Abstract
The 26S proteasome is a multisubunit enzyme composed of a cylindrical catalytic core (20S) and a regulatory particle (19S) that together perform the essential degradation of cellular proteins tagged by ubiquitin. To date, however, substrate trajectory within the complex remains elusive. Here we describe a previously unknown functional unit within the 19S, comprising two subunits, Rpn1 and Rpn2. These toroids physically link the site of substrate recruitment with the site of proteolysis. Rpn2 interfaces with the 20S, whereas Rpn1 sits atop Rpn2, serving as a docking site for a substrate-recruitment factor. The 19S ATPases encircle the Rpn1-Rpn2 stack, covering the remainder of the 20S surface. Both Rpn1-Rpn2 and the ATPases are required for substrate translocation and gating of the proteolytic channel. Similar pairing of units is found in unfoldases and nuclear transporters, exposing common features of these protein nanomachines.
Organisms from all domains of life contain ATP-dependent proteases that enforce both protein quality control and biological regulation by removing their targets in a timely manner1–4. The proteasome is the major such regulatory protease in eukaryotic cells, whereas several simpler analogs exists in archaea, bacteria and certain organelles. The uniqueness of the eukaryotic 26S proteasome regards its selectivity toward proteins covalently tagged with ubiquitin as a degradation signal. The 26S proteasome comprises two main components: the proteolytic 20S core particle (CP) and the multisubunit ATPase-containing 19S regulatory particle (RP)5–7. The 20S CP is a barrel-shaped stack of four heptameric rings organized in α7-β7-β7-α7 fashion, formed from seven similar yet distinguishable α and β subunits. In latent 20S CP, the channels through either of the two outer α-rings are primarily closed, hindering access of substrates to the internal proteolytic chamber enclosed by the two inner β-rings8–12. Various activators attach to the surface of the α-ring and facilitate substrate entry by forcing open the channel into the lumen of the 20S CP7,13,14. However, to degrade most globular or ubiquitinated substrates, appending of the 19S RP to the outer surfaces of the α-rings is required6,10,13,15,16.
The 19S RP binds ubiquitinated substrates and subsequently deubiquitinates, unfolds and translocates them through the opened entry pore into the catalytic chamber of the 20S CP where they are degraded. Whereas these functions are well recognized, little is known about the fine structure of the 19S RP or the delegation of tasks between its subunits. The 19S RP can be separated into two stable subcomplexes: lid and base5,6,15. The base is the most proximal of the two to the catalytic core, and as such is thought to carry out the final regulatory steps in the substrate trajectory toward proteolysis within the 20S chamber. Two large leucine-rich proteins, Rpn1 (also know as S2) and Rpn2 (also known as S1), together with six ATPases (Rpt1–6) are the essential components of the base; however, their spatial arrangement and the division of labor between them remain poorly defined. In the current view, largely based on archaeal and bacterial analogs, the six Rpt ATPases are thought to attach to the exposed surface of the α-ring, where they help to gate the channel and translocate substrates into the catalytic core1–4,12,15,17,18. How this arrangement accommodates the two largest proteasomal subunits within the base, Rpn1 and Rpn2, and what role it assigns either of these approximately 100 kDa proteins remain an enigma. A bioinformatic model predicting that Rpn1 and Rpn2 fold into α-helical solenoids with toroidal shape19 is particularly intriguing, as all non-ATPase proteasome activators, such as 11S Reg (also known as PA28), PA26 and PA200 (also known as Blm10) have a repetitive α-helical structure20 that associates directly with the 20S surface4,7,12,13,21,22. Inspired by these examples, we set out to experimentally determine the structure of Rpn1 and Rpn2, assess their interaction with the 20S CP and define their role within the base using proteasomes purified from Saccharomyces cerevisiae as a model.
RESULTS
Rpn1 and Rpn2 toroids interact with the 20S
Purified native Rpn1 and Rpn2 proteins were imaged by atomic force microscopy (AFM) in solution (Fig. 1a). Nonaveraged images of single particles show both proteins to be toroids (compact solenoids of reduced pitch) with a well-resolved indentation at their center (Fig. 1a). Statistical analysis of particle volume or height shows that each sample consists of a single population of particles 2.5 ± 0.3 nm (Rpn2) or 4.0 ± 0.9 nm (Rpn1) high and ~5 nm in diameter (Fig. 1b and Supplementary Fig. 1 online). The homogeneity of these samples was confirmed by analytical ultracentrifugation (AUC). Fitting the sedimentation data indicated that all soluble particles in either sample are monomers (experimental molecular weight of 109,524 ± 178 Da versus a sequence-calculated weight of 109,429 Da for Rpn1, and 104129 ± 129 Da versus 104,234 Da for Rpn2; Fig. 1c). A donut is an unusual and striking structure for a monomeric protein, although it is elegantly compatible with the axial profile of the 20S barrel and the base of which Rpn1 and Rpn2 are a part.
Figure 1.
