Abstract
DNA inverted repeats (IRs) are hotspots of genomic instability in both prokaryotes and eukaryotes. This feature is commonly attributed to their ability to fold into hairpin- or cruciform-like DNA structures interfering with DNA replication and other genetic processes. However, direct evidence that IRs are replication stall sites in vivo is currently lacking. Here, we show by 2D electrophoretic analysis of replication intermediates that replication forks stall at IRs in bacteria, yeast, and mammalian cells. We found that DNA hairpins, rather than DNA cruciforms, are responsible for the replication stalling by comparing the effects of specifically designed imperfect IRs with varying lengths of their central spacer. Finally, we report that yeast fork-stabilizing proteins, Tof1 and Mrc1, are required to counteract repeat-mediated replication stalling. We show that the function of the Tof1 protein at DNA structure-mediated stall sites is different from its previously described effect on protein-mediated replication fork barriers.
Keywords: fork stalling, DNA palindrome, genome instability
The DNA repeats capable of forming unusual secondary structures are common elements of various genomes and were implicated in mutagenesis and gross chromosomal rearrangements leading to human disease (reviewed in refs. 1 and 2). Inverted repeats (IRs) are among the best-studied examples of such DNA motifs. IRs are abundant in both prokaryotic and eukaryotic genomes (3, 4). They can form DNA hairpins in single-stranded state or DNA cruciforms in double-stranded state (5, 6). IRs, and other structure-forming sequences, have been shown to block DNA replication in vitro (7–11). In vivo, the most remarkable biological property of IRs is their propensity to induce genomic instability in a wide variety of organisms. In bacteria, IRs are highly unstable, which instability is known to depend on DNA replication (12). In eukaryotes, they lead to dsDNA breaks (13–16) and chromosomal fragility (17), stimulate homologous recombination (13, 18–20), and induce gross chromosomal rearrangements, such as translocations and deletions (13, 16, 21–26).
It was hypothesized that formation of stable secondary DNA structures by IRs may cause replication stalling that is, in turn, responsible for genome instability (27–29). However, there is no direct evidence of the replication stalling at IRs in vivo. Furthermore, it was not clear what secondary structure, DNA hairpin, or DNA cruciform could be responsible for the replication fork blockage. Here, we looked at the replication fork progression through long IRs in prokaryotes and eukaryotes, using 2D gel electrophoretic analysis of replication intermediates. We show that replication fork stalling at long IRs in vivo is a universal phenomenon that occurs in bacteria, yeast, and mammalian cells. We further addressed the mechanism of fork arrest by specifically modifying the IRs so that we differentially affected the formation of hairpin and cruciform structures in vivo. Our data indicate that fork stalling is caused by DNA hairpins likely formed by IRs during the lagging strand synthesis.
The stabilization of stalled replication forks is fundamental for preventing genomic instability in eukaryotes (reviewed in ref. 30). This function primarily depends on the replication checkpoint proteins of the ATR pathway, including yeast Mrc1p (mammalian claspin) (reviewed in ref. 31). More recently, it was shown that stalled replication forks are additionally stabilized in a checkpoint-independent manner where the key players were yeast Tof1p (mammalian timeless) and Mrc1p (32, 33). Interestingly, Tof1p in a complex with the Csm3 protein is also implicated in the fork arrest at the protein-mediated replication fork barriers (34, 35). Therefore, we studied the role of fork-stabilizing proteins at DNA structure-mediated replication stalls. We found that both Tof1 and Mrc1 proteins alleviate the replication fork stalling at DNA hairpins. This result implies that the function of Tof1 protein is fundamentally different at hairpin-mediated replication stalls versus protein-mediated fork barriers.
Results
Replication Fork Stalling at DNA IRs Is Structure-Mediated in Both Prokaryotes and Eukaryotes.
