Abstract
Large-scale gene expression studies can now be routinely performed on macroamounts of cells, but it is unclear to which extent current methods are valuable for analyzing complex tissues. In the present study, we used the method of serial analysis of gene expression (SAGE) for quantitative mRNA profiling in the mouse kidney. We first performed SAGE at the whole-kidney level by sequencing 12,000 mRNA tags. Most abundant tags corresponded to transcripts widely distributed or enriched in the predominant kidney epithelial cells (proximal tubular cells), whereas transcripts specific for minor cell types were barely evidenced. To better explore such cells, we set up a SAGE adaptation for downsized extracts, enabling a 1,000-fold reduction of the amount of starting material. The potential of this approach was evaluated by studying gene expression in microdissected kidney tubules (50,000 cells). Specific gene expression profiles were obtained, and known markers (e.g., uromodulin in the thick ascending limb of Henle's loop and aquaporin-2 in the collecting duct) were found appropriately enriched. In addition, several enriched tags had no databank match, suggesting that they correspond to unknown or poorly characterized transcripts with specific tissue distribution. It is concluded that SAGE adaptation for downsized extracts makes possible large-scale quantitative gene expression measurements in small biological samples and will help to study the tissue expression and function of genes not evidenced with other high-throughput methods.
The high amount of information accumulated over the last few years in the nucleotide sequence databases calls for approaches that fully use these resources to study cell function and regulation on a genomic scale. Four years ago, two high-throughput methods for quantitative monitoring of gene expression were published simultaneously (1, 2). The DNA microarray approach (1) consists of parallel hybridization of labeled targets to immobilized probes. On the other hand, the method called serial analysis of gene expression (SAGE) (2) relies on sequencing short diagnostic sequence tags. Both methods demonstrated the feasibility of genome-wide expression studies, at least in unicellular organisms (3, 4).
In higher organisms, the heterogeneity of most tissues makes desirable the availability of high-throughput methods suitable for purified cell populations. Indeed, analysis of defined cell types is a prerequisite to establish links between molecular (i.e., mRNA expression level) and physiological phenotypes and, thereby, to progress toward the elucidation of genes function. However, the DNA micro- or macroarray approaches (1, 4–6), as well as the SAGE method (2, 3), were designed and most often are used to study macroamounts of biological material [1–5 μg of poly(A) RNAs, corresponding to 0.5–2.5 × 107 mammalian cells]. The present study therefore was undertaken to seek conditions enabling to perform large-scale gene expression studies on small tissue samples.
SAGE is particularly well suited for organisms whose genome is not completely sequenced, because it does not require a hybridization probe for each transcript and allows new genes to be discovered. This method relies on two experimentally corroborated principles (2, 3). First, a short nucleotide sequence tag (10 bp) isolated from a defined region of a transcript is sufficient for its unequivocal identification. Second, concatenation of several tags into a single clone, characterized by DNA sequencing, greatly increases the efficiency of data acquisition. SAGE involves several steps, including mRNA purification, then generation, isolation, and PCR amplification of cDNA tags. We postulated that increasing the efficiency of these different steps should make it possible to reduce the amount of tissue required to generate a SAGE library. This possibility was explored by using the mouse kidney as a model tissue. Indeed, the different epithelial cells of the mammalian kidney vary greatly in abundance and are distributed along successive nephron portions with specific physiological properties. We therefore set up a modified SAGE assay compatible with the analysis of small biological samples and referred it to as SAGE adaptation for downsized extracts (SADE). We show here that SADE provides representative gene expression profiles in microdissected nephron segments.
Methods
Tissue Samples.
Experiments were carried out on male C57 BL/6J mice (8–10 weeks old; Charles River Breeding Laboratories). After anesthesia (sodium pentobarbital, 140 μg/g body weight), the kidneys were quickly removed and frozen in liquid nitrogen for subsequent RNA isolation. For experiments on isolated nephron segments, the left kidney was perfused with Hanks' modified microdissection solution and then with the same solution containing 0.15% collagenase (Boehringer Mannheim) and removed rapidly, and microdissection was performed at 4°C (7). Medullary thick ascending limbs of Henle's loop (MTALs) and outer medullary collecting ducts (OMCDs) were obtained from the inner stripe and outer stripe of the outer medulla.
Primers and Linkers.
Biotinylated oligo(dT)20 were obtained from Boehringer Mannheim. Other oligonucleotides were from Genset (Evry, France). SAGE linker 1 was formed through hybridization of oligonucleotides 1A (5′-TTTTGCCAGGTCACTCAAGTCGGTCATTCATGTCAGCACAGGGAC-3′) and 1B (5′-GATCGTCCCTGTGCTGACATGAATGACCGACTTGAGTGACCTGGCA-3′). SAGE linker 2 was obtained by hybridizing oligonucleotides 2A (5′-TTTTTGCTCAGGCTCAAGGCTCGTCTAATCACAGTCGGAAGG- GAC-3′) and 2B (5′-GATCGTCCCTTCCGACTGTGATTAGACGAGCCTTGAGCCTGAGCAA-3′). Oligonucleotides 1B and 2B included two modifications (5′ phosphorylation and 3′ C7 amino modification). All four oligonucleotides were gel-purified. The sequence of cognate PCR primers for linkers 1 and 2 were 5′-GCCAGGTCACTCAAGTCGGTCATT-3′ and 5′-TGCTCAGGCTCAAGGCTCGTCTA-3′, respectively. DNA sequencing was performed by using T7 primer. The sequence of reverse transcriptase–PCR (RT-PCR) primers is available from the authors on request.