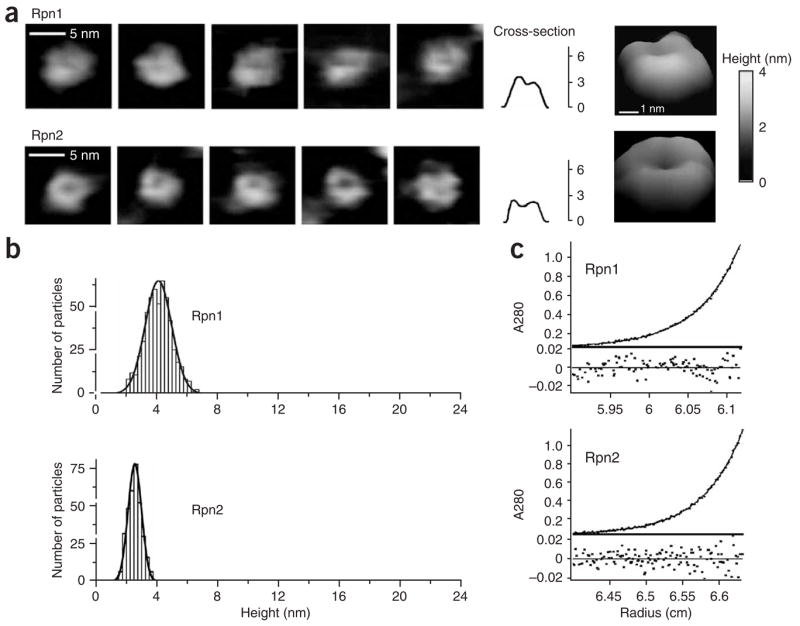
Toroidal shape of Rpn1 and Rpn2 monomers. (a) Nonaveraged AFM images of native Rpn1 and Rpn2 particles in solution. Horizontal sections are provided for the leftmost complexes, followed by a representative tilted enlargement, with a height bar (right). Multiparticle fields are shown in Supplementary Figure 3. (b) Statistical analysis of Rpn1 and Rpn2 based on height distribution shows each sample to contain a single population with an average height of 4.0 ± 0.9 nm and 2.5 ± 0.3 nm, respectively (see an example of the procedure in Supplementary Fig. 4). (c) Analytical ultracentrifugation sedimentation-equilibrium profile of Rpn1 (1.74 μM) or Rpn2 (1.95 μM) on a Beckman XL-A with an An50 Ti rotor at 16,000 r.p.m. Fitting the data to an ideal single-species model (Supplementary Methods) gives an experimental molecular weight of 109,524 ± 178 Da for Rpn1 and 104,129 ± 129 Da for Rpn2, corresponding within experimental error to the sequence-calculated monomeric weight (109,429 Da and 104,234 Da, respectively). Below, the residuals of the fit as a function of radial position.
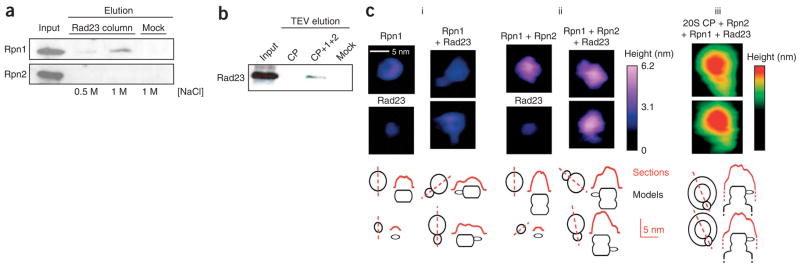
To establish the proximity of Rpn1 and Rpn2 to the proteasomal core, we assayed for a direct association by cross-linking and gel-shift assays. Either protein was cross-linked in the presence of 20S CP, and the resulting mixture was resolved by gel electrophoresis. We detected no changes in the migration pattern of Rpn1 upon cross-linking (Fig. 2a, lane 3); however, a new broad band seemed to trail roughly 25–35 kDa above the migration pattern of Rpn2 only when cross-linked in the presence of 20S CP (Fig. 2a, lane 7). This addition corresponds to the molecular weight of individual CP subunits, all of which are in the 25–35 kDa range15. Tandem mass spectrometry (MS/MS) analysis of this cross-linked protein band revealed the presence of five α subunits alongside Rpn2 (Fig. 2a, highlighted in red, and Supplementary Fig. 2 online). In comparison, similar MS/MS analysis of the corresponding gel region above Rpn1 yielded nothing but Rpn1-derived peptides (Fig. 2a, lane 3, and Supplementary Fig. 2). Likewise, we did not detect cross-linked subunits of 20S CP in the corresponding gel slice of 20S CP alone (Fig. 2a, lanes 4 and 8, and Supplementary Fig. 2) indicating that migration of individual α subunits in the 100–130 kDa region is a direct consequence of interaction with Rpn2. Evidently, Rpn2 can attach directly to the 20S CP without the need for any other 19S component, showing a unique and specific affinity for the α-ring surface.
