W, Plisetskaya E M. Comp Physiol Biochem. 1997;117:201–206. [Google Scholar]
- 25.Ooi G T, Orlowski C C, Brown A L, Becker R E, Unterman T G, Rechler M M. Mol Endocrinol. 1990;4:321–328. doi: 10.1210/mend-4-2-321. [DOI] [PubMed] [Google Scholar]
- 26.Orlowski C C, Brown A L, Ooi G T, Yang Y W-H, Tseng Y-H, Rechler M M. Endocrinology. 1990;126:644–652. doi: 10.1210/endo-126-1-644. [DOI] [PubMed] [Google Scholar]
- 27.Binkert C, Landwehr J, Mary J-L, Schwander J, Heirich G. EMBO J. 1989;8:2497–2502. doi: 10.1002/j.1460-2075.1989.tb08386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourner M J, Busby W H, Jr, Siegel N R, Krivi G G, McCusker R H, Clemmons D R. J Cell Biochem. 1992;48:215–226. doi: 10.1002/jcb.240480212. [DOI] [PubMed] [Google Scholar]
- 29.Margot J B, Binkert C, Mary J-L, Landwehr J, Heinrich G, Schwander J. Mol Endocrinol. 1989;3:1053–1060. doi: 10.1210/mend-3-7-1053. [DOI] [PubMed] [Google Scholar]
- 30.Feyen J H M, Evans D B, Binker C, Heinrich G F, Geisse S, Kocher H P. J Biol Chem. 1991;266:19469–19474. [PubMed] [Google Scholar]
- 31.Wang J F, Harrada E, Hill D J, Ruckingham K D, Philips I, Becks G P. J Endocrinol. 1991;130:129–140. doi: 10.1677/joe.0.1300129. [DOI] [PubMed] [Google Scholar]
- 32.Slootweg M C, Ohlsson C, Salles J-P, deVries C P, Netelenbos J C. Endocrinology. 1995;136:4210–4217. doi: 10.1210/endo.136.10.7545101. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Nagpal M L, Lin T, Shimasaki S, Ling N. Mol Endocrinol. 1994;8:69–76. doi: 10.1210/mend.8.1.7512196. [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw S L, D'Ercole A J, Han V K M. Endocrinology. 1999;140:575–584. doi: 10.1210/endo.140.2.6498. [DOI] [PubMed] [Google Scholar]
- 35.Arai T, Busby W, Jr, Clemmons D R. Endocrinology. 1996;137:4571–4575. doi: 10.1210/endo.137.11.8895319. [DOI] [PubMed] [Google Scholar]
- 36.Russo V C, Bach L A, Fosang A J, Baker N L, Werther G A. Endocrinology. 1997;138:4855–4867. doi: 10.1210/endo.138.11.5472. [DOI] [PubMed] [Google Scholar]
- 37.Upton F Z, Szabo J C, Wallance J C, Ballard F J. J Mol Endocrinol. 1990;5:77–84. doi: 10.1677/jme.0.0050077. [DOI] [PubMed] [Google Scholar]
- 38.Delhanty P J D, Han V K M. J Mol Endocrinol. 1992;9:31–38. doi: 10.1677/jme.0.0090031. [DOI] [PubMed] [Google Scholar]
- 39.Landwehr J, Kaupmann K, Heinrich G, Schwander J. Gene. 1993;124:281–286. doi: 10.1016/0378-1119(93)90406-s. [DOI] [PubMed] [Google Scholar]
- 40.Schoen T J, Mazuruk K, Waldbillig R J, Potts J, Beebe D C, Chader G J, Rodriguez I R. J Mol Endocrinol. 1995;15:49–59. doi: 10.1677/jme.0.0150049. [DOI] [PubMed] [Google Scholar]
- 41.Park, R., Shepherd, B. S., Nishioka, R. S., Grau, G. E. & Bern, H. A. (2000) Gen. Comp. Endocrinol, in press. [DOI] [PubMed]