Generation of SAGE Libraries.
Total RNAs were extracted as described (8). Poly(A) RNAs were selected on oligo(dT)-cellulose spin columns (CLONTECH). SAGE libraries were constructed by using a protocol modified from the original method (2). The main modifications we have introduced are the use of Sau3AI (instead of NlaIII) as anchoring enzyme, T7 DNA polymerase for blunt-ending cDNA tags, and new PCR primers for ditags amplification. Briefly, cDNA was synthesized from 5 μg of poly(A) RNAs with the cDNA synthesis system kit (Life Technologies, Gaithersburg, MD) by following the manufacturer's recommendations for procedure I, using 170 pmol of biotinylated oligo(dT)20. Second-strand cDNA synthesis was performed in the presence of [α-32P]dCTP (0.5 μCi/nmol). The double-strand cDNA was digested with Sau3AI, and the 3′ end was isolated through binding to 1 mg of paramagnetic streptavidin beads (Dynal), whereas the unbound fraction (5′ end) was analyzed by gel electrophoresis and liquid scintillation counting. The streptavidin-bound cDNA was divided into two fractions. Each fraction was ligated to 10 pmol of either linker 1 or linker 2, then digested with BsmFI. The released cDNA tags were blunt-ended for 10 min at 42°C in a 50-μl reaction volume containing 20 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 25 mM NaCl, 10 mM DTT, 400 μM dNTP, and 10 units of T7 DNA polymerase (Pharmacia Biotech). The two fractions then were ligated to each other by using T4 DNA ligase. PCR (95°C, 30 sec; 58°C, 30 sec; 70°C, 45 sec: 26–28 cycles) was carried out on 1% of ligation reaction in a 100-μl reaction volume containing 20 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 4 mM DTT, 100 μM dNTP, 50 pmol of primers 1 and 2, and 5 units of Taq polymerase. The products from 10–12 reactions were pooled. The 110-bp DNA fragment was purified by agarose gel electrophoresis and submitted to preparative PCR (120–150 reactions performed as described above, except that the amount of primers was reduced to 25 pmol and the number of cycles was 12). The PCR sample was digested with Sau3AI, and ditags were purified and concatenated as described (2). Concatemers ≥350 bp were recovered and ligated into BamHI-cut pBluescript II (Stratagene), and the resulting product was used to transform Escherichia coli XL-2 blue ultracompetent cells (Stratagene).
Sequencing reactions were performed in our laboratory or by Genome Express (Grenoble, France) by using Big Dye terminator sequencing chemistry (Applied Biosystems) and run on 377-XL Applied Biosystems automated sequencers. Electrophoregrams were checked for ambiguous base calls and to correct misreads.
Micromethod for Serial Analysis of Gene Expression.
Besides modifications described above to the SAGE method, we set up an alternative assay suitable both for macrosamples (250 mg of tissue) and microsamples (0.5 mg of tissue, or 50,000 cells). We describe here only the method used for obtaining libraries from microdissected kidney tubules, because library generation from larger samples requires only to scale up the reaction volumes and amount of enzymes. Pools of microdissected tubules were centrifuged for 5 min at 1,000 × g. Poly(A) RNAs were isolated through hybridization to oligo(dT)25 covalently bound to magnetic beads by using Dynabeads mRNA direct kit (Dynal). Briefly, 100 μl of lysis binding buffer supplemented, as well as all solutions used for subsequent steps carried out on beads, with 20 μg/ml glycogen (Boehringer Mannheim) was added on the cell pellet. The lysate was transferred onto 100 μg of oligo(dT)25 beads, and the sample was incubated for 10 min at room temperature. The beads were rinsed twice with washing buffer containing lithium dodecyl sulfate, three times with washing buffer, and three times with RT buffer, resuspended in 12.5 μl of the same buffer, and transferred at 37°C. First- and second-strand cDNA syntheses were performed as described above, except that reaction volumes were halved and that the second-strand reaction synthesis was incubated overnight at 16°C. Initial libraries were generated by using mouse murine leukemia virus RT (M-MLV RT). However, Superscript II M-MLV RT (Life Technologies) provided ≈4-fold higher cDNA yields and, therefore, is strongly recommended for very small samples (≤50,000 cells). After double-strand cDNA synthesis, the beads were rinsed four times with 200 μl of TEN buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/1 M NaCl/0.1 mg/ml BSA) and three times with 200 μl of Sau3AI digestion buffer also containing 0.1 mg/ml BSA, and the cDNA was digested with Sau3AI. After removal of the Sau3AI-released fraction, the beads were rinsed three times with TEN buffer and three times with TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA) containing 0.1 mg/ml BSA and divided into two aliquots for ligation to 0.5 pmol of either linker 1 or 2. Subsequent steps were carried out as described above, except that the first series of PCR consisted of 29–31 cycles and was carried out on 4% of the ditags.