Figure 2.
Rpn1 and Rpn2 assemble as a unit onto the α-ring of the 20S CP. (a) Cross-linking to 20S CP in a gel-shift assay. Rpn1 or Rpn2 were mixed with 20S CP, cross-linked with glutaraldehyde (GA) and resolved by 8% SDS-PAGE. The shift in migration of Rpn2 (lane 7) is due to cross-linking to individual CP subunits. CP subunits are 25–35 kDa, and thus they do not appear in the gel region (lanes 4 and 8). Proteins identified by MS/MS in the bracketed region of lane 7 are red in the complementary cartoon (details in Supplementary Fig. 2). No similar interaction was detected for Rpn1 with CP (lane 3). (b) Interaction of Rpn1 and Rpn2. Cross-linked products of Rpn1 and Rpn2 mixture were resolved by 6% SDS-PAGE (left, lane 3). Rpn1 associates with immobilized 20S only in the presence of Rpn2 (right; one-seventh of starting material is shown for reference).
All known ring-like non-ATPase activator complexes that attach to the 20S surface (for example, PA26, PA28/11S reg, PA200/Blm10) are roughly twice the molecular mass of Rpn2. In this context, given that we could not detect any interaction of Rpn1 with the 20S (Fig. 2a), we investigated the possibility that Rpn1 and Rpn2 interact with the 20S core as a unit. Indeed, Rpn1 attached to immobilized 20S CP only in the presence of Rpn2 (Fig. 2b, right), pointing to a direct and specific association of Rpn1-Rpn2 as a unit within the proteasome complex. Additionally, direct interaction of Rpn1 with Rpn2 was evident by cross-linking (Fig. 2b, left). Thus, the proteasome emerges as a layered structure, with Rpn1 placed distally to Rpn2, as the latter comes into direct contact with the outer α-ring of the 20S core.
A concentric stack of rings
A stratified arrangement in which each layer is made up of discrete subunits lends itself to single-particle analysis to determine the relative location of each subunit. As a foundation, we plated purified 20S core particles in liquid on a mica surface for analysis by AFM. Most 20S CP molecules in our sample were oriented as upright barrels, conveniently exposing an α-ring to the AFM probe, yielding homogenous fields of particles 15 ± 0.3 nm high and ~12 nm in diameter (Fig. 3a,b and Supplementary Figs. 3–5 online), consistent with previous AFM studies11,23,24. Typically of latent 20S CP10,11, most particles were imaged in the closed state (Fig. 3a, i; the first four particles from left to right). Having characterized the structural properties of Rpn1, Rpn2 and 20S CP samples, we set out to monitor changes to the population distribution and the architecture of individual particles upon mixing these components. When adding Rpn1 to a 20S CP sample, only the discrete populations corresponding to the unassociated species of Rpn1 and 20S were apparent (Fig. 3b), and no structural changes to individual particles were discernible (Fig. 3a, ii). Thus, the two do not form a stable complex in solution as deduced also from Figure 2a. In contrast, a new population of particles, 2.5 nm higher than the 20S barrels alone, was observed when Rpn2 was added to the 20S sample (Fig. 3b, iii). Individual images show that these extensions protrude from the center of the α-ring and are similar in dimension to those of a single Rpn2 molecule (Fig. 3a,b, iii). The population of extended 20S particles was even more pronounced when both Rpn1 and Rpn2 were added to 20S samples (Fig. 3a,b, iv). Core particles with a distinct chimney 5.4 nm high by 5 nm wide were easily recognized as a separate species alongside 20S and excess Rpn1 and Rpn2 (Fig. 4 and Supplementary Fig. 3). Once again, the dimensions of this protrusion are similar to those of the Rpn1-Rpn2 heterodimer that resulted upon mixing the two proteins in absence of 20S CP (Fig. 3a,b, v). These results confirm that Rpn1 and Rpn2 interact as a unit with the 20S CP, their internal 2 nm cavities presumably flush with the similarly sized entry pore through the α-ring9.
Figure 3.
Rpn1-Rpn2 stack extends the 20S proteolytic channel. (a) AFM images of: 20S CP (i), 20S–Rpn1 (ii), 20S–Rpn2 (iii) and 20S–Rpn2-Rpn1 (iv) and a separate mixture of Rpn1-Rpn2 (v). Dimensions of all particles are summarized in Supplementary Figure 1. Horizontal sections are provided for the leftmost complexes followed by a representative tilted enlargement, with a height bar (right). Multiparticle fields are shown in Supplementary Figure 3. (b) Statistical analysis of all samples based on height distribution identifies the species present in each mixture. A new population of extended 20S CP is found only in the presence of Rpn2 (iii) or Rpn1-Rpn2 (iv). An excess of starting reagents was always present is these mixtures.
Figure 4.

