Ras GTPases mediate signaling pathways in cell proliferation, development and apoptosis. They undergo isoprenylation at a C-terminal CaaX signal (a usually represents aliphatic and X any amino acid) followed by proteolysis of aaX and carboxymethylation. In the case of H-ras, a subsequent dual palmitoylation of cysteines adjacent to the site of farnesylation produces a mature anchor for plasma membrane targeting.[1,2] Atomistic informations, such as the structure of membrane-bound ras and the free energy of complex formation, are vital in research efforts geared towards designing ras isoform selective anti-cancer agents. The most common experimental techniques are not yet able to provide such atomistic data. Here we present computational results on the free energy profile for the transfer of the H-ras membrane anchor from water to a bilayer of DMPC lipids. We find that there is no significant barrier for insertion and that once a few carbon atoms of the ras lipid chains cross the membrane-water interface, the free energy displays a steeply downhill profile. Insertion into the hydrocarbon core of the ras lipids and the interfacial localization of the backbone together produce up to 30 kcal/mol gain in free energy. Additionally, using the recently reported computationally-derived structures of full-length H-ras in a DMPC bilayer,[3] we explain how a small free energy difference would enable modulation of H-ras membrane binding by the linker and the catalytic domain.
Molecular dynamics (MD) simulations of the H-ras anchor (residues 180–186) were carried out using the CHARMM27 force field[4] and the program NAMD.[5] Details of the simulation have been published elsewhere.[3] The potential of mean force (PMF) was computed using the adaptive-biasing-force (ABF) method.[6] The reaction coordinate was defined by the distance along the membrane normal between the geometric centers of the lower leaflet phosphorous atoms and the peptide backbone C and O atoms (see Supporting information).
The thermodynamic path depicted in scheme I was used to describe the insertion process. The main idea is that the hydrophobic effect[7] drives the spontaneous transfer of the highly non-polar peptide from a region of high dielectric (water, εs) to an intermediate one (bilayer-water interface, εi), and subsequently to the hydrophobic core (core, εnp). The first two processes could involve structural reorganization, such as stretching of the ras lipid tails and adjustment of the backbone, respectively.
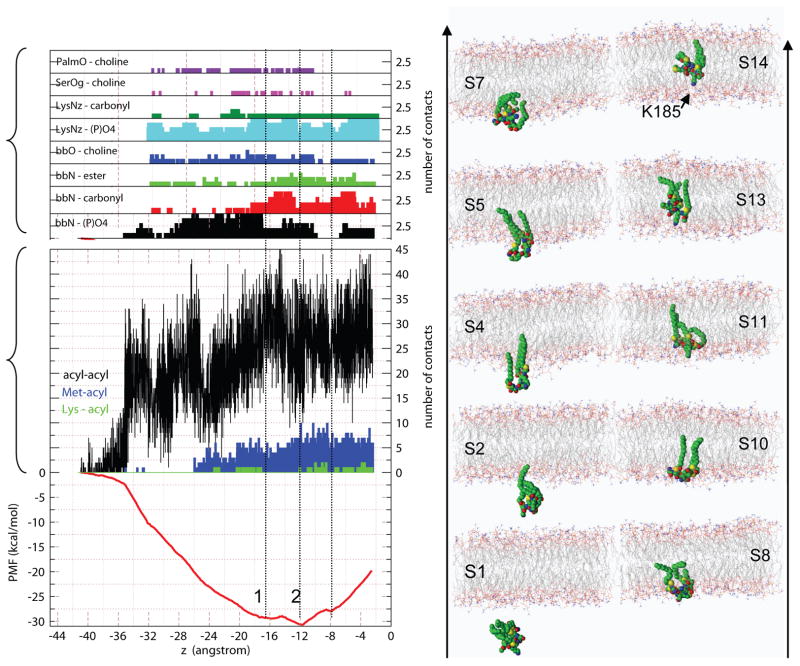
Fig. 1 shows the free energy profile (PMF). The initial flat region prior to peptide-membrane contact shows that there is no significant barrier between the aqueous phase and the water-membrane interface. This remarkable result suggests that insertion is spontaneous and non-specific, which may be compared with the spontaneous insertion of mono-S-acylated and -isoprenylated lipopeptides into lipid vesicles.[8] Furthermore, when ras was microinjected into a mammalian cell, it first binds to all cellular membranes non-specifically before it gets sorted in the Golgi.[9] In addition, the initial flat profile is expected from a chemical standpoint for a peptide located within about three layers of waters (~9 Å) from the membrane surface, which was the case in the simulations. If further away, an entropy-controlled barrier – arising from thermal fluctuations and the peptide’s reorganization so as to present its hydrophobic face to the bilayer – might be encountered. For example, it has been shown that antimicrobial peptides bind to the interface and subsequently penetrate the bilayer if their hydrophobic faces, and not their positively charged faces, were directed toward the interface.[10] Thus, the reorganization in water is not fully accounted for, although the agreement between the computed and experimental free energy change (ΔG, see below) suggests that its effect on thermodynamic stability is small.
