Abstract
Background
Recently, the type 2 diabetes medication, rosiglitazone, has come under scrutiny for possibly increasing the risk of cardiac disease and death. To investigate the effects of rosiglitazone on the diabetic heart, we performed cardiac transcriptional profiling and imaging studies of a murine model of type 2 diabetes, the C57BL/KLS-leprdb/leprdb (db/db) mouse.
Methods and Findings
We compared cardiac gene expression profiles from three groups: untreated db/db mice, db/db mice after rosiglitazone treatment, and non-diabetic db/+ mice. Prior to sacrifice, we also performed cardiac magnetic resonance (CMR) and echocardiography. As expected, overall the db/db gene expression signature was markedly different from control, but to our surprise was not significantly reversed with rosiglitazone. In particular, we have uncovered a number of rosiglitazone modulated genes and pathways that may play a role in the pathophysiology of the increase in cardiac mortality as seen in several recent meta-analyses. Specifically, the cumulative upregulation of (1) a matrix metalloproteinase gene that has previously been implicated in plaque rupture, (2) potassium channel genes involved in membrane potential maintenance and action potential generation, and (3) sphingolipid and ceramide metabolism-related genes, together give cause for concern over rosiglitazone's safety. Lastly, in vivo imaging studies revealed minimal differences between rosiglitazone-treated and untreated db/db mouse hearts, indicating that rosiglitazone's effects on gene expression in the heart do not immediately turn into detectable gross functional changes.
Conclusions
This study maps the genomic expression patterns in the hearts of the db/db murine model of diabetes and illustrates the impact of rosiglitazone on these patterns. The db/db gene expression signature was markedly different from control, and was not reversed with rosiglitazone. A smaller number of unique and interesting changes in gene expression were noted with rosiglitazone treatment. Further study of these genes and molecular pathways will provide important insights into the cardiac decompensation associated with both diabetes and rosiglitazone treatment.
Introduction
Cardiovascular disease is the leading cause of morbidity and mortality in patients with type 2 diabetes [1]. While atherosclerotic coronary artery disease is highly prevalent in many diabetic patients, the occurrence of nonischemic cardiomyopathy (“diabetic cardiomyopathy”) suggests that other processes such as microangiopathy, metabolic factors, or myocardial fibrosis [1] might be involved. Thiazolidinediones (TZDs) such as rosiglitazone are insulin sensitizers that may also have beneficial properties for diabetic cardiomyopathy. These agonists bind to a subfamily of nuclear hormone receptors, the peroxisome proliferator-activated receptors (PPARs, including α, β/δ and γ isoforms, of which the γ isoform is the most common target), and activate transcription factors that modulate gene expression, ultimately leading to increased insulin sensitivity in peripheral tissues through poorly defined pathways [2], [3]. However, a recent meta-analysis of clinical trial outcomes for the PPARγ agonist rosiglitazone found a significant increase in the risk of myocardial infarction, and a borderline significant risk of death from cardiovascular causes [4]. The reasons for this increased risk are unclear, and a better understanding of the effects of rosiglitazone on the diabetic heart is urgently needed.
Despite the large number of studies on rosiglitazone and diabetes, the transcriptional network changes by which PPARγ agonists and other TZDs promote insulin sensitivity are not well characterized. A number of groups have studied gene expression in the db/db mouse liver [5], as well as transcriptional changes induced by TZDs in adipocytes, kidney, aorta, and pancreatic islet cells in various mouse models [6], [7], [8], [9], but none has studied the heart directly. Moreover, determining PPAR-agonist target tissues is complicated by the fact that the three PPAR isoforms are not distributed equally across all tissues. In adipose tissue, PPARγ predominates and promotes differentiation and lipid storage [10], resulting in suppression of lipolysis by insulin and reduction of plasma free fatty acid concentrations. In the cardiovascular system, PPARγ is expressed in vascular smooth muscle and vascular endothelium [11], but is believed to be in low abundance in cardiomyocytes [12].
Animal models of type 2 diabetes present an optimal system for studying the effects of rosiglitazone. A well-established murine model is the C57BL/KLS-leprdb/leprdb (db/db) mouse that has a mutation in the leptin receptor. Homozygous db/db mice become obese by 3–4 wk of age and develop hyperglycemia at 4–8 wk. Serum insulin levels increase as early as 10–14 days, peak at 6–8 wk, then decrease afterward. These mice continue to be hyperinsulinemic throughout life and ultimately develop cardiomyopathy (contractile dysfunction), as evidenced from metabolic experiments using cultured db/db cardiomyocytes [13], echocardiographic studies [14], isolated working heart preparations [15], and direct in situ ventricular pressure measurements [16]. Recent studies by our lab and others have delineated the longitudinal structural and metabolic cardiomyopathic changes in the db/db mouse using cardiac magnetic resonance (CMR) and Fluorine-18–2-fluoro-2-deoxy-d-glucose ([18F]FDG) PET scanning [17].
To address the many questions concerning the molecular and functional effects of rosiglitazone on the diabetic heart, we used CMR imaging and echocardiagraphic studies to assess the development of cardiomyopathy in untreated and rosiglitazone-treated db/db mice, as well as normal db/+ mice. These studies were followed by cardiac gene expression profiling of the diabetic and control phenotypes to understand the underlying molecular changes in heart tissue. A comparison of the cardiac transcriptomes of rosiglitazone-treated db/db mice with untreated db/db and normal db/+ mice thus provides insights into the effects of rosiglitazone on the heart, either through direct or indirect mechanisms, as well as clues to its potential toxicity.
Methods
Animal and sample tissue preparation
Homozygous db/db mice (Jackson Laboratories, Bar Harbor, ME) were maintained on a normal chow diet and housed in a room with a 12∶12-h light-dark cycle and an ambient temperature of 22°C. For the treatment group, homozygous db/db mice were maintained on 5 mg/kg/day (approximately 0.225 mg/mouse/day) rosiglitazone (GlaxoSmithKline, London, UK) that was mixed with normal chow. Unless otherwise stated, heterozygous db/+ littermates were used as control animals. All protocols were approved by the Administrative Panel on Laboratory Animal Care at Stanford University and were carried out in accordance with the guidelines of the American Association for Accreditation of Laboratory Animal Care. At the end of the study, animals were euthanized with a lethal dose of isoflurane. Immediately after death, wet heart weight (HW) and body weight (BW) were measured. Whole hearts, pancreas, and liver were harvested and preserved in TRIzol reagent (Invitrogen, San Diego, CA) for subsequent mRNA isolation, and a subset of hearts were fixed in 10% formalin for histological evaluation.
Insulin tolerance testing
Insulin tolerance testing was performed on mice after a 6-h fast. At the time of testing, a bolus of human regular insulin (Eli Lilly, Indianapolis, IN; 0.75 IU/kg) was injected intraperitoneally. In blood derived from a tail nick, glucose levels were then determined with a FreeStyle blood glucose monitoring system (Abbott Laboratories, Abbott Park, IL) at baseline and 30, 60, and 120 min after injection.
Insulin Enzyme-Linked ImmunoSorbent Assay (ELISA)
After a 6-h fast, roughly 200 µl of blood was obtained from each animal via retroorbital bleeding. Samples were placed in 500- µl tubes containing EDTA (Becton-Dickinson, Franklin Lakes, NJ) and centrifuged at 4°C and 13,200 rpm for 10 min. Approximately 50–75 µl of supernatant (serum) was then collected for further processing. Serum insulin concentrations were measured with a Mercodia mouse insulin enzyme immunoassay kit (Alpco Diagnostics, Salem, NH).