Raw AFM topograph of a 20S + Rpn2 + Rpn1 mixture in solution. An example of an AFM field (600 × 600 nm) of a 20S + Rpn2 + Rpn1 mixture in solution was collected in the height mode showing randomly oriented particles. The height of each pixel is represented by a color palette, according to the adjacent height bar. A tilted view (side plot) of 200 × 200 nm fragment of the same field is shown (right). Visual inspection of raw fields, using the height scale as a guideline, allows us to distinguish easily between the different complexes. The highest complexes (22 nm), representing 20S–Rpn2-Rpn1, are marked with a red arrow; assembly intermediates representing 20S–Rpn2 are marked with yellow arrows and free 20S CP is marked with green arrows. The low and narrow particles, labeled *, represent an excess of Rpn1 and Rpn2.
The novel alignment of Rpn1-Rpn2 relative to the 20S CP pore encouraged us to test whether they regulate traffic through the gated channel into the central cavity. Indeed, Rpn1-Rpn2 enhanced peptidase rates of free 20S CP with a half-maximum activation observed at a ratio of 1.5:1 (Fig. 5a). The maximal activation attained was four-fold, less than the roughly ten-fold activation reported for the base or 19S RP9,10,15,16,25. As the only known mode of proteasome activation is exerted via channel opening, we tested whether Rpn1-Rpn2 can stimulate constitutively open 20S CP (α3α7ΔN). Predictably, we observed no such effect on the peptide hydrolysis rate (Fig. 5b). In contrast, Rpn1-Rpn2 did not influence proteolysis rates of casein (Fig. 5c), a protein that is rapidly proteolyzed by the open-channel 20S CP but only minimally by the latent form10. We conclude that binding of Rpn1-Rpn2 to the 20S CP partially regulates channel gating by facilitating the entry of small substrates, although this unit is apparently insufficient on its own to enhance general proteolysis. For full proteolytic activation, the Rpt ATPases are required10,16,17,26.
Figure 5.
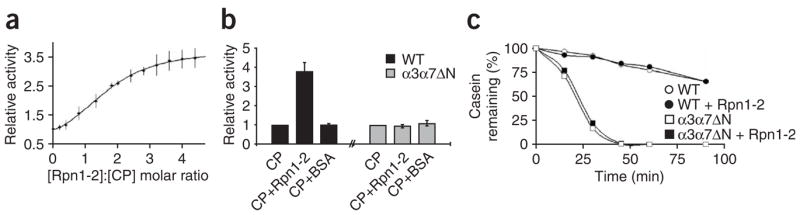
Rpn1-Rpn2 unit (labeled Rpn1-2) influences traffic through the proteolytic channel. (a) 20S CP peptidase activity increases with increasing Rpn1-Rpn2 concentration. Half-maximal activation is measured at a 1.5:1 molar ratio of Rpn1-Rpn2 to CP. (b) Rpn1-Rpn2 does not influence previously open-channel 20S CP. Peptidase activity of purified wild-type or open-channel (α3α7ΔN) CP were measured and compared upon addition of a three-fold molar excess of Rpn1-Rpn2 or BSA. (c) Degradation rates of casein by equal amounts of 20S CP purified from wild type or α3α7ΔN, alone or in the presence of Rpn1-Rpn2. Error bars represent s.d. of three independent experiments.
Toward a structure of the 19S regulatory particle
To verify such a startling arrangement, placing Rpn1-Rpn2 at the center of the 20S α-ring, we isolated intact 26S proteasome holoenzymes and dissected them along a biochemical gradient with the expectation that peripheral subunits would detach first. Indeed, layer after layer of peripheral subunits was shed—first the lid, then Rpn10 and the Rpt ATPases, and finally the Rpn1-Rpn2 stack, unearthing the naked 20S CP (Fig. 6a). Each resulting subcomplex concentrates into separate fractions along the gradient: 26S holoenzyme (fraction 13), Base-CP (fraction 14), Rpn1-Rpn2–CP (fraction 16) and latent 20S CP (fraction 17). This pattern was corroborated by immunoblotting for proteasome subunits (Fig. 6b). The composition of the previously unseen Rpn1-Rpn2-CP species was further confirmed by MS/MS analysis (Fig. 6c). AFM images support the conclusion that comigrating subunits are complexed into distinct proteasome subspecies (Fig. 6d, above). The distribution of these proteasome-breakdown species in each fraction can be identified and quantified (Supplementary Fig. 6 online) on the basis of the dimensions and architecture obtained for isolated complexes of defined composition (Figs. 1 and 3). It is noteworthy that images of stripped-down particles found in fractions 15 and 16 are experimentally indistinguishable from the newly identified reconstituted complexes (Fig. 6d, below). By finding conclusive proof that Rpn1 and Rpn2 are indeed found naturally in a stable complex with 20S CP, we could proceed to determine the relationship of the Rpt ATPases with this previously unknown complex.
Figure 6.