SAGE Data Analysis.
Sequence files were analyzed by using sage software (2). Tags corresponding to linker sequences were discarded, and those originating from duplicate ditags were counted only once (2). For tag identification, the tag list of each library was matched against a mouse tag database extracted by sage software from GenBank release 111. For an expressed sequence tag (EST) match to be considered correct, we checked that the EST displayed the most 3′ Sau3AI site of the cDNA. This was achieved by checking that the EST had a poly(A) tail and/or analyzing the corresponding consensus sequence of the database of The Institute for Genome Research. Assessment of significant differences between two SAGE libraries was made by using Monte Carlo simulation analysis (9). A P value of 0.05 or less was considered significant.
RT-PCR.
Totals RNAs were extracted from the whole kidney tissue by the method of Chomczynski and Sacchi (8) or from isolated tubules by using a microadaptation of this method (7). RT-PCR was carried out on 1 ng of total kidney RNAs or 0.3 mm of kidney tubules. These amounts correspond to ≈102 cells (10, 11). The primers were selected from mouse full-length cDNA or EST sequences available from GenBank. Reverse transcription and PCR were performed sequentially in the same reaction in the presence of [α-32P]dCTP (0.5 μCi/nmol) to directly label the product (7). Amplification consisted of 25 PCR cycles, providing for each target DNA amounts that could be detected by autoradiography, but not by ethidium bromide staining (end product concentration <1 nM). These experimental conditions allow exponential accumulation of DNA to occur throughout the PCR and, hence, the accurate comparison of signals obtained from different samples (7). Aliquots (20 μl) of RT-PCR samples were electrophoresed through a 2% agarose slab gel. The gel was then fixed in 10% acetic acid, vacuum-dried, and exposed for autoradiography.
Results and Discussion
Gene Expression Pattern in the Kidney.
Gene expression profiling first was carried out at the whole-kidney level. The 12,154 SAGE tags sequenced corresponded to 4,800 different tags; 1,200 were recorded two to several hundred times, whereas the remaining 3,600 tags were detected only once. This distribution fits with the overall pattern of gene expression in mammalian cells, wherein only a few percent of mRNA species reaches high copy number, whereas most mRNAs display faint levels (12).
The tags sampled with a frequency of >1% had all GenBank entries, and most of them (7 of 11) matched to mitochondrial genes [cytochrome c oxidase polypeptide II (5.4%) and I (1.5%); NADH-ubiquinone oxidoreductase chain 2 (3.7%), 1 (1.6%), and 4 (1.6%); cytochrome b (1.5%); ATP synthase α chain (1.2%)]. The very high levels for tags corresponding to mitochondrial transcripts is likely of physiological relevance, because the kidney has extensive oxidative metabolism and contributes to 10% of bodily ATP production.
Table 1 shows the data for the most abundant tags ascribed to nuclear transcripts. A significant fraction (25%) of these tags corresponds to mRNAs known to be specific for, or highly enriched in, proximal tubular cells, i.e., the predominant kidney cell type. Besides tissue specificity, the prevalence of such tags is consistent with results obtained for mRNA levels in the kidney by using different approaches (13–16). Other abundant tags matching genes highly expressed in the proximal tubule include those for the Na/glucose type II transporter (n = 12), the parathyroid hormone (PTH)/PTH-rp receptor (n = 11), and aquaporin-1 (n = 8). Table 1 also shows that high levels for mRNAs widely and strongly expressed (e.g., ferritins and nuclear-encoded cytochromes, glyceraldehyde-3-phosphate dehydrogenase, heat shock protein 70, β-actin) are also predicted from our data. The presence of tags for α-globin likely comes from the fact that blood cells were not washed out from the kidney, whereas that for rRNA suggests that more stringent conditions should be used to select poly(A) RNAs (see below).
Table 1.