Nonesterified fatty acids (NEFA)
Plasma nonesterified fatty acid concentrations were measured using a HR series NEFA-HR(2) colorimetric assay kit (Wako Chemicals USA, Richmond, VA).
Left ventricular functional analysis with echocardiogram
Echocardiography was performed by a blinded investigator (ZL) using the Siemens-Acuson Sequioa C512 system equipped with a multi-frequency (8–14 MHz) 15L8 transducer. Analysis of M-mode images was performed using the Siemens built-in software. Left ventricular end diastolic diameter (EDD) and end-systolic diameter (ESD) were measured and used to calculate fractional shortening (FS) by the following formula: FS = [EDD-ESD]/EDD. LV volume at end diastolic (EDV) and end-systole (ESV) were calculated by the bullet method as follows: EDV = 0.85×CSA(d)×L(d), ESV = 0.85×CSA(s)×L(s), where CSA(d) and (s) are endocardial area in end-diastole and end-systole, respectively, obtained from short-axis view at the level of the papillary muscles. L(d) and L(s) are the LV length (apex to mid-mitral annulus plane) in end-diastole and end-systole, respectively, obtained from the parasternal long-axis view. LV ejection fraction (EF%) was calculated as: EF% = (EDV−ESV)×100/EDV.
Cardiac magnetic resonance imaging
To prepare for scanning, induction of anesthesia was accomplished with 2% isoflurane and 1 l/min oxygen. Respiratory rate was monitored and used to manually calibrate the maintenance dose of isoflurane at 1.25–1.5%. Platinum needle ECG leads were inserted subcutaneously in the right and left anterior chest wall. Respiration was monitored with a pneumatic pillow sensor positioned along the abdomen. Body temperature was maintained at 36–37°C by a flow of heated air thermostatically controlled by a rectal temperature probe. Heart rate (HR), respiratory rate, and body temperature were recorded every 4 min during image acquisition. Magnetic resonance images were acquired by a blinded investigator (PY) with a 4.7-T magnet (Bruker BioSpin, Fremont, CA) controlled by a Varian Inova Console (Varian, Palo Alto, CA), using a transmit-receive quadrature volume coil with an inner diameter of 3.5 cm. For particularly obese animals, a larger coil with an inner diameter of 6 cm was utilized. Image acquisition was gated to the ECG R-wave (Small Animal Instruments, Stony Brook, NY). Coronal and axial scout images were used to position a two-dimensional imaging plane along the short axis of the left ventricular (LV) cavity. Gated gradient echo sequences were then used to acquire sequential short-axis slices spaced 1 mm apart from apex to base. For each sequence, 12 cine frames encompassing one cardiac cycle were obtained at each slice level with the following sequence parameters: acquisition time (TR) = 100–140 ms, echo time (TE) = 2.8–3.5 ms, number of repeats (NEX) = 8, field of view (FOV) = 30×30 mm, matrix = 128×128, flip angle = 60°. For each short-axis slice, planimetry measurements of LV myocardial area were conducted off-line by tracing the epicardial and endocardial borders at end systole and end diastole with MRVision software (MRVision, Winchester, MA). For these purposes the papillary muscles were considered part of the LV cavity. Anteroseptal and posterior wall thickness measurements were performed on a midventricular slice at the level of the papillary muscles at end diastole. LV mass (LVM) was derived from the sum of the differences between the end-diastolic epicardial and endocardial areas from apex to base, adjusted for the specific gravity of myocardial tissue (1.055 g/ml). LV end-diastolic (LVEDV) and end-systolic (LVESV) volumes were calculated as the sum of the endocardial areas of each slice from the apex to the LV outflow tract at end diastole and end systole, respectively. LV ejection fraction (LVEF) was calculated as (LVEDV−LVESV)/LVEDV. Cardiac output (CO) was calculated as (LVEDV−LVESV)/(HR).
RNA isolation and quality control
Total RNA was isolated separately from five heart samples for each condition using TRIzol reagent (Invitrogen, San Diego, CA) according to the manufacturer's instructions for a total of 15 distinct RNA samples. Total RNA was purified using RNeasy columns (Qiagen, Chatsworth, CA). RNA concentration was measured by spectrophotometry, and RNA integrity assessed with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) with 6000 Nano Chips according to the manufacturer's instructions. RNA was judged as suitable for array hybridization only if samples exhibited intact bands corresponding to 18S and 28S ribosomal RNA subunits and had a RNA Integrity Number (RIN) greater than six. Universal Reference RNAs for mouse (Stratagene, La Jolla, CA) were purchased as microarray reference controls.
Labeling reaction and hybridization
Using Low RNA Input Fluorescent Linear Amplification Kits (Agilent Technologies, Santa Clara, CA, USA), cDNA was reverse transcribed from each RNA sample, and cRNA was then transcribed and fluorescently labeled from each cDNA sample. The fifteen mouse heart tissue cRNA samples were labeled with Cy5 and the Universal Mouse reference cRNA was labeled with Cy3, yielding a total of 5 biological replicates per condition. The resulting cRNA was purified using an RNeasy kit (Qiagen, Valencia, CA, USA) followed by quantification of the cRNA by spectroscopy using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). 825 ng Cy3- and Cy5- labeled and amplified cRNA was mixed and fragmented according to the Agilent technology protocol. cRNA was hybridized to 4×44K whole human genome microarray slides from Agilent (Part G4112F) according to the manufacturer's instructions. The hybridization was carried in a rotating hybridization chamber in the dark at 65°C for 17 h.
Washing, scanning, and feature extraction
Slides were washed with Gene Expression Wash Buffer 1 and 2 (Agilent Technologies, Santa Clara, CA) followed by Acetonitrile. A final wash in Stabilization and Drying Solution was performed to prevent Cy-5 degradation by ozone. The array was scanned using an Agilent G2505B DNA microarray scanner under extended Dynamic range. The image files were extracted using the Agilent Feature Extraction software version 9.5.1 applying LOWESS background subtraction and dye-normalization. Further analyses were performed with BRB ArrayTools Version 3.4 (National Cancer Institute) and TIGR MeV software (http://www.tm4.org/).
Real-time quantitative PCR for confirmation of microarray results
RNA to cDNA conversion was performed with a SuperScript First-Strand cDNA synthesis kit (Invitrogen). The cDNA was then used as a template in a TaqMan real-time PCR assay with the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). All samples were run in triplicates. Specialized premade gene expression assay reagents for potassium channel, subfamily K, member 1 (Kcnk1; catalog no. Mm00434624_m1), matrix metalloproteinase 3 (Mmp3; Mm00440295_m1), beta-1,4-N-acetyl-galactosaminyl transferase 1 (B4galnt1; Mm00484653_m1), carboxyl ester lipase (Cel; Mm00486975_m1), and N-acylsphingosine amidohydrolase 2 (Asah2; Mm00479659_m1), purchased from Applied Biosystems, were used for these experiments. Threshold cycles were placed in the logarithmic portion of the amplification curve, and each sample was referenced to 18S RNA (part no. 4319413E) amplification to control for the total amount of RNA. Fold differences between samples (relative quantification) were calculated with the delta-delta method [S1/S2 = 2−(T1–T2)], where S1 and S2 represent samples 1 and 2 and T1 and T2 denote the threshold cycles for S1 and S2.
Histologic analysis of cardiac tissue
At the time of death, a select group of hearts were fixed in 10% formalin, cut at the midventricular level, and embedded in paraffin blocks. The blocks were then sectioned into short-axis slices, which were subsequently stained with hematoxylin and eoxin according to standard protocols. High-power fields magnified to 40× from the midportion of the LV free wall were photographed.