Two functional units within the base. (a,b) Isolated 26S proteasome holoenzymes were fractionated along a phosphate gradient. Eluted fractions were resolved by SDS-PAGE and stained for protein (a) or immunoblotted for proteasome subunits (b). Proteasomes of specific subunit composition concentrate into separate fractions. Similar fractionation was observed for the open-channel mutant (Supplementary Fig. 7 online). (c) The isolated complex from fraction 16 was subjected to MS/MS analysis. Two protein bands migrating slightly higher than 100 kDa were identified by MS/MS as Rpn1 and Rpn2. No peptides derived from any other Rpn or Rpt subunit were detected in the sample. (d) AFM images of representative particles identified in each fraction (fr.): 14, proteasomes sans lid (Base–CP complexes); 15–16, the newly identified 20S-extended species (Rpn1-Rpn2–CP, and Rpn2–CP); 17, stripped core particle (20S CP). Above, top views and horizontal sections. Below, the predominant isolated species are indistinguishable from reconstituted complexes of defined composition. (e) Peptidase activity of proteasome subspecies from each fraction (taken from a,b) with and without 0.02% (w/v) SDS. The change relative to basal levels is shown as a bar graph for each fraction. (f) The rate of casein proteolysis by proteasomes from each fraction was measured. Error bars represent s.d. of three independent experiments.
The current study mandates reassessment of subunit layout within the 19S RP. The dome-like base15 and the newly identified Rpn2-Rpn1 stack (Fig. 3a) share a similar height, about 6 nm, but they differ in their diameters (12 nm and 5 nm, respectively; Supplementary Fig. 1). The difference in subunit composition between the Rpn1-Rpn2 extension and a complete base is the lack of the Rpt ATPases. Thus, localizing Rpn1 and Rpn2 to the central pillar of the base delegates the Rpt ATPases—unexpectedly—to a peripheral location. AFM analysis of fraction 14, which contains all the components of the base including the ATPases (Fig. 6a,b), identifies 20S complexes capped with a 6 × 13 nm dome (Fig. 6d, above, and Supplementary Fig. 6), indistinguishable from the dimensions of the previously characterized base-CP15 (Fig. 6d, below). Comparison of the dimensions of the Rpn1-Rpn2 unit with those of the base (Fig. 6d) shows that the Rpt ATPases are positioned on the same stratum as Rpn1-Rpn2, but in a peripheral position. Moreover, preliminary analysis of molecules imaged in fractions 14–15 revealed the presence of ring-shaped particles with dimensions corresponding to those expected for such a ring of ATPases (detailed characterization of these rings will be provided in a future study). A requirement of intact Rpn2 both for proteasome peptidase activity and for binding of regulatory particles to the 20S core particle is also supported by genetic evidence obtained with rpn2 mutants in vivo27,28. Thus, the dome-like base of the 19S RP emerges as a ring of ATPases modeled around an inner Rpn1-Rpn2 stack; both units interact with the surface of the 20S CP α-ring. Sheltering a cavity over this base, the lid completes the well-recognized image of the 19S RP15,29–31.
Two functional units within the base
An interesting feature of the proteasome as it emerges from this study is that structural layers correspond with distinct levels of proteolytic activation. Stripped 20S CP found in fraction 17 was latent; its peptidase activity could be enhanced by an order of magnitude with SDS (Fig. 6e). In contrast, peptidase activity of complexes present in fractions 15 and 16 could be SDS-activated merely three-fold, pointing to a degree of preactivation. This is in agreement with the high abundance of Rpn1-Rpn2–extended 20S species found in these fractions (Supplementary Fig. 6). Consistently, 26S holoenzymes present in fraction 13 were, as expected, fully activated, and therefore did not lend themselves to additional activation (Fig. 6e). The necessity for ATPases becomes clear when measuring protein degradation by the same complexes. The Rpn1-Rpn2 extension to the proteolytic channel did not accelerate proteolysis of casein (Fig. 6f; fractions 15 and 16), as observed for the reconstituted complexes (Fig. 5c). Only proteasome species retaining the peripheral layer of Rpt subunits degraded casein efficiently (Fig. 6f; fraction 13 and 14). The additive contribution of each layer to the function of the 19S RP finally explains how channel opening is an event that does not strictly call for ATP hydrolysis10–12,16,18,21,32, whereas unfolding and rapid proteolysis of proteins requires functional Rpt ATPases15,16,26,33–35.