Abundant tags for nuclear transcripts in the mouse kidney
| N | % | Tag | Databank entry |
|---|---|---|---|
| 355 | 2.92 | CATGACATCC | Plasma glutathione peroxidase (U13705)* |
| 189 | 1.55 | CTGGATGAGA | Kidney androgen-regulated protein (M22810)† |
| 135 | 1.11 | TGCATGCCCT | Ferritin light chain (J04716) |
| 132 | 1.09 | ATCGACACTT | 5.8S rRNA (K01367) |
| 69 | 0.57 | CCCACTTATG | Ferritin heavy chain (J03941) |
| 63 | 0.52 | AGCATTGAGC | Renal type II Na/Pi cotransporter (L33878)† |
| 60 | 0.49 | ACCAGGACCT | No match |
| 58 | 0.48 | GTGACTGGGT | Cytochrome c oxidase subunit IV (X54691) |
| 53 | 0.44 | TGACGTGCCG | Glyceraldehyde-3-phosphate dehydrogenase (M32599) |
| 47 | 0.39 | TCTACACGAA | Est 519201 5′ from mouse kidney (AA106199) |
| 42 | 0.35 | CCGTCAACTT | α-Globin (L75940) |
| 40 | 0.33 | GGGAAGTACG | Argininosuccinate synthetase (M31690)* |
| 38 | 0.31 | CTTAACTTCC | Na, K-ATPase γ-subunit (Q04646) |
| 38 | 0.31 | TAACTGTGGA | ESTs |
| 36 | 0.30 | CCACAGGTCT | Transcobalamin II (AF090686)* |
| 35 | 0.29 | AATGCCCTCA | Acidic ribosomal phosphoprotein P1 (U29402) |
| 34 | 0.28 | ATGTTATTCA | Cytochrome c oxidase subunit VIII precursor (U37721) |
| 34 | 0.28 | TGCAGAAGCG | Cellular glutathione peroxidase (X03920) |
| 31 | 0.25 | TGACTCCCTC | B2 repetitive sequence |
| 30 | 0.25 | ATCATGTTTA | ESTs, similar to rat aldolase B (M10149)* |
| 30 | 0.25 | CAGAAGCAGT | Endogenous murine leukemia virus sequence (L08395) |
| 29 | 0.24 | TATCTGCTGG | Cytochrome β-558 (M31775) |
| 27 | 0.22 | AGGATGACTG | Renal-specific organic cation transporter (AB005451)† |
| 26 | 0.21 | AATGATGAGG | Heat shock protein 70 (M19141) |
| 22 | 0.18 | AGGCAGTGTT | ESTs |
| 22 | 0.18 | TCATTATATA | EST, similar to human aspartoacylase (P45381) |
| 21 | 0.17 | CACATTGAGG | Ketohexokinase (Y09335) |
| 20 | 0.16 | AGCAAGCAGG | β-Actin (X03672) |
| 20 | 0.16 | AGGTGGTGAC | γ-Glutamyl transpeptidase (U30509)* |
| 20 | 0.16 | TGACGCCCTC | B2 repetitive sequence |
| 20 | 0.16 | TTGTTTGCCA | ESTs, similar to mouse H+ ATP synthase subunit c (L19737) |
The table lists tags detected ≥20 times (mitochondrial tags were excluded from the report) in the kidney SAGE library (12,154 tags sequenced). Note that several tags correspond to mRNAs enriched in the kidney proximal tubule (*) or specific (†) for this nephron portion. Additional data are available at http://www-dsv.cea.fr/thema/get/sade.html
Proximal tubular cells account for 60% of the kidney cell mass, with the remaining fraction corresponding to a variety of minor cell types. These anatomical features explain why whole-kidney SAGE data are chiefly informative for proximal tubular markers. Exploring less abundant cells requires a different approach. Because all nephron segments can be isolated by microdissection, we undertook to set up a microadaptation of SAGE suitable for tiny amounts of cells.
Scaling Down the SAGE Method.
To check for possible improvement of the SAGE method, we undertook a quantitative approach. Fig. 1 Left shows results of a typical SAGE experiment carried out on the kidney. Starting from 500 mg of tissue, 1 mg of total RNAs was obtained routinely, 5 μg of poly(A) RNAs was recovered after oligo(dT) chromatography, and ≈2 μg of cDNA (quantified through radioactivity incorporation) was synthesized. Gel electrophoresis analysis indicated an average cDNA length of ≈1.5 kb. A restriction enzyme with a 4-bp recognition site (SAGE-anchoring enzyme) produces DNA fragments whose mean length is 256 bp (44). Consequently, cleavage of 1.5-kb long cDNAs should enable us to recover a 3′ (biotinylated) fraction corresponding to 15–20% (0.3 μg in the current experiment) of the cDNA mass. As shown in Fig. 1, the amount obtained was 100-fold lower than predicted. This indicates that quantitative cDNA recovery should greatly improve the SAGE procedure.
Figure 1.
Overview of differences between SAGE and SADE. (Left) Typical yields of different steps used for construction of a SAGE library. Total RNAs are extracted by using the acid guanidinium thiocyanate-phenol-chloroform method (8), and poly(A) RNAs are isolated by oligo(dT)-cellulose chromatography. Synthesis of cDNA is initiated with a biotinylated oligo(dT) primer. The cDNA then is cleaved with the SAGE anchoring enzyme (Sau3AI), and its 3′-end is isolated by binding to streptavidin beads. After recovery and ligation of cDNA tags to form ditags, PCR generates the expected fragments (110 bp), together with shorter parasitic products. (Right) mRNAs are directly isolated from the tissue lysate by binding to oligo(dT) covalently bound to magnetic beads. Synthesis and cleavage of cDNA then are performed on beads. The high yield of the procedure makes it possible to generate sufficient ditags for predominant amplification of the expected product (see text for details).