Statistical Analysis
For all tests, one-way ANOVA was performed. All P values <0.05 were considered significant. For microarray analysis, the Significance Analysis of Microarrays (SAM) algorithm was used to identify genes with statistically significant differences in expression among the conditions. SAM is a statistically rigorous test that incorporates a FDR calculation to correct for multiple testing errors. Gene Ontology group overrepresentation analysis was performed using Fisher's exact test with FDR correction through High Throughput GoMiner software.
Results
Insulin resistance in rosiglitazone-treated, untreated and control mice
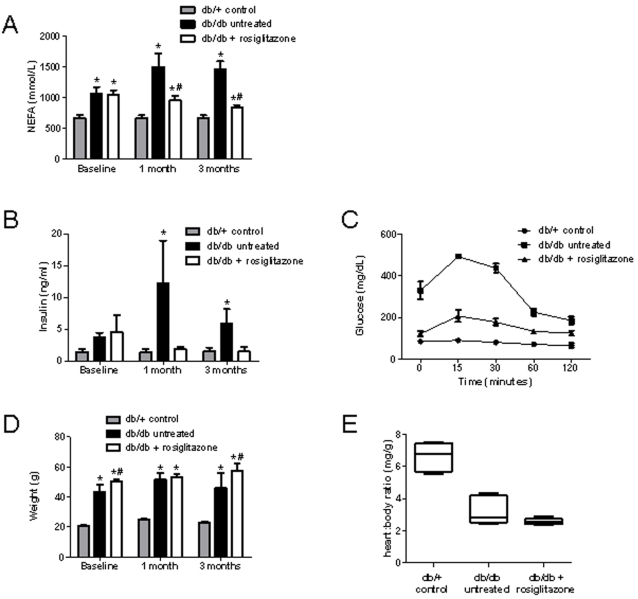
We analyzed a number of metabolic parameters to confirm insulin resistance in our db/db murine model of diabetes before proceeding with microarrays. Insulin resistance is strongly associated with obesity, and one mechanism may be the generation of metabolic messengers such as free fatty acids by adipose tissue that inhibit insulin action on muscle [18], [19]. We measured plasma non-esterified fatty acid (NEFA) and insulin levels to follow insulin resistance in our three groups of mice. Figure 1a shows plasma NEFA levels in the three groups of mice. Compared to untreated db/db mice, NEFA levels in the rosiglitazone-treated db/db mice decreased dramatically. Fasting insulin levels ( Figure 1b ) and insulin tolerance testing ( Figure 1c ) further confirmed the improved insulin sensitivity with rosiglitazone treatment.
Figure 1. Insulin resistance and mean body/heart weights of rosiglitazone-treated, untreated, and control mice.
(A) Plasma NEFA levels at 0, 1, and 3 months in untreated db/db mice (n = 10), rosiglitazone-treated db/db mice (n = 10), and db/+ mice (n = 10). NEFA levels were higher in both the treated and untreated db/db groups at baseline when compared to db/+ control mice. Compared to untreated db/db mice, NEFA levels in the rosiglitazone-treated db/db group decreased significantly during the first month (960.9 vs. 1501.0 mmol/L, P<0.05), and decreased further by the third month to near db/+ control levels. (B) Fasting insulin levels in the three groups at 0, 1, and 3 months. At baseline, both treated and untreated db/db groups had increased levels of insulin compared to db/+ control. Insulin levels dramatically increased in untreated db/db mice at 1 month, but progressively decreased in rosiglitazone-treated mice. (C) Insulin tolerance testing at 3 months. Serum glucose levels after insulin injection were significantly higher in untreated db/db mice when compared to db/+ mice; the rosiglitazone-treated db/db group had moderately elevated glucose levels after insulin administration. These results corroborate the NEFA and insulin studies that demonstrate improved insulin sensitivity with rosiglitazone treatment. (D) Mean body weights of the three groups of mice at 0, 1, and 3 months. Weights of both treated and untreated db/db mice were markedly higher than db/+ controls at baseline. Furthermore, rosiglitazone-treated mice had generally higher mean body weights when compared to untreated mice. (E) Mean heart/body weight ratios at sacrifice (4 months after treatment initiation). No significant difference between treated and untreated groups (3.27 vs. 2.58, P = 0.13), though there does appear to be a downward trend in the data. Heart/body weight ratio was significantly higher in the db/+ group compared to both db/db groups. Values are mean±SEM. *P<0.05 vs. age-matched db/+; #P<0.05 vs. age-matched untreated db/db.
Body and heart morphometric analysis
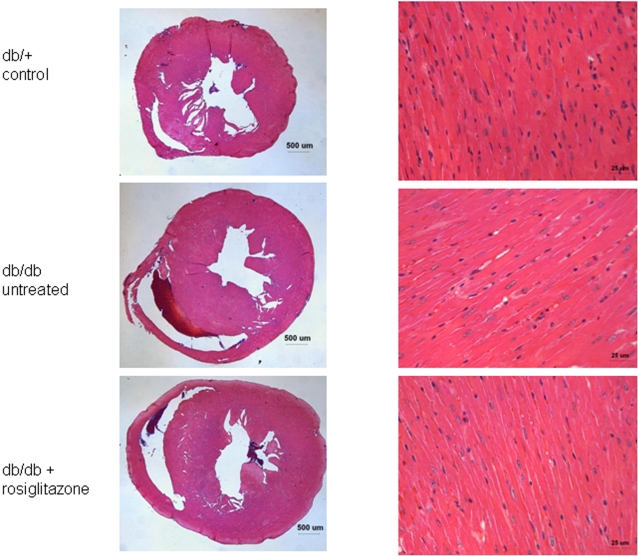
In addition to the obesity associated with type 2 diabetes, rosiglitazone and other TZDs are widely known to cause further weight gain due to fluid retention and altered adipogenesis [20]. Mean body weights of both treated and untreated db/db mice were markedly higher than db/+ controls at baseline ( Figure 1d ). Furthermore, rosiglitazone-treated mice had generally higher mean body weights when compared to untreated mice, as expected. The heart/body weight ratio was significantly higher in the db/+ group when compared to the treated and untreated db/db groups ( Figure 1e ). However, histological analysis did not reveal obvious differences at the microscopic level ( Figure 2 ).
Figure 2. High-power light microscopy slides of myocardial tissue from rosiglitazone-treated and untreated db/db mice, and db/+ controls.
Individual cardiomyocytes from all three groups appear qualitatively similar based on morphology and size, though the untreated db/db cardiomyocytes exhibit very mild thickening. No increases in inflammatory cells, cell death, fibrosis, or other processes were observed in the two db/db groups compared to db/+ controls.
Left ventricular functional analysis with echocardiogram
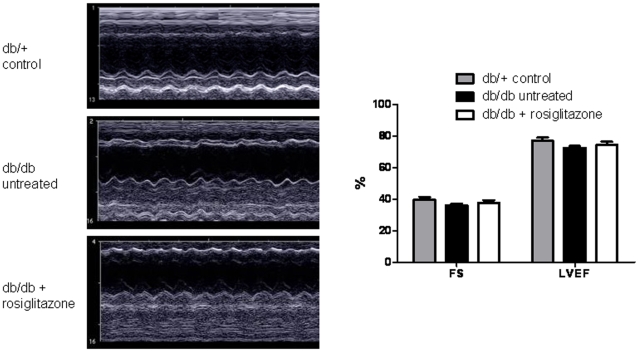
Previous groups have observed a significant decrease in left ventricular ejection fraction (LVEF) and fractional shortening (FS) in db/db mice when compared with db/+ controls [14], [21]. In one study, the FS was reduced by as much as 16% at 12 weeks of age [14]. However, more accurate CMR studies of cardiac contractility have not confirmed the dramatic differences seen in the echocardiographic studies, observing only an approximately 2% reduction in LVEF at 22 weeks of age [17]. Furthermore, clinical echocardiographic studies of type 2 diabetic patients treated with rosiglitazone found no significant changes in LVEF after 52 weeks of treatment [22]. To investigate cardiac contractility and verify these previous findings, we assessed cardiac contractility in the three mouse groups using echocardiography ( Figure 3 ). The mean LVEF and FS levels of the untreated db/db group were not significantly different from those in the db/+ group, though there does appear to be a downward trend in contractility in the db/db group. Further, we found no significant LVEF or FS differences between the rosiglitazone-treated db/db group and untreated db/db group, confirming the results of the clinical study that found no evidence of rosiglitazone-induced changes in cardiac contractility.