Figure 1.
(left), ABF calculated PMF for the insertion of H-ras anchor into a DMPC bilayer plotted against the distance of C and O atoms’ center-of-geometry from the bilayer center (z, see Supporting Information). Also shown are the polar (top) and carbon-carbon (middle) peptide-DMPC contacts. bbO = carbonyl, bbN = amide, PalmO = palmitoyl carbonyls. (right), Snapshots illustrating peptide localizations in ten of the fourteen sections of independent ABF runs (see Supporting information). Sections S1 and S2 are associated with the initial flat region and section S11 with the start of rise in the PMF.
Formation of a few initial contacts between ras and DMPC acyl carbons triggers a steep descent of the PMF (Fig. 1, left). This critical number of contacts, 5–7, corresponds to a free energy gain of ~ −2.5 kcal/mol. Remarkably, the same number of contacts was previously estimated to be essential for a subsequent “spontaneous” insertion in MD time scales.[3, 11]
To underline that the computed free energy captures all relevant contributions, we compare it with two experimental observations. The heptane-water partition equilibrium free energy for a 16 carbon free fatty acid (FFA) was determined to be ~9.0 kcal/mol.[12] Shahinian and Silvius determined effective dissociation constants (Kdeff) for the association of lipopeptides with 90:10 egg yolk PC/POPE vesicles.[13] They measured, depending on the fluorophore and amino acid composition, a Kdeff of 0.196, 0.60 and 0.126 μM (or, using ΔG=− RT ln Kdeff, 9.51, 8.82 and 9.78 kcal/mol) for palmitoyl, and 12.5 and 18.5 μM (6.68 and 6.95 kcal/mol) for farnesyl. Note, however, that the hexadecyl chain used here may have stronger membrane affinity than farnesyl. The insertion free energy for a combination of two palmitoyl and one hexadecyl would be ~ −27.0 kcal/mol using the FFA data of Kampf et al. [12] and, ~ −25.6 kcal/mol using the mean values of Kdeff. [13] Both are remarkably close to our computed overall free energy of −29.5 to −30.6 kcal/mol. The small discrepancy can be attributed to the additional hydrogen bonding and vdW contributions. An example of the former is the interaction of the backbone with the DMPC carbonyls (see below); and the latter by the interaction of Met182 side chain with the tails (Fig. 1), which contributes ~ −1.24 kcal/mol.[11, 14].
During the insertion, other vdW interactions, including the snorkeling of Lys185 side chain, do also form until a free energy minimum (1, PMF = −29.5 kcal/mol) is reached at z = −15.9 Å. However, a narrower and deeper minimum (2, PMF = −30.6 kcal/mol) appears at z = − 11.7 Å. The 1.1 kcal/mol (~2 kBT) difference between the two minima (roughly equivalent to the energy of a single hydrogen bond) originates from small changes in the interaction of the backbone with the bilayer; the backbone amide interacts with the phosphates in minimum 1, but with the glycerol carbonyls in 2 (Fig. 1).
The trend of the profile reverses following the translocation of the backbone into the hydrocarbon core (Fig. 1), which has been estimated to cost ~5.6 kcal/mol.[11, 15] Note that in addition to the hydrogen bonds by the amide groups (Figs. 1&2), contacts by the peptide carbonyls suggest small contributions whose disappearances coincide with the rise of the PMF. The hydroxyl group of Ser183 interacts with the DMPC carbonyls (Fig. 2) while the charge on Lys185 is “solvated” by 2–6 phosphate oxygen atoms even after the backbone has lost its hydrogen bonds (Figs. 1&2). Thus, the polar and charged residues in the anchor maintain the peptide’s orientation.
Figure 2.

Hydrogen bonds between H-ras anchor and DMPC. The peptide is in stick and the DMPC lipids involved in hydrogen bonding are in ball-and-stick (the rest in gray lines). The orange dotted lines represent hydrogen bonds.
The large free energy gain by the insertion of the triply lipidated H-ras anchor implies that spontaneous dissociation is unlikely, as suggested before.[13] Contribution of the individual lipid modifications to affinity may be inferred from general microscopy results which showed that the cytosolic pool of H-ras G12V is much smaller than the doubly lipid modified N-ras G12V, or variants of H-ras G12V with either Palm181 or Palm184 removed.[16]
Two structural models have been computationally predicted – and experimentally supported – for the GDP- (H-ras-GDP) and GTP-bound (H-ras-GTP) full-length H-ras in a DMPC bilayer.[3] One of the main differences between the two structures was the insertion depth of the anchor. [3, 17] The average z-location of the anchor backbone was 12 and 16 Å in H-ras-GDP and H-ras-GTP, which almost exactly match the location of minima 1 and 2, respectively (Fig. 1). One consequence of this was that H-ras-GTP increased membrane thickness in its vicinity (the change in the average phosphorous atom location across the bilayer, DPP = 2.7 Å) while H-ras-GDP reduced it (DPP = −1.4 Å). Summarizing these data, Fig. 3 illustrates that ras can switch between the two modes of membrane binding with a cost or gain in free energy that can be compensated for by formation or destruction of as few as one hydrogen bond or a couple of vdW contacts. This fine balance of free energy and the associated conformational change, which is modulated by the linker and the catalytic domain, may enable ras to undergo alterations in membrane affinity,[16, 18] lateral segregation[19] and nanoclustering.[3, 17, 20]
Figure 3.