The alternative method that we set up (Fig. 1 Right) is essentially a single-tube assay from obtaining of tissue lysate to cDNA tag recovery and is referred to as SADE. Poly(A) RNAs are isolated directly from the tissue lysate through binding to oligo(dT) covalently bound to magnetic beads, and the cDNA is synthesized immediately. Starting from 250 mg of tissue, 3.6 μg of cDNAs were obtained. The 4-fold increase of cDNA amount, as compared with the original method, suggests improved yield for mRNA recovery. Moreover, the cDNA fraction still present on beads after digestion with the anchoring enzyme (20%) exactly matches that predicted (see above). At this stage, a 400-fold difference is observed for SADE and SAGE yields. Finally, PCR amplification of SADE products generates the expected 110-bp fragment (linkers + ditag), clearly enriched as compared with shorter fragments corresponding to linkers alone.
The increased efficiency achieved with SADE for cDNA tag generation suggested that gene expression profiling could be performed on small tissue samples. This possibility was checked by preparing libraries from 250 and 0.5 mg of the same kidney sample and sequencing 1,200 tags in each library, corresponding to 764 and 820 different tags, respectively (data not shown). A significant correlation (r = 0.88) was obtained between the abundance of the same tags in the two libraries. The reliability of SADE also was checked by comparing the data obtained from 0.5 mg of kidney with 1,200 tags sequenced from a conventional SAGE kidney library. The number of tags differentially represented (P < 0.05 by Monte Carlo analysis) was similar to that obtained when comparing 1,200 tags of two SAGE libraries generated from the same kidney sample (n = 14 and 12, respectively). A systematic difference between SAGE and SADE concerns rRNA tags, which reach up to 1% of SAGE tags (Table 1) but were always absent from SADE libraries. This demonstrates that SADE is reliable both in terms of mRNA yields and purity.
Gene Expression Profiles in Isolated Nephron Segments.
To demonstrate the potential of SADE, libraries then were generated from microdissected nephron segments. We studied the MTAL, which is nearly homogeneous at the cell level, and the OMCD, which consists of three cell types (principal, α-, and β-intercalated cells in a 6:3:1 ratio) (17). Libraries were obtained from 150 mm of tubules (50,000 cells), and 7,500 tags were sequenced in each case.
In the MTAL library, the amount of mitochondrial tags was even higher than in the kidney library (data not shown). Thus, all nine most abundant tags (range: 5.8–0.6% of total tags) corresponded to transcripts encoded by the mitochondrial genome. Turning to tags for nuclear transcripts, Table 2 shows that 60% (20 of 34) of the most abundant tags were enriched significantly in the MTAL library. This supports the notion that a specific gene expression profile was obtained. Three tags correspond to uromodulin (Tamm–Horfall protein), which is synthesized specifically in the thick ascending limb of Henle's loop (18) and known as the major protein in normal urine. Alignment of mouse uromodulin cDNA and ESTs demonstrates that alternative polyadenylation signals and splice sites account for the generation of several transcripts from the same mRNA. The total amount of uromodulin tags (n = 89) predicts a very high mRNA expression level. On the other hand, several abundant tags correspond to genes that allow, either directly or indirectly, a high rate of NaCl reabsorption in the MTAL. The former class includes transporters [Na,K-ATPase subunits (n = 21–25), ClC-K1 chloride channel (n = 13), Na-K-2Cl transporter (n = 8)], and the latter enzymatic activities that are linked to ATP production (ATP synthase and cytochrome c oxidase subunits, creatine kinase B, adenine nucleotide translocase-2). This gene expression pattern agrees with the fact that MTAL cells display a very high rate of NaCl reabsorption and have especially high Na,K-ATPase activity (19). Five tags were not identified, because they either had no GenBank entry or corresponded to orphan ESTs.
Table 2.