Substrate recruitment
Another key property of the base (generally attributed to Rpn1) is its ability to bind ubiquitin-like (Ubl) domains present in cargo-delivery proteins such as Rad23 (refs. 36–39). Despite their sequence and structural similarities, Rpn1 and Rpn2 were clearly distinguishable in their ability to bind Rad23 (Fig. 7a). Rpn1 was sufficient to recruit Rad23 to the proteasome when stacked onto Rpn2, even in the absence of the remaining 19S RP subunits (Fig. 7b). Attachment of Rad23 to Rpn1 was easily visualized by AFM and distinguishable from isolated components (Fig. 7c, i). Reflecting the selectivity of Rad23 for Rpn1, only a single Rad23 was found on each stack (Fig. 7c, ii). Therefore, the ‘rump base’ comprising Rpn1 and Rpn2 can be used to orient substrate binding within the proteasome. We found that the Rad23 protrusion was situated at the circumference of Rpn1, close to the central axis of the base (Fig. 7c, iii). The rational behind the dissimilar binding properties of Rpn1 and Rpn2 for Ubl-domains is now made clear: Rpn2 docks onto the center of the α-ring, exposing the distal Rpn1 for the recruitment of substrates or auxiliary factors (Fig. 8a,b). Although the results presented herein indicate that Rpn1 sits atop Rpn2, we do not preclude that, within the 19S RP, either protein may have regions that extend outward beyond their ring-like frame, exposing some regions of Rpn2 for the binding of additional subunits.
Figure 7.
Rpn1-Rpn2 recruits ubiquitin-like domains to the epicenter of the base. (a) Rpn1 or Rpn2 were pulled down by immobilized Rad23 on Sepharose beads or empty beads as a negative control (Mock). (b) The Rpn1-Rpn2 unit is sufficient to recruit Rad23 to the proteasome. Protein A–immobilized 20S CP, with or without the Rpn1-Rpn2 extension, was preincubated with Rad23, washed and TEV protease–eluted for immunoblotting for associated proteins. IgG beads were used as negative control (Mock). (c) The site of Rad23 attachment at the base. (i) Upon mixing of Rpn1 with Rad23, 24% ± 7% of Rpn1 particles were decorated with a single molecule of Rad23 and easily distinguishable from free Rpn1 or Rad23 (left). No interactions of Rpn2 with Rad23 were observed (not shown). (ii) A single Rad23 molecule protrudes outward from the Rpn1-Rpn2 stack. Depending on the orientation of Rpn1-Rpn2 on the mica surface, Rad23 protrudes either from the proximal (below) or distal ring (above). (iii) In presence of Rad23, Rpn1-Rpn2–CP complex is decorated with a single Rad23 molecule (14 out of 19 particles in 5 fields). Horizontal sections map the site of Rad23 attachment to the top ring (Rpn1) only.
Figure 8.
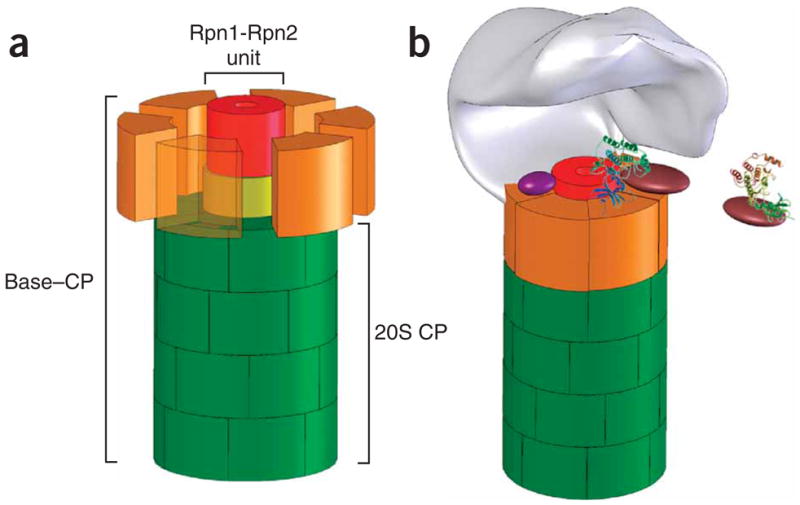
A model for how Rpn1-Rpn2 mediates substrate reception and translocation through the proteasome. (a) The central unit within the base. In a schematic structural model of the base (roughly drawn to scale), Rpn2 (yellow) attaches to the center of the 20S CP surface (green). Rpn1 (red) sits atop Rpn2. Rpt ATPases (orange; dispersed, with one transparent for clarity) attach to the 20S surface, wrapping around Rpn1-Rpn2 to complete the dome-like base. The model is a simplification based on the most structured regions of these proteins that are picked up in our assays. We do not exclude that both Rpn1 and Rpn2 may have extended flexible regions for interaction with auxiliary factors that protrude beyond the ring-like frame. (b) The 19S RP revisited. A lid subcomplex (schematically shown in gray) entombs a cavity within the 19S by attaching asymmetrically to the base15,29–31. The resulting interface between the lid and base accommodates interchanging proteasome auxiliary factors and substrate delivery proteins (for example, purple and brown), many of which are nominally part of the base. The protruding rim of the Rpn1-Rpn2 stalk may serve as a docking site for substrate-recruitment factors, relaying substrates to the ATPase domain for unfolding and translocation through the extended proteolytic channel. Both the ATPase domain and the newly identified central unit partake in translocation, although the precise trajectory is yet to be charted.