Figure 3. Left ventricular functional analysis with echocardiogram.
The mean left ventricular ejection fraction (LVEF) and fractional shortening (FS) of the untreated db/db group (72.4±2.3% and 36.2±1.7%, respectively) were not significantly different from the db/+ group (76.9±4.3%, P = 0.11 and 39.7±3.6%, P = 0.11, respectively), though there appears to be a downward trend in contractility between these two groups. Further, there were no significant LVEF or FS differences between the rosiglitazone-treated db/db group (74.3±3.9% and 37.8±3.1%, respectively) and untreated db/db group (P = 0.43 and 0.40, respectively). Values are mean±SEM.
Cardiac magnetic resonance imaging
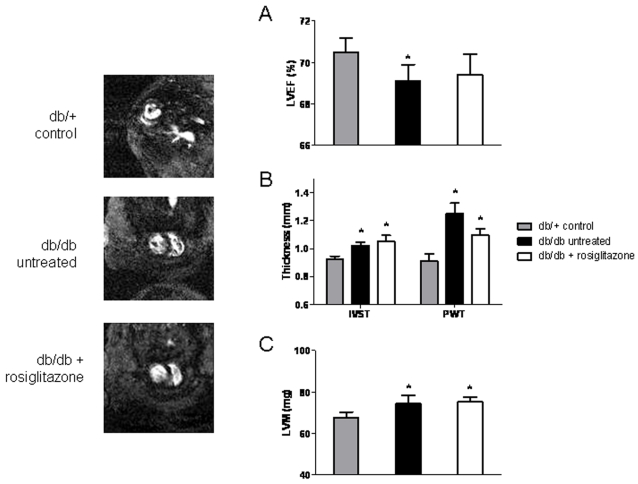
Using cardiac magnetic resonance (CMR) scanning, our group has previously shown progressive cardiomyopathic changes in db/db mice when compared to db/+ controls [17]. Left ventricular mass (LVM), interventricular septal thickness (IVST) and posterior wall thickness (PWT) were significantly increased in db/db mice, while LVEF was only marginally decreased. For the CMR imaging in this new study ( Figure 4 ), we included rosiglitazone-treated db/db mice in addition to the untreated db/db and db/+ groups, and scanned them at 8 weeks after treatment initiation to look for cardiomyopathic changes. CMR scans revealed significant, albeit subtle, cardiomyopathic changes in untreated db/db mice compared to db/+ controls ( Figure 4 ). However, rosiglitazone treatment resulted in no significant improvements in cardiac contractility, which confirmed our echocardiographic studies.
Figure 4. Cardiac magnetic resonance imaging.
CMR imaging at 8 weeks after treatment initiation of rosiglitazone-treated db/db mice (n = 4), untreated db/db mice (n = 4) and db/+ groups (n = 4). Left ventricular mass (LVM), interventricular septal thickness (IVST) and posterior wall thickness (PWT) were significantly increased in both treated and untreated db/db mice relative to db/+ controls, while LVEF was decreased (though not significantly in the rosiglitazone-treated group). No significant differences in LVM, IVST, PWT, or LVEF between rosiglitazone-treated and untreated db/db mice were detected (P = 0.82, 0.66, 0.14, and 0.88, respectively). Values are mean±SEM. *P<0.05 vs. age-matched db/+.
Gene expression analysis
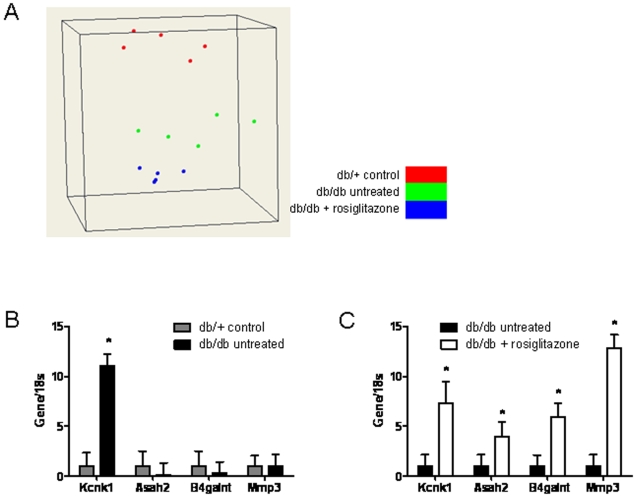
Whole-heart RNA from five mice from each of the three groups after four months with or without treatment was used for microarray analysis. Overall, there were significant transcriptional differences between the db/+ and both db/db groups, while differences between rosiglitazone-treated and untreated db/db groups were much less dramatic. Figure 5a shows a 3-dimensional scaling plot based on principal component analysis of the expression data, which graphically demonstrates that rosiglitazone did not restore db/db transcriptomes to the normal db/+ state.
Figure 5. Multidimensional scaling plot of the expression data and real-time PCR analysis.
(A) 3-dimensional scaling plot provides a graphical representation of high-dimensional expression data in low dimensions. Each point within a “cloud” represents a single microarray, and the similarity within a set of microarrays is indicated by their physical proximity to one other. As evidenced from the figure, each group (db/+ control, untreated db/db, and rosiglitazone-treated db/db) clusters into a distinct grouping, indicating that each group has a similar transcriptome that can be distinguished from the two other groups. Taqman real-time PCR of four selected genes normalized to 18s shows (B) significant upregulation of Kcnk1 in untreated db/db vs. db/+ mice and (C) significant upregulation of all four genes in rosiglitazone-treated vs. untreated db/db mice. These data confirm the altered regulatory patterns of these four genes in the microarray data. Values are mean±SEM. *P<0.05 vs. control.
Genes that exhibited significant differential expression across the three groups were identified using the SAM statistical algorithm (selected genes are listed in Tables 1 , 2 ; full gene txt files are included as Supplemental Gene Lists S1, S2). Interesting genes that were significantly upregulated in the untreated db/db group compared to control include PPARγ (Pparg, the receptor for rosiglitazone), FK506 binding proteins (Fkbp5 and Fkbp10), several potassium channel proteins (Kcnk1 and Kcnd3), two matrix metalloproteinases (Mmp3 and Mmp8), a cyclin-dependent kinase inhibitor (Cdkn1), and myostatin (Gdf8) ( Table 1 ). Specific downregulated genes of interest include the anti-apoptotic gene B-cell lymphoma–leukemia 2 (Bcl2), several tumor necrosis factor-alpha (TNFα) genes and transforming growth factor (TGFβ) genes, vascular endothelial growth factor (Vegfc), adiponectin (Adipoq), and apelin (Apln). We confirmed the microarray results of 4 selected genes with real-time PCR ( Figures 5b, 5c ). When we compared the db/db rosiglitazone treated group to control, we found that upregulated genes include the previously-mentioned Mmp3 and Kcnk1 transcripts, as well as a potassium channel-interacting gene (Kcnip4), ryanodine receptor (Ryr1), Myosin IIIB (Myo3b), Patched homolog 1, and apelin ( Table 2 ).