A cartoon illustrating two modes of membrane binding by ras and their connection with membrane structural deformation and insertion free energy. Note that membrane perturbations at the lower leaflet are damped by the catalytic domain.
The presented structural and energetic data, which were not accessible to common experimental techniques, provide new opportunities for the design of H-ras selective anti-cancer drugs such as ligands that inhibit the conformational change associated with the membrane binding of the activated protein. Note that changes in protein conformation can open previously uncharacterized binding sites; targeting these sites have led to useful drugs, such as HIV integrase inhibitor Raltegravir. Finally, the work lays the foundation for future computational and experimental investigations of the thermodynamics of membrane binding by lapidated proteins. We are currently investigating the contribution of individual lipid-modifications and entropy to membrane affinity.
Supplementary Material
Scheme I: A thermodynamic model for the insertion of the ras peptide into a DMPC bilayer.
Abbreviations
- PMF
potential of mean force
- DMPC
1,2-dimyristoyl-sn-Glycero-3-phosphocholine
- MD
molecular dynamics
Footnotes
A.A.G. acknowledges financial support from the Commission for the Promotion of Young Academics of the University of Zurich. We thank Prof. J. F. Hancock, Drs. M. Hanzal-Bayer, D. Abankwa, D. Hamelberg and I. Ivanov for discussions and critical reading of the manuscript, the San Diego Super Computer Center and the Center for Theoretical Biological Physics for computational resources. Additional support has been provided by the National Science Foundation, National Institutes of Health, Howard Hughes Medical Institute, National Biomedical Computation Resource, and Accelrys Inc.
References
- 1.Resh MD. Nat Chem Biol. 2006;2:584. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez L, Magee AI, Marshall CJ, Hancock JF. Embo J. 1989;8:1093. doi: 10.1002/j.1460-2075.1989.tb03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. J Med Chem. 2007;50:674. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 4.MacKerell AD, et al. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. (see Supporting Information) [DOI] [PubMed] [Google Scholar]
- 5.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Henin J, Chipot C. J Chem Phys. 2004;121:2904. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]; b) Rodriguez-Gomez D, Darve E, Pohorille A. J Chem Phys. 2004;120:3563. doi: 10.1063/1.1642607. [DOI] [PubMed] [Google Scholar]; c) Chipot C, Henin J. J Chem Phy. 2005:123. doi: 10.1063/1.2138694. [DOI] [PubMed] [Google Scholar]; e) Henin J, Pohorille A, Chipot C. J Am Chem Soc. 2005;127:8478. doi: 10.1021/ja050581y. [DOI] [PubMed] [Google Scholar]
- 7.Chandler D. Nature. 2005;437:640. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 8.Silvius JR, l'Heureux F. Biochemistry. 1994;33:3014. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 9.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. Science. 2005;307:1746. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd CM, Vogel HJ, Tieleman DP. Biochem J. 2003;370:233. doi: 10.1042/BJ20021255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorfe AA, Pellarin R, Caflisch A. J Am Chem Soc. 2004;126:15277. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- 12.Kampf JP, Cupp D, Kleinfeld AM. J Biol Chem. 2006;281:21566. doi: 10.1074/jbc.M602067200. [DOI] [PubMed] [Google Scholar]
- 13.Shahinian S, Silvius JR. Biochemistry. 1995;34:3813. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 14.Wimley WC, Creamer TP, White SH. Biochemistry. 1996;35:5109. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 15.White SH. FEBS Lett. 2003;555:116. doi: 10.1016/s0014-5793(03)01153-0. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Mol Cell Biol. 2005;25:6722. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abankwa D, Gorfe AA, Hancock JF. Sem Cell Dev Biol. 2007 doi: 10.1016/j.semcdb.2007.08.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Mol Cell Biol. 2004;24:6799. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. Nat Cell Biol. 2001;3:368. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]; b) Plowman SJ, Hancock JF. Biochim Biophys Acta. 2005;1746:274. doi: 10.1016/j.bbamcr.2005.06.004. [DOI] [PubMed] [Google Scholar]; c) Hancock JF. Nat Rev Mol Cell Biol. 2006;7:456. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Nicolau DV, Jr, Burrage K, Parton RG, Hancock JF. Mol Cell Biol. 2006;26:313. doi: 10.1128/MCB.26.1.313-323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Plowman SJ, Muncke C, Parton RG, Hancock JF. Proc Natl Acad Sci U S A. 2005;102:15500. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scheme I: A thermodynamic model for the insertion of the ras peptide into a DMPC bilayer.