DISCUSSION
Mapping substrate recruitment to the surface of the base assigns the initial preparation of substrates to the cavernous interface between the base and the lid (Fig. 8b). In this study, we used Rad23 as a paradigm for Ubl domains, but in fact Rpn1-Rpn2 may generally engage ubiquitin domain–containing proteins, many of which are cargo-delivery proteins or deubiquitinating enzymes (DUBs). It may not be a coincidence that a designated platform within the regulatory complex (Rpn1-Rpn2) occurred in eukaryotes simultaneously with the advent of ubiquitin as a targeting module. Concentrating several auxiliary factors, within the same space and in the vicinity of the Rpt ATPases, helps to synchronize anchoring, editing of the polyubiquitin tag and substrate unfolding (Fig. 8b). A potential substrate-reception platform at the epicenter of the base suggests that substrate unraveling also occurs in the same plane, on the surface of the Rpt ATPases (Fig. 8b). Indeed, unfolding by the regulatory complexes of other ATP-dependent proteases, disaggregating or unfolding machines such as Hsp100 (ClpB), ClpA/X, HslU and PAN is often carried out by designated flexible α-helical domains35,40–45. This common feature of unfoldases is in stark contrast to the mechanism of chaperonins, which engulf their substrates and refold them within an inner chamber46,47.
Physical separation between the initial binding site and the entry pore into the 20S CP necessitates a mechanism to mediate between the two functional domains. Rpn1-Rpn2 fulfils this role by coordinating substrate recruitment and translocation into the proteolytic chamber, all within one structural unit. As documented previously16,17,26, and as apparent from Figures 5 and 6, the eukaryotic Rpt ATPases are largely responsible for substrate translocation and channel gating. We propose that they share these responsibilities with Rpn1-Rpn2: the ATPases impose nucleotide-dependent conformational changes onto the enveloped Rpn1-Rpn2, synchronizing the functions of the two units within the base. The asymmetrical solenoid structure of the Rpn1-Rpn2 extension may funnel substrates toward the gate at the entrance to the 20S CP. A similar arrangement of an outer ATPase ring engulfing a flexible inner non-ATPase domain is used by ClpX to glide substrates toward the ClpP protease. This bacterial ATP-dependent protease contains six α-helical ZBD domains that latch onto substrate at the epicenter of the ATPase ring. In response to nucleotide binding and hydrolysis, the domains rearrange to form a ‘molecular stent’ right through the 5-nm internal cavity within ClpX42,43. This dramatic conformational change threads substrates from the site of binding and unfolding on one surface of this rudimentary regulatory particle to the proteolytic chamber that attaches to the other. With a similar order of subassemblies in the nuclear transport machinery, HEAT repeat β-karyopherins also undergo conformational changes driven by their partner, RanGTPase48,49. The inherent flexibility of repetitive α-helical structures may be a common tool to propagate conformational changes from nucleotide binding and hydrolysis in one domain to reversible protein-protein interactions in another.
METHODS
Cross-linking
We used freshly prepared glutaraldehyde (GA) in buffer (150 mM NaCl, HEPES 50 mM, pH 7.5) to cross-link isolated samples of Rpn1, Rpn2, 20S CP or appropriate mixtures. Reactions between CP and Rpn1-Rpn2 were performed at a ratio of 1:2.5. Cross-linking of Rpn1, Rpn2 or a 1:1 ratio of Rpn1:Rpn2 were performed at a total protein concentration of 220 nM. Proteins were preincubated for 15 min, at which time GA was added to a final concentration of 10 mM. Reactions were quenched with excess ethanolamine 10 min later. We then separated the samples by SDS-PAGE (8%, Fig. 2a; 6%, Fig. 2b) and immunoblotted with anti–RGS-His6. Cross-linked products were excised, trypsin digested and subjected to MS/MS analysis for protein identification (Smoler Protein Center, Technion).
Purification of CP–Rpn2-Rpn1 subcomplex from whole-cell extract
We purified 26S proteasome holoenzymes from wild type (BY4741) as published25. The Rpn1-Rpn2–CP subcomplex was purified by depleting these proteasome samples of ATP and separating them on an 8-ml ceramic hydroxyapatite column (BioRad) along a 10–400 mM potassium phosphate gradient at pH 7.2. As a control, we found that, when applied independently, Rpn1 and Rpn2 elute earlier in the gradient and do not overlap with the elution profile of any proteasomal subspecies.
Atomic force microscopy imaging
We performed AFM imaging in a tapping mode in liquid using a NanoScope IIIa (Veeco) microscope, as previously described11,50. Protein samples were adsorbed on freshly cleaved mica in the appropriate buffer and, after thermal equilibration, were scanned in the height mode (only the trace direction shown). To allow for automatic height measurements, the images of whole fields were subjected to the line-wise plane correction using the LMS third-order fit function, followed by height analysis within the grain analysis function (SPIP; Supplementary Methods online). We did not use any other image processing except occasional removal of scan-line marks (NanoScope IIIa software, v. 5.12).