Table 1. Untreated db/db vs. db/+ control mice.
| Gene Symbol | RefSeq ID | UGCluster | Name | Score(d) | Fold Change |
| Kcnk1 | NM_008430 | Mm.10800 | Potassium channel, subfamily K, member 1 | 7.82 | 26.03 |
| Gdf8 | NM_010834 | - | growth differentiation factor 8 (Gdf8) | 5.12 | 15.93 |
| Egfbp2 | NM_010115 | - | epidermal growth factor binding protein type B (Egfbp2) | 4.77 | 9.47 |
| Fkbp5 | NM_010220 | Mm.276405 | FK506 binding protein 5 | 9.76 | 6.11 |
| Mmp8 | NM_008611 | Mm.16415 | Matrix metallopeptidase 8 | 5.22 | 5.93 |
| Edn3 | NM_007903 | Mm.9478 | Endothelin 3 | 6.59 | 5.72 |
| Cdkn1a | NM_007669 | Mm.195663 | Cyclin-dependent kinase inhibitor 1A (P21) | 5.13 | 3.74 |
| Kcnj4 | NM_008427 | Mm.140760 | Potassium inwardly-rectifying channel, subfamily J, member 4 | 5.06 | 2.68 |
| Map3k6 | NM_016693 | Mm.36640 | Mitogen-activated protein kinase kinase kinase 6 | 9.39 | 2.54 |
| Map3k15 | BC031147 | Mm.386889 | Mitogen-activated protein kinase kinase kinase 15 | 5.88 | 2.10 |
| Fkbp10 | NM_010221 | Mm.3894 | FK506 binding protein 10 | 5.21 | 2.09 |
| Mmp3 | NM_010809 | Mm.4993 | Matrix metallopeptidase 3 | 4.30 | 1.90 |
| Kcnd3 | NM_019931 | Mm.44530 | Potassium voltage-gated channel, Shal-related family, member 3 | 5.93 | 1.77 |
| Ryr3 | XM_619795 | Mm.436657 | Ryanodine receptor 3 | 5.45 | 1.74 |
| Pparg | NM_011146 | Mm.3020 | Peroxisome proliferator activated receptor gamma | 5.51 | 1.42 |
| Tnfrsf22 | NM_023680 | Mm.261384 | Tumor necrosis factor receptor superfamily, member 22 | 4.06 | 1.40 |
| Aifm2 | NM_153779 | Mm.286309 | Apoptosis-inducing factor, mitochondrion-associated 2 | 5.62 | 1.39 |
| Il4ra | NM_001008700 | Mm.233802|Mm.441865 | interleukin 4 receptor, alpha (Il4ra) | 4.96 | 1.37 |
| Sod1 | BC057074 | Mm.431677 | superoxide dismutase 1, soluble (cDNA clone IMAGE:5697222), partial cds. | 4.02 | 1.34 |
| Adipor2 | NM_197985 | Mm.291826 | Adiponectin receptor 2 | 4.42 | 1.22 |
| Myo9a | AK029836 | Mm.249545 | Myosin IXa | −4.70 | −1.26 |
| Casp7 | NM_007611 | Mm.35687 | Caspase 7 | −4.46 | −1.27 |
| Casp2 | NM_007610 | Mm.3921|Mm.433648 | caspase 2 (Casp2) | −5.10 | −1.34 |
| Vegfc | NM_009506 | Mm.1402 | Vascular endothelial growth factor C | −6.20 | −1.39 |
| Il13ra1 | NM_133990 | Mm.24208 | Interleukin 13 receptor, alpha 1 | −4.24 | −1.40 |
| Gja1 | NM_010288 | Mm.378921 | Gap junction membrane channel protein alpha 1 | −4.85 | −1.49 |
| Il13ra1 | NM_133990 | Mm.24208 | Interleukin 13 receptor, alpha 1 | −4.23 | −1.50 |
| Myo1b | NM_010863 | Mm.3390 | Myosin IB | −6.11 | −1.57 |
| Tnfaip8 | NM_134131 | Mm.27740 | Tumor necrosis factor, alpha-induced protein 8 | −4.65 | −1.71 |
| Adrb2 | NM_007420 | Mm.5598 | Adrenergic receptor, beta 2 | −4.91 | −1.81 |
| Tnfaip8l1 | NM_025566 | Mm.2312 | Tumor necrosis factor, alpha-induced protein 8-like 1 | −5.52 | −1.88 |
| Il16 | NM_010551 | Mm.10137 | Interleukin 16 | −24.20 | −2.42 |
| Myo1d | AK037051 | Mm.151948 | Myosin ID | −4.19 | −2.45 |
| Apln | NM_013912 | Mm.29262 | Apelin | −4.51 | −2.54 |
| Bcl2 | NM_009741 | 257460|Mm.462969 | B-cell leukemia/lymphoma 2 (Bcl2), transcript variant 1 | −7.36 | −2.56 |
| Tnfrsf13c | NM_028075 | Mm.240047 | Tumor necrosis factor receptor superfamily, member 13c | −6.33 | −4.54 |
| Adipoq | NM_009605 | Mm.3969 | Adiponectin, C1Q and collagen domain containing | −5.31 | −4.59 |
Selected genes that were differentially expressed across untreated db/db mice and db/+ controls. All of the genes presented in the tables have statistically significant expression changes. Listed are the most important parameters, including the standard identifications of the gene, a statistical measure of significance (“Score(d)”), and magnitude of the change (“Fold Change”).
Table 2. Rosiglitazone-treated vs. untreated db/db mice.
| Gene Symbol | RefSeq ID | UGCluster | Name | Score(d) | Fold Change |
| Kcnip4 | NM_030265 | Mm.160172 | Kv channel interacting protein 4 | 4.46 | 4.76 |
| Ptch1 | NM_008957 | Mm.228798 | Patched homolog 1 | 6.67 | 3.32 |
| Ryr1 | NM_009109 | Mm.439745 | Ryanodine receptor 1, skeletal muscle | 5.27 | 3.17 |
| Kcnk1 | NM_008430 | Mm.10800 | Potassium channel, subfamily K, member 1 | 6.38 | 2.84 |
| Myo3b | AK033795 | Mm.99648|Mm.458853 | adult male epididymis cDNA, RIKEN full-length enriched library, clone:9230110G05 | 4.46 | 2.58 |
| Mmp3 | NM_010809 | Mm.4993 | Matrix metallopeptidase 3 | 4.70 | 2.03 |
| Tnfsf18 | NM_183391 | Mm.276823 | Tumor necrosis factor (ligand) superfamily, member 18 | 3.82 | 1.86 |
| Apln | NM_013912 | Mm.29262 | Apelin | 3.52 | 1.80 |
| Anxa13 | NM_027211 | Mm.237985 | Annexin A13 | 4.04 | 1.64 |
| St3gal5 | NM_011375 | Mm.38248 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 5.56 | 1.62 |
| Col22a1 | XM_193814 | Mm.322500 | Collagen, type XXII, alpha 1 | 3.65 | 1.45 |
| Col4a1 | NM_009931 | Mm.738 | Procollagen, type IV, alpha 1 | 4.98 | 1.26 |
| Kras | NM_021284 | Mm.383182 | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 4.40 | 1.23 |
| B4galnt1 | BC022180 | Mm.386762 | Beta-1,4-N-acetyl-galactosaminyl transferase 1 | 7.83 | 1.44 |
| Cel | NM_009885 | Mm.236017 | Carboxyl ester lipase | 6.31 | 1.38 |
| Pparbp | NM_013634 | Mm.12926 | Peroxisome proliferator activated receptor binding protein | 3.68 | 1.29 |
| Tnfrsf10b | NM_020275 | Mm.193430 | Tumor necrosis factor receptor superfamily, member 10b | 3.54 | 1.27 |
| Asah2 | NM_018830 | Mm.104900 | N-acylsphingosine amidohydrolase 2 | 4.38 | 1.21 |
| Myh9 | NM_022410 | Mm.29677 | Myosin, heavy polypeptide 9, non-muscle | 4.03 | 1.19 |
| Bdkrb1 | NM_007539 | Mm.377078 | Bradykinin receptor, beta 1 | −6.72 | −1.96 |
| Hsf1 | NM_008296 | Mm.347444 | Heat shock factor 1 | −5.70 | −1.17 |
| Col23a1 | AK162470 | Mm.154093|Mm.392367 | 12 days embryo embryonic body between diaphragm region and neck cDNA, RIKEN full-length enriched library, clone:9430076L03 | −5.70 | −1.13 |
Selected genes that were differentially expressed across rosiglitazone-treated and untreated db/db mice.