Abundant tags for nuclear transcripts in MTAL: Comparison with whole-kidney data
| Tag | MTAL | Kidney | Databank entry |
|---|---|---|---|
| TTCATGGTTC | 38 | 2* | Uromodulin (Tamm–Horsfall) (L33406) |
| TAAGATGAGA | 38 | 4* | ESTs, similar to mouse uromodulin (L33406) |
| TTGATATTTG | 25 | 10* | ESTs, similar to Hu Na,K-ATPase α1-subunit (J05096) |
| GTTCTCACCC | 25 | 11* | ESTs |
| GAAACTCTCT | 24 | 1* | Creatine kinase B (M74149) |
| GTGACTGGGT | 23 | 35* | Cytochrome c oxidase subunit IV (X54691) |
| TTGTGTCAGT | 22 | 7* | Na,K-ATPase β1-subunit (X61433) |
| CTTAACTTCC | 21 | 23 | Na,K-ATPase γ-subunit (Q04646) |
| ACCGACCGCA | 21 | 8* | Integral membrane protein 2B1 (U76253) |
| GCTCATTGGA | 18 | 1* | ESTs |
| CGCAGTGGCA | 17 | 10 | ESTs, similar to human ubiquinol cytochrome c reductase core I (L16842) |
| TCTCCATATC | 17 | 4* | ESTs, similar to mouse ATP synthase β-subunit (AF0030559) |
| AAGAAATACA | 17 | 7* | Adenine nucleotide translocase-2 (U27316) |
| GCCTGGAGAA | 17 | 11 | ESTs, similar to mouse ribosomal protein L41 (U93862) |
| GCTTTCAGCA | 14 | 0* | ESTs, similar to human extracellular proteinase inhibitor homologue (X63187) |
| ACTCTGGAGT | 14 | 8 | Follistatin-like protein (L75822) |
| TTGTTTGCCA | 13 | 12 | ESTs, similar to mouse H+ ATP synthase subunit c (L19737) |
| CTTTCTCTAT | 13 | 1* | ESTs, similar to uromodulin (L33406) |
| AACCCACCAG | 13 | 0* | Kidney chloride channel CIC-K1 (AF124848) |
| AAATAAAGTT | 13 | 2* | Lactate dehydrogenase 2, B chain (X51905) |
| CTGGGTACTA | 12 | 4* | Lymphocyte differentiation antigen (M18184) |
| ACAAAGTTTG | 12 | 7 | ATP synthase α-subunit (L01062) |
| TTTGCCGGCA | 11 | 9 | Ubiquitin (X51703) |
| GCGGCGAGGT | 11 | 1* | No match |
| TGACGTGCCG | 11 | 32* | Glyceraldehyde-3-phosphate dehydrogenase (M32599) |
| TGCGTATGGC | 10 | 7 | Ribosomal protein L28 (X74856) |
| AGGAGCTGGC | 10 | 9 | H+ ATP synthase subunit c (L19737) |
| ATGCTAGTCT | 10 | 10 | Ribosomal protein S29 (L31609) |
| CCCACTGCAC | 10 | 9 | No match |
| GCATTTGCCA | 10 | 0* | No match |
| GTCGTTCTGG | 10 | 5 | Protein synthesis elongation factor 1α (M22432) |
| AACTGTGCAG | 10 | 2* | ESTs, similar to human ribosomal protein L17 (X52839) |
| AATGATGAGG | 10 | 16 | Heat shock protein 70 (M19141) |
| TTCCATCCCT | 10 | 9 | Cytochrome c oxidase subunit VIa (L06465) |
The table lists tags detected ≥10 times (mitochondrial tags were excluded from the report) in the MTAL library (7,438 tags sequenced). For comparison, data obtained at the whole-kidney level also are indicated and normalized to a similar number of tags. Asterisks indicate significant differences, assessed by Monte Carlo trial, between abundance in MTAL and kidney libraries. Additional data are available at http://www-dsv.cea.fr/thema/get/sade.html
In the OMCD library, mitochondrial tags were also present in high amounts, with seven of the eight most abundant tags corresponding to mitochondrial transcripts (data not shown). Among the most abundant tags for nuclear transcripts (Table 3), two match mRNAs encoding proteins essential for collecting duct functions, i.e., regulation of water reabsorption by vasopressin and of Na transport by aldosterone (17). Thus, the most abundant tag (n = 129) corresponds to the vasopressin-sensitive water channel (aquaporin-2). On the other hand, type 2 11β-hydroxysteroid dehydrogenase ensures aldosterone-dependent regulation of Na transport by transforming glucocorticoids into inactive compounds. Kidney-specific cadherin has been shown to be a basolateral membrane protein in a subpopulation of collecting duct cells (20). Tag for the band 3 anion exchanger, a marker for collecting duct α-intercalated cells, also was abundant (n = 8). However, the most interesting observation of our study may be that the number of unidentified abundant tags is twice higher in the OMCD than in the MTAL library. This high amount of uncharacterized tags is likely related to the paucity of each collecting duct cell type. The OMCD consists of three different cell types, and there is only one OMCD for four to six nephrons. Hence, screening of whole-kidney cDNA libraries requires very large-scale sequencing to explore transcripts specific for a subpopulation of collecting duct cells. By contrast, our microassay offers the opportunity to find new markers for such cell types.
Table 3.