Enzymatic properties of proteasome subcomplexes
To quantify the peptidase activity in each fraction, we incubated approximately equimolar samples from all column fractions for 10 min at 30 °C with 0.1 mM fluorogenic peptide Suc-LLVY-AMC. Aliquots of same samples were independently incubated for 10 min at 30 °C with 0.1 mM Suc-LLVY-AMC and 0.02% SDS. The percent activation was calculated by dividing the peptidase activity in the presence of SDS by that measured in its absence. Data shown is averaged from three independent purifications, with error bars representing s.d. in each case.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank D. Cassel, O. Kleifeld and T. Rosenzweig for comments and critically reading the manuscript. A. Kajava is acknowledged for advice. N. Reis provided technical assistance. We thank D. Fass for assisting with AUC runs and analysis. We thank Y. Matiuhin (Technion) for Rad23 constructs. This work was funded by grants from the Israel Academy of Science/Israel Science Foundation (ISF), The USA-Israel Binational Science Foundation (BSF) and the Malat Family Foundation (via the Technion VP for research) to M.H.G., the NIH R01 grant (M.G.), and the Enhancement Research Grant and San Antonio Cancer Institute (SACI) support for P.A.O. R.R. was partially supported by an anonymous scholarship award (via the Technion Graduate School).
Footnotes
AUTHOR CONTRIBUTIONS
R.R. performed biochemical experiments. M.G. and P.A.O. carried out AFM imaging. M.G. and M.H.G. designed and supervised experiments. All authors discussed the results and participated in writing the manuscript.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Maupin-Furlow JA, et al. Proteasomes from structure to function: perspectives from Archaea. Curr Top Dev Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- 2.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.Zwickl P, Baumeister W, Steven A. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]
- 5.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 8.Groll M, et al. Structure of 20S proteasome from yeast at a 2.4 resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 9.Groll M, et al. A gated channel into the core particle of the proteasome. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 10.Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 11.Osmulski PA, Gaczynska M. Nanoenzymology of the 20S proteasome: proteasomal actions are controlled by the allosteric transition. Biochemistry. 2002;41:7047–7053. doi: 10.1021/bi0159130. [DOI] [PubMed] [Google Scholar]
- 12.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9/Signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 16.Liu C-W. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler A, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 18.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Kajava AV. What curves α-solenoids? Evidence for an α-helical toroid structure of Rpn1 and Rpn2 proteins of the 26S proteasome. J Biol Chem. 2002;277:49791–49798. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 21.Ortega J, et al. The axial channel of the 20 S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGγ proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorn IT, Eschrich R, Seemuller E, Guckenberger R, Tampe R. High-resolution AFM-imaging and mechanistic analysis of the 20S proteasome. J Mol Biol. 1999;288:1027–1036. doi: 10.1006/jmbi.1999.2714. [DOI] [PubMed] [Google Scholar]
- 24.Thess A, et al. Specific orientation and two-dimensional crystallization of the proteasome at metal-chelating lipid interfaces. J Biol Chem. 2002;277:36321–36328. doi: 10.1074/jbc.M202145200. [DOI] [PubMed] [Google Scholar]
- 25.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the S. cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the β7 subunit and activator complexes stabilize nascent 20S proteasomes and promote their maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 28.Isono E, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickell S, et al. Structural analysis of the 26S proteasome by cryoelectron tomography. Biochem Biophys Res Commun. 2007;353:115–120. doi: 10.1016/j.bbrc.2006.11.141. [DOI] [PubMed] [Google Scholar]
- 30.Kurucz E, et al. Assembly of the Drosophila 26 S proteasome is accompanied by extensive subunit rearrangements. Biochem J. 2002;365:527–536. doi: 10.1042/BJ20011520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walz J, et al. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 32.Iwanczyk J, et al. Structure of the Blm10–20S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strickland E, Hakala K, Thomas PJ, DeMartino GN. Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26S proteasome. J Biol Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- 34.Braun BC, et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 35.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 36.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 37.Seeger M, et al. Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J Biol Chem. 2003;278:16791–16796. doi: 10.1074/jbc.M208281200. [DOI] [PubMed] [Google Scholar]
- 38.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 39.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Choi JM, Tsai FTF. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 42.Thibault G, Tsitrin Y, Davidson T, Gribun A, Houry WA. Large nucleotide-dependent movement of the N-terminal domain of the ClpX chaperone. EMBO J. 2006;25:3367–3376. doi: 10.1038/sj.emboj.7601223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zolkiewski M. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol. 2006;61:1094–1100. doi: 10.1111/j.1365-2958.2006.05309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 46.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 47.Horwich AL, Weber-Ban EU, Finley D. Chaperone rings in protein folding and degradation. Proc Natl Acad Sci USA. 1999;96:11033–11040. doi: 10.1073/pnas.96.20.11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Petosa C, et al. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 50.Osmulski PA, Gaczynska M. Atomic force microscopy of the proteasome. Methods Enzymol. 2005;398:414–425. doi: 10.1016/S0076-6879(05)98034-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.