While identifying individual genes that are differentially regulated in each of the conditions is informative, it is also useful to ascertain which cellular processes are up- or down- regulated; therefore, we performed the Gene Ontology pathway overrepresentation analysis using Fisher's exact test. Lipid and protein metabolism, fatty acid beta-oxidation, cell death, apoptosis, peroxisome organization, and biogenesis were significantly upregulated in untreated db/db mice when compared to control db/+ mice (Supplemental Tables S1, S2, S3, S4). Among major pathways that were significantly downregulated are cell proliferation and cycle, immune response, blood vessel and vasculature development, and anti-apoptosis. Interestingly, comparing the rosiglitazone-treated db/db mice to control mice, we did not see a reversal of the pathways that were upregulated in the untreated db/db group. The additional pathways that were significantly upregulated in the rosiglitazone-treated db/db hearts, however, included secretion, exocytosis, protein and intracellular transport, cellular protein metabolism, and sphingolipid metabolism.
Discussion
Few studies have studied the global cardiac gene expression changes resulting from rosiglitazone treatment. Understanding these molecular changes will help elucidate the potential mechanism(s) for the increased cardiac disease observed in diabetic patients treated with this drug. Our microarray results demonstrate that rosiglitazone does not reverse many of the significant gene expression and pathway changes that occur in untreated diabetic hearts, such as apoptosis and lipid metabolism. Furthermore, our microarray analysis indicates that rosiglitazone did not upregulate important insulin-regulated glucose transporters such as Glut4 in the heart, a finding that has been reported previously in isolated rat cardiomyocytes [23]. However, we observed a number of unique genes which appear to be differentially regulated between rosiglitazone-treated and untreated animals that may give mechanistic insights into rosiglitazone's effects on the diabetic heart.
We were initially puzzled to find that PPARγ expression was upregulated in the hearts of untreated diabetic mice, as it has been reported to be low in cardiomyocytes [12], [24], [25]. However, several studies have shown PPARγ expression to be important to cardiac metabolism. One study found that rosiglitazone-treated Zucker fatty rat hearts had improved glucose uptake as well as improved contractility compared to lean rat hearts [19]. Another study found that mice lacking PPARγ in cardiomyocytes exhibited mild cardiac hypertrophy [26]. These findings may reflect an indirect effect of rosiligitazone on the heart, as demonstrated by a study in which metabolic changes in PPARγ-treated db/db hearts were thought to be secondary to changes in the supply of exogenous substrates [21]. It appears that cardiac PPARγ expression may play an important though poorly understood role in cardiac physiology, and it is possible that modulation of PPARγ signaling may affect the response to cardiac ischemia.
Matrix metalloproteinases are known to play a critical role in atherosclerosis and cardiovascular tissue remodeling, mediating the balance between matrix accumulation and degradation [27]. We found that rosiglitazone caused significant upregulation of Mmp3, which encodes the matrix metalloproteinase stromelysin. Mmp3 is typically expressed by macrophages within the atherosclerotic plaques found in coronary artery disease [28], and has been shown to promote plaque rupture, myocardial infarction and aneurysm [29]. It is therefore possible that the in vivo upregulation of Mmp3 by rosiglitazone will promote plaque rupture, leading to increased rates of myocardial infarction, as well as increasing tissue remodeling after ischemia.
In both untreated and rosiglitazone-treated db/db mice, we found several potassium channel-related genes that were highly upregulated, including Kcnip4 and Kcnk1. These findings support the hypothesis that cardiomyocytes from db/db hearts exhibit electrophysiological alterations such as attenuated outward K+ currents [21], [30]. In general, glycolytic ATP production is important for the normal function of cardiac membrane ion channels and pumps, and inhibition of glycolysis causes altered intracellular ion concentrations that may affect cardiac action potentials. Specifically, Kcnip4 modulates A-type potassium channels and has been shown to increase expression of cardiac A-type Kv-4 channels [31]. Kcnk1 encodes a two-pore potassium channel, TWIK-1, which plays a major role in setting the resting membrane potential in many cell types [32]. Although it is unclear to what degree cardiac arrhythmias might have played a role in the mortality shown in the meta-analysis of rosiglitazone trials [4], the changes we observed in cardiomyocyte potassium channel expression raise some concerns.
Rosiglitazone treatment also upregulated four genes related to sphingolipid metabolism: B4galnt1, Cel, Asah2, and St3gal5. Found in eukaryotic plasma membranes, sphingolipids have emerged as a class of lipid mediators believed to be important for endogenous modulation of cardiovascular function [33], [34]. Generation of sphingolipid metabolites such as ceramide, sphingosine, and sphingosine-1-phosphate can activate smooth muscle cell proliferation, endothelial cell differentiation, apoptosis and cell death, migration, vasoconstriction, and dilation. Specifically, ceramide is an important mediator of lipotoxicity in the heart, and accumulation of this metabolite has been associated with cardiomyocyte apoptosis [35]. Recent evidence indicates that the TZD pioglitazone causes increased de novo ceramide synthesis in rat hearts [36], although another group has reported decreased ceramide accumulation and subsequent reduction in apoptosis [37]. Regardless of the specific mechanism involved, our microarray data suggest a significant role for sphingolipid metabolism in the rosiglitazone-treated diabetic heart which we are actively investigating further.
The downregulation of the hormone adiponectin in the diabetic state has been well established in adipocytes and serum [38], but it was not until 2005 that Pineiro et al. showed evidence that cardiomyocytes also synthesize adiponectin [39]. Confirming a recent report by Natarajan et al. [40], our results demonstrate a reduction of adiponectin in the diabetic heart, though we did not observe restoration of its expression after rosiglitazone treatment. In contrast, the signaling peptide apelin, a recently discovered regulator of cardiac and vascular function [41], was downregulated in the untreated db/db group but was restored to normal levels after rosiglitazone treatment. Apelin is expressed in the endothelium of heart, kidney, and lung, and its receptor (APJ) is expressed by myocardial cells and some vascular smooth muscle cells. Apelin has inotropic effects on cardiac contractility and has been shown to decrease systemic vascular resistance, and is therefore likely beneficial during heart failure. In fact, apelin signaling may augment inhibition of the renin-angiotensin system [42], which is known to exacerbate heart failure when left unchecked. Though not well understood, the altered expression of apelin we observed in the diabetic heart in response to rosiglitazone may present a novel pathway for the study of diabetic cardiomyopathy.