Abundant tags for nuclear transcripts in OMCD: Comparison with whole-kidney data
| Tag | OMCD | Kidney | Databank entry |
|---|---|---|---|
| GTGGCAGTGG | 129 | 11* | ESTs, similar to rat aquaporin-2 (D13906)† |
| TGGCAGTGGG | 41 | 0* | No match |
| TTATAATTTG | 39 | 1* | ESTs |
| TGACTCCCTC | 22 | 19 | B2 repetitive sequence |
| ACCGACCGCA | 19 | 8* | Integral membrane protein 2B1 (U76253) |
| AAGTTTAAAT | 18 | 1* | Thymosin β-4 (X16053) |
| AGCAAGCAGG | 18 | 12 | β-Actin (X03672) |
| CAAAAAGCTA | 17 | 2* | ESTs, similar to rat ribosomal protein L11 (X62146) |
| GTTCTCACCC | 15 | 11 | ESTs |
| CAGAAGAAGT | 14 | 3* | Endogenous murine leukemia virus (M17326) |
| CAGAAGCAGT | 14 | 19 | Endogenous murine leukemia virus sequence (L08395) |
| TGACCAAGGC | 13 | 1* | 11β-hydroxysteroid dehydrogenase type 2 (X90647) |
| AAATAAAGTT | 13 | 2* | Lactate dehydrogenase 2, B chain (X51905) |
| CTGGTGTCCT | 13 | 2* | Endothelial monocyte-activating polypeptide I (U41341) |
| AGAAGCAGTG | 12 | 0* | No match |
| ACATTCCTTA | 12 | 0* | EST 693054 (AA253657)† |
| ACTCTGGAGT | 12 | 8 | Follistatin-like protein (L75822) |
| GCTTTCAGCA | 12 | 0* | ESTs, similar to human extracellular proteinase inhibitor homologue (X63187) |
| TTCCATCCCT | 12 | 9 | Cytochrome c oxidase subunit VIa (L06465) |
| TGATGCCCTC | 12 | 5* | B2 repetitive sequence |
| GGTGCCACCC | 11 | 6 | Insulinoma/ribosomal protein S15 gene (M33330) |
| GTGACTGGGT | 11 | 36* | Cytochrome c oxidase subunit IV (X54691) |
| CAAGCAGCCC | 11 | 4* | Kidney-specific cadherin (AF016271) |
| AACAACCCAA | 10 | 1* | ESTs |
| TACACACACA | 10 | 0* | ESTs |
| AGCCAGCAGA | 10 | 0* | No match |
| TCTCCATATC | 10 | 4 | ESTs, similar to mouse ATP synthase β-subunit (AF030559) |
| GCCTGGAGAA | 10 | 11 | ESTs, similar to mouse ribosomal protein L41 (U93862) |
| GCTCATTGGA | 10 | 1* | ESTs |
| TGCGGTGACT | 10 | 1* | Ribosomal protein L7 (X57961) or S24 (X60289) |
| AGGAATCCAG | 10 | 9 | TDD5 (U52073) |
| TGACGCCCTC | 10 | 12 | B2 repetitive sequence |
The table lists tags detected ≥10 times (mitochondrial tags were excluded from the report) in the OMCD library (7,563 tags sequenced). For comparison, data obtained at the whole-kidney level also are indicated and normalized to a similar number of tags. Additional data are available at http://www-dsv.cea.fr/thema/get/sade.html
Significant difference, as determined by Monte Carlo trial, between abundance in OMCD and kidney libraries.
† EST match confirmed by sequencing the corresponding cDNA IMAGE clone.
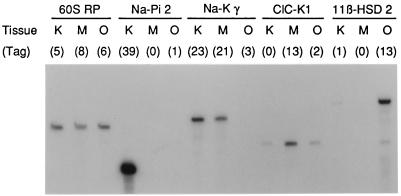
To check for representative tag sampling in the various libraries, a set of selected mRNAs was studied further by RT-PCR. As shown in Fig. 2, SAGE and RT-PCR data were consistent, confirming that reliable gene expression profiles were obtained.
Figure 2.
Comparative analysis of gene expression by SAGE and RT-PCR in the kidney (K), MTAL (M), and OMCD (O). RT-PCR (25 PCR cycles) was performed in the presence of [α-32P]dCTP by using RNA amounts corresponding to 102 cells. The DNA fragments were electrophoresed through a 2% agarose gel and detected by autoradiography. The expected size of the predominant PCR product was as follows: 60S ribosomal protein (60S RP), 392 bp; type II Na-Pi transporter (Na-Pi 2), 272 bp; Na,K-ATPase γ-subunit (Na-K γ), 413 bp; ClC-K1, 330 bp; type II 11β-hydroxysteroid dehydrogenase (11β-HSD 2), 461 bp. Abundance of SAGE tags (normalized to 7,500 total tags) in each library is shown above the gel. The RT-PCR experiment is representative of three that gave similar results.
Previous attempts for profiling gene expression in the kidney were carried out by large-scale sequencing of cDNA clones. Takenaka et al. (15) sequenced 1,000 clones of a mouse proximal tubule library. Because this library was generated by using cDNAs cleaved with MboI, an isoschizomer of the SAGE anchoring enzyme (Sau3AI) used in our study, it is of interest to compare our whole-kidney SAGE data with those obtained by Takenaka et al. (15). In the proximal tubule library, eight identified cDNAs were sampled ≥4 times. All of them also were detected by us (range: 0.03–1.7%), and two (kidney androgen-regulated protein and ferritin heavy chain) had exactly the same frequency as in our whole-kidney library. These data agree with the fact that the proximal tubule accounts for ≈60% of the kidney mass.