In conclusion, this study has mapped the genomic expression patterns in the hearts of the db/db murine model of diabetes and the effects of rosiglitazone on these patterns. Overall, as we expected, the db/db gene expression signature was markedly different from control, but to our initial surprise was not significantly reversed with rosiglitazone. In fact, many of the transcriptional changes induced by diabetic disease on the heart appeared to be further exacerbated by rosiglitazone. In particular, we have highlighted a number of rosiglitazone modulated genes and pathways that may play a role in the pathophysiology of the increase in cardiac mortality seen in the meta-analysis by Nissen et al. The cumulative upregulation of (1) a matrix metalloproteinase gene that has previously been implicated in plaque rupture, (2) potassium channel genes involved in membrane potential maintenance and action potential generation, and (3) sphingolipid and ceramide metabolism related genes, all give cause for concern over rosiglitazone's safety. Interestingly, a second meta-analysis has suggested that pioglitazone, also a TZD, may not have the same negative cardiovascular effects as rosiglitazone [43]. We believe the overall expression changes caused by rosiglitazone in the heart will likely also be found with pioglitazone treatment, though with some differences. This is based on previous microarray studies of adipocytes [44] and hepatocytes [45] that found similar, but not identical, expression profiles after treatment with pioglitazone, rosiglitazone, and other TZDs. Future investigations of how these two drugs differ at the transcriptional level in the heart will further elucidate the networks that govern diabetic cardiovascular function and disease.
Supporting Information
Upregulated Gene Ontology pathways with rosiglitazone-treated db/db mice.
(0.03 MB RTF)
Downregulated Gene Ontology pathways with rosiglitazone-treated db/db mice.
(0.02 MB RTF)
Upregulated Gene Ontology pathways in untreated db/db mice.
(0.04 MB RTF)
Downregulated Gene Ontology pathways in untreated db/db mice.
(0.05 MB RTF)
txt file of microarray results from untreated db/db vs. db/+ analysis
(0.15 MB TXT)
txt file of microarray results from db/db+rosiglitazone vs db/db analysis.
(0.04 MB TXT)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this work was supplied in part by research grants from Burroughs Wellcome Foundation Career Award in Medical Scientist (JCW). KDW is a recipient of a Stanford University Bio-X Graduate Student Fellowship and Society of Nuclear Medicine Student Fellowship. This study was designed by JCW in collaboration with his academic colleagues. This investigation did not have separate industry funding. The funding sources had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
References
- 1.Bauters C, Lamblin N, Mc Fadden EP, Van Belle E, Millaire A, et al. Influence of diabetes mellitus on heart failure risk and outcome. Cardiovascular Diabetology. 2003;2:1–1. doi: 10.1186/1475-2840-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vamecq J, Latruffe N. Medical significance of peroxisome proliferator-activated receptors. The Lancet. 1999;354:141–148. doi: 10.1016/S0140-6736(98)10364-1. [DOI] [PubMed] [Google Scholar]
- 3.Olefsky JM. Treatment of insulin resistance with peroxisome proliferator-activated receptor gamma agonists. The Journal of Clinical Investigation. 2000;106:467–472. doi: 10.1172/JCI10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England Journal of Medicine. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 5.Heishi M, Ichihara J, Teramoto R, Itakura Y, Hayashi K, et al. Global gene expression analysis in liver of obese diabetic db/db mice treated with metformin. Diabetologia. 2006;49:1647–1655. doi: 10.1007/s00125-006-0271-y. [DOI] [PubMed] [Google Scholar]
- 6.Makino H, Miyamoto Y, Sawai K, Mori K, Mukoyama M, et al. Altered gene expression related to glomerulogenesis and podocyte structure in early diabetic nephropathy of db/db mice and its restoration by pioglitazone. Diabetes. 2006;55:2747–2756. doi: 10.2337/db05-1683. [DOI] [PubMed] [Google Scholar]
- 7.Albrektsen T, Frederiksen KS, Holmes WE, Boel E, Taylor K, et al. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes. 2002;51:1042–1051. doi: 10.2337/diabetes.51.4.1042. [DOI] [PubMed] [Google Scholar]
- 8.Keen HL, Ryan MJ, Beyer A, Mathur S, Scheetz TE, et al. Gene expression profiling of potential PPARgamma target genes in mouse aorta. Physiological Genomics. 2004;18:33–42. doi: 10.1152/physiolgenomics.00027.2004. [DOI] [PubMed] [Google Scholar]
- 9.Parton LE, Diraison F, Neill SE, Ghosh SK, Rubino MA, et al. Impact of PPARgamma overexpression and activation on pancreatic islet gene expression profile analyzed with oligonucleotide microarrays. American Journal of Physiology Endocrinology and Metabolism. 2004;287:E390–E404. doi: 10.1152/ajpendo.00016.2004. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED, Spiegelman BM. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. The Journal of Biological Chemistry. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 11.Marx N, Schönbeck U, Lazar MA, Libby P, Plutzky J. Peroxisome proliferator-activated receptor gamma activators inhibit gene expression and migration in human vascular smooth muscle cells. Circulation Research. 1998;83:1097–1103. doi: 10.1161/01.res.83.11.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly DP. PPARs of the heart: three is a crowd. Circulation Research. 2003;92:482–484. doi: 10.1161/01.RES.0000064382.46274.95. [DOI] [PubMed] [Google Scholar]
- 13.Aasum E, Hafstad AD, Severson DL, Larsen TS. Age-dependent changes in metabolism, contractile function, and ischemic sensitivity in hearts from db/db mice. Diabetes. 2003;52:434–441. doi: 10.2337/diabetes.52.2.434. [DOI] [PubMed] [Google Scholar]
- 14.Semeniuk LM, Kryski AJ, Severson DL. Echocardiographic assessment of cardiac function in diabetic db/db and transgenic db/db-hGLUT4 mice. American Journal of Physiology Heart and Circulatory Physiology. 2002;283:H976–H982. doi: 10.1152/ajpheart.00088.2002. [DOI] [PubMed] [Google Scholar]
- 15.Belke DD, Larsen TS, Gibbs EM, Severson DL. Altered metabolism causes cardiac dysfunction in perfused hearts from diabetic (db/db) mice. American Journal of Physiology Endocrinology and Metabolism. 2000;279:E1104–E1113. doi: 10.1152/ajpendo.2000.279.5.E1104. [DOI] [PubMed] [Google Scholar]
- 16.Buchanan J, Mazumder PK, Hu P, Chakrabarti G, Roberts MW, et al. Reduced cardiac efficiency and altered substrate metabolism precedes the onset of hyperglycemia and contractile dysfunction in two mouse models of insulin resistance and obesity. Endocrinology. 2005;146:5341–5349. doi: 10.1210/en.2005-0938. [DOI] [PubMed] [Google Scholar]
- 17.Yue P, Arai T, Terashima M, Sheikh AY, Cao F, et al. Magnetic resonance imaging of progressive cardiomyopathic changes in the db/db mouse. American Journal of Physiology Heart and Circulatory Physiology. 2007;292:H2106–H2118. doi: 10.1152/ajpheart.00856.2006. [DOI] [PubMed] [Google Scholar]
- 18.Boden G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes. 1997;46:3–10. [PubMed] [Google Scholar]
- 19.Sidell RJ, Cole MA, Draper NJ, Desrois M, Buckingham RE, et al. Thiazolidinedione treatment normalizes insulin resistance and ischemic injury in the zucker fatty rat heart. Diabetes. 2002;51:1110–1117. doi: 10.2337/diabetes.51.4.1110. [DOI] [PubMed] [Google Scholar]