The microassay that we set up includes the following modifications to the SAGE procedure: (i) single-step method for mRNA extraction from tissue lysate; (ii) use of a RT lacking RNase-H activity; (iii) use of a different anchoring enzyme (see above); (iv) modification of procedure for blunt-ending cDNA tags; and (v) design of primers optimized for PCR amplification. All these modifications probably are not of similar importance for successful library generation from a small amount of cells. Datson et al. (21) were able to obtain a SAGE library from ≈105 hippocampal cells by changing only the mRNA extraction method. In this case, biotinylated oligo(dT) primers bound to streptavidin-coated tubes were used for mRNA purification, and a library allowing 1,800 tags to be sequenced was prepared. Our study demonstrates clear-cut different efficiencies for methods based on covalently bound oligo(dT) and biotin-streptavidin interaction. The two SADE libraries presented here were obtained from 50,000 cells, but we also were successful using 15,000 cells (data not shown).
In conclusion, this study describes a microassay for serial analysis of gene expression. Its validity was established by profiling mRNA levels in kidney tubules isolated by microdissection. One obvious application of this approach is to provide the expression level of known and unknown transcripts in well delineated tissue fragments. The current limit of our assay is a few tens of thousand cells.
Acknowledgments
We thank Drs. C. de Rouffignac and M. D. Legrand for their enthusiasm for all aspects of this work. We also thank Dr. G. Baverel for critical reading of the manuscript and Mrs. Y. Sallent for secretarial assistance.
Abbreviations
- MTAL
medullary thick ascending limb of Henle's loop
- OMCD
outer medullary collecting duct
- SAGE
serial analysis of gene expression
- SADE
SAGE adaptation for downsized extracts
- RT-PCR
reverse transcriptase–PCR
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 5.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Nat Biotech. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 6.Gress T M, Hoheisel J D, Lennon G G, Zehetner G, Lehrach H. Mamm Genome. 1992;3:609–619. doi: 10.1007/BF00352477. [DOI] [PubMed] [Google Scholar]
- 7.Chabardès D, Firsov D, Aarab L, Clabecq A, Bellanger A C, Siaume-Perez S, Elalouf J M. J Biol Chem. 1996;271:19264–19271. doi: 10.1074/jbc.271.32.19264. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1991. [Google Scholar]
- 11.Vandewalle A. Am J Physiol. 1986;246:F427–F436. doi: 10.1152/ajprenal.1984.246.4.F427. [DOI] [PubMed] [Google Scholar]
- 12.Hastie N D, Bishop J O. Cell. 1976;9:761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- 13.Maser R L, Magenheimer B S, Calvet J P. J Biol Chem. 1994;269:27066–27073. [PubMed] [Google Scholar]
- 14.Meseguer A, Watson C S, Catterall J F. Mol Endocrinol. 1989;3:962–967. doi: 10.1210/mend-3-6-962. [DOI] [PubMed] [Google Scholar]
- 15.Takenaka M, Imai E, Kaneko T, Ito T, Moriyama T, Yamauchi A, Hori M, Kawamoto S, Okubo K. Kidney Int. 1998;53:562–572. doi: 10.1046/j.1523-1755.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann C M, Wagner C A, Busch A E, Markovich D, Biber J, Lang F, Murer H. Pflügers Arch. 1995;430:830–836. doi: 10.1007/BF00386183. [DOI] [PubMed] [Google Scholar]
- 17.Morel F, Doucet A. In: The Kidney: Physiology and Pathophysiology. Seldin D W, Giebisch G, editors. New York: Raven; 1992. pp. 1049–1086. [Google Scholar]
- 18.Hession H, Decker J M, Sherblom A P, Kumar S, Yue C C, Mattaliano R J, Tizard R, Kawashima E, Schmeissner U, Heletky S, et al. Science. 1987;237:1479–1484. doi: 10.1126/science.3498215. [DOI] [PubMed] [Google Scholar]
- 19.Katz A I, Doucet A, Morel F. Am J Physiol. 1979;237:F114–F120. doi: 10.1152/ajprenal.1979.237.2.F114. [DOI] [PubMed] [Google Scholar]
- 20.Thomson R B, Igarashi P, Biemesderfer D, Kim R, Abu-Alfa A, Soleimani M, Aronson P S. J Biol Chem. 1995;270:17594–17601. doi: 10.1074/jbc.270.29.17594. [DOI] [PubMed] [Google Scholar]
- 21.Datson N A, van der Perk-de Jong J, van den Berg M P, de Kloet E R, Vreugdenhil E. Nucleic Acids Res. 1999;27:1300–1307. doi: 10.1093/nar/27.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]