- 20.Nesto RW, Bell D, Bonow RO, Fonseca V, Grundy SM, et al. Thiazolidinedione use, fluid retention, and congestive heart failure: a consensus statement from the American Heart Association and American Diabetes Association. Diabetes Care. 2004;27:256–263. doi: 10.2337/diacare.27.1.256. [DOI] [PubMed] [Google Scholar]
- 21.Carley AN, Semeniuk LM, Shimoni Y, Aasum E, Larsen TS, et al. Treatment of type 2 diabetic db/db mice with a novel PPARgamma agonist improves cardiac metabolism but not contractile function. American Journal of Physiology Endocrinology and Metabolism. 2004;286:E449–E455. doi: 10.1152/ajpendo.00329.2003. [DOI] [PubMed] [Google Scholar]
- 22.Dargie HJ, Hildebrandt PR, Riegger GnAJ, McMurray JJV, McMorn SO, et al. A randomized, placebo-controlled trial assessing the effects of rosiglitazone on echocardiographic function and cardiac status in type 2 diabetic patients with New York Heart Association Functional Class I or II Heart Failure. Journal of the American College of Cardiology. 2007;49:1696–1704. doi: 10.1016/j.jacc.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 23.Gilde AJ, van der Lee KAJM, Willemsen PHM, Chinetti G, van der Leij FR, et al. Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circulation Research. 2003;92:518–524. doi: 10.1161/01.RES.0000060700.55247.7C. [DOI] [PubMed] [Google Scholar]
- 24.Liu LS, Tanaka H, Ishii S, Eckel J. The new antidiabetic drug MCC-555 acutely sensitizes insulin signaling in isolated cardiomyocytes. Endocrinology. 1998;139:4531–4539. doi: 10.1210/endo.139.11.6310. [DOI] [PubMed] [Google Scholar]
- 25.Bishop-Bailey D. Peroxisome proliferator-activated receptors in the cardiovascular system. British Journal of Pharmacology. 2000;129:823–834. doi: 10.1038/sj.bjp.0703149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circulation Research. 2005;97:372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
- 27.Rockman MV, Hahn MW, Soranzo N, Loisel DA, Goldstein DB, et al. Positive selection on MMP3 regulation has shaped heart disease risk. Current Biology. 2004;14:1531–1539. doi: 10.1016/j.cub.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 28.Henney AM, Wakeley PR, Davies MJ, Foster K, Hembry R, et al. Localization of stromelysin gene expression in atherosclerotic plaques by in situ hybridization. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8154–8158. doi: 10.1073/pnas.88.18.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyzade S, Zhang S, Wong Y-k, Day INM, Eriksson P, et al. Influences of matrix metalloproteinase-3 gene variation on extent of coronary atherosclerosis and risk of myocardial infarction. Journal of the American College of Cardiology. 2003;41:2130–2137. doi: 10.1016/s0735-1097(03)00482-0. [DOI] [PubMed] [Google Scholar]
- 30.Shimoni Y. Inhibition of the formation or action of angiotensin II reverses attenuated K+ currents in type 1 and type 2 diabetes. The Journal of Physiology. 2001;537:83–92. doi: 10.1111/j.1469-7793.2001.0083k.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JE, Shi W, Wang HS, McDonald C, Yu H, et al. Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circulation Research. 1996;79:659–668. doi: 10.1161/01.res.79.4.659. [DOI] [PubMed] [Google Scholar]
- 32.Lesage F, Lazdunski M. Molecular and functional properties of two-pore-domain potassium channels. American Journal of Physiology Renal, Fluid and Electrolyte Physiology. 2000;279:F793–F801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee S, Kolmakova A, Miller M. The role of the phospholipid sphingomyelin in heart disease. Current Opinion in Investigational Drugs. 2006;7:219–228. [PubMed] [Google Scholar]
- 34.Levade T, Augé N, Veldman RJ, Cuvillier O, Nègre-Salvayre A, et al. Sphingolipid mediators in cardiovascular cell biology and pathology. Circulation Research. 2001;89:957–968. doi: 10.1161/hh2301.100350. [DOI] [PubMed] [Google Scholar]
- 35.Unger RH, Orci L. Lipoapoptosis: its mechanism and its diseases. Biochimica et Biophysica Acta. 2002;1585:202–212. doi: 10.1016/s1388-1981(02)00342-6. [DOI] [PubMed] [Google Scholar]
- 36.Baranowski M, Blachnio A, Zabielski P, Gorski J. Pioglitazone induces de novo ceramide synthesis in the rat heart. Prostaglandins & Other Lipid Mediators. 2007;83:99–111. doi: 10.1016/j.prostaglandins.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhou YT, Grayburn P, Karim A, Shimabukuro M, Higa M, et al. Lipotoxic heart disease in obese rats: implications for human obesity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1784–1789. doi: 10.1073/pnas.97.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hug C, Lodish HF. The role of the adipocyte hormone adiponectin in cardiovascular disease. Current Opinion in Pharmacology. 2005;5:129–134. doi: 10.1016/j.coph.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Pineiro R, Iglesias MaJ, Gallego Ra, Raghay K, Eiras S, et al. Adiponectin is synthesized and secreted by human and murine cardiomyocytes. FEBS Letters. 2005;579:5163–5169. doi: 10.1016/j.febslet.2005.07.098. [DOI] [PubMed] [Google Scholar]
- 40.Natarajan R, Salloum FN, Fisher BJ, Kukreja RC, Fowler AA. Hypoxia inducible factor-1 upregulates adiponectin in diabetic mouse hearts and attenuates post-ischemic injury. Journal of Cardiovascular Pharmacology. 2008;51:178–187. doi: 10.1097/FJC.0b013e31815f248d. [DOI] [PubMed] [Google Scholar]
- 41.Chen MM, Ashley EA, Deng DXF, Tsalenko A, Deng A, et al. Novel role for the potent endogenous inotrope apelin in human cardiac dysfunction. Circulation. 2003;108:1432–1439. doi: 10.1161/01.CIR.0000091235.94914.75. [DOI] [PubMed] [Google Scholar]
- 42.Iwanaga Y, Kihara Y, Takenaka H, Kita T. Down-regulation of cardiac apelin system in hypertrophied and failing hearts: Possible role of angiotensin II-angiotensin type 1 receptor system. Journal of Molecular and Cellular Cardiology. 2006;41:798–806. doi: 10.1016/j.yjmcc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Lincoff AM, Wolski K, Nicholls SJ, Nissen SE. Pioglitazone and risk of cardiovascular events in patients with type 2 diabetes mellitus: a meta-analysis of randomized trials. JAMA. 2007;298:1180–1188. doi: 10.1001/jama.298.10.1180. [DOI] [PubMed] [Google Scholar]
- 44.Sears DD, Hsiao A, Ofrecio JM, Chapman J, He W, et al. Selective modulation of promoter recruitment and transcriptional activity of PPARgamma. Biochemical and Biophysical Research Communications. 2007;364:515–521. doi: 10.1016/j.bbrc.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Zhang L, Sun Y, Muskhelishvili L, Blann E, et al. Differences in hepatotoxicity and gene expression profiles by anti-diabetic PPAR gamma agonists on rat primary hepatocytes and human HepG2 cells. Molecular Diversity. 2006;10:349–360. doi: 10.1007/s11030-006-9038-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Upregulated Gene Ontology pathways with rosiglitazone-treated db/db mice.
(0.03 MB RTF)
Downregulated Gene Ontology pathways with rosiglitazone-treated db/db mice.
(0.02 MB RTF)
Upregulated Gene Ontology pathways in untreated db/db mice.
(0.04 MB RTF)
Downregulated Gene Ontology pathways in untreated db/db mice.
(0.05 MB RTF)
txt file of microarray results from untreated db/db vs. db/+ analysis
(0.15 MB TXT)
txt file of microarray results from db/db+rosiglitazone vs db/db analysis.
(0.04 MB TXT)