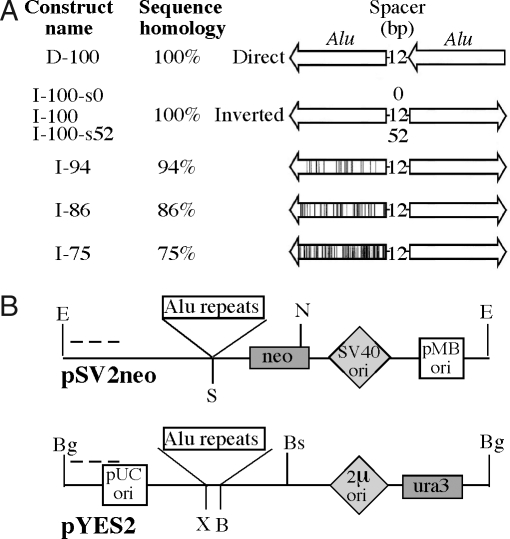
The most frequently occurring long IRs in the human genome are inverted Alu repeats (36). In yeast, these repeats are hotspots of homologous recombination and chromosomal fragility (13, 16, 18, 37). We, thus, analyzed the effects of Alu IRs on the replication fork progression in bacterial, yeast, and primate cells by using 2D electrophoresis of replication intermediates (38). Our basic constructs (Fig. 1) consisted of two identical 320-bp-long human Alu elements placed in either direct or inverted orientations relative to each other and separated by 12-bp-long spacers. In addition, Alu IRs of 94%, 86%, and 75% homology between the repetitive halves were used to assess the effect of sequence divergence on the replication fork progression. We also varied the length of the central spacer from 0 to 52 bp for the IR with 100% sequence homology. (Fig. 1A).
Fig. 1.
Alu repeat plasmid constructs. (A) Alu repeat constructs used (arrows point to the 3′ ends of the Alu sequences) (20). (B) (Upper) Alu repeats cloned into the SmaI site (S) of the pSV2neo plasmid. The EcoRI (E), NcoI (N) fragment was analyzed by 2D electrophoresis. (Lower) Alu repeats cloned into the multiple cloning site of pYES2, between XhoI (X) and BamHI (B) sites. The BglI (Bg), BseRI (Bs) fragment was analyzed by 2D electrophoresis. Dashed lines indicate hybridization probes.
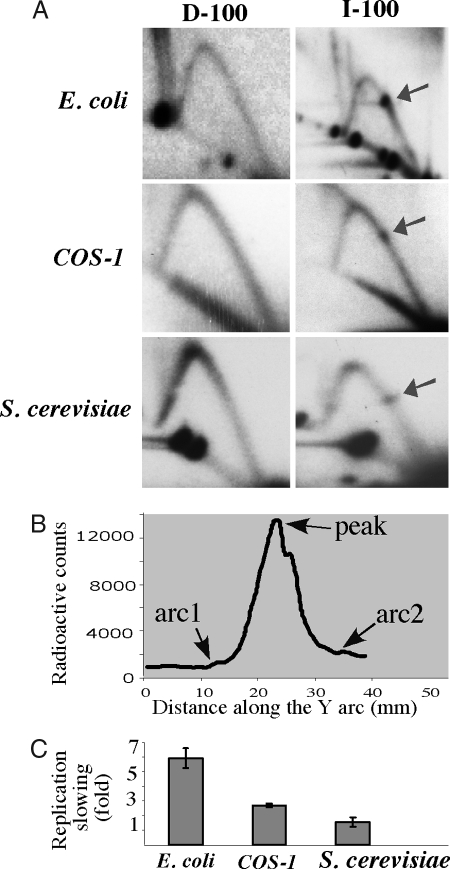
Inverted Alu repeats caused a profound replication blockage in Escherichia coli. (Fig. 2A). A quantitative analysis of this block, defined as the ratio between the maximum radioactive count at the stall site and that at the smooth arc (Fig. 2B), showed that IR slowed DNA replication 6-fold (Fig. 2C). Although this replication stalling is quite severe, it is not complete, as the Y arc continues past the bulge. Direct Alu repeats that have the same sequence composition as the inverted Alus, but cannot adopt secondary structures, did not affect the replication fork progression. Thus, replication blockage caused by the IRs is likely a result of their secondary structure, rather than sequence-specific protein binding.
Fig. 2.
Replication forks stall at inverted Alu repeats in E. coli, S. cerevisiae, and COS-1 cells (A) 2D gels showing replication through direct (D-100) and inverted (I-100) Alu repeats with 100% sequence homology in E. coli, COS-1 cells, and S. cerevisiae. Arrows indicate replication stall sites. (B) Radioactive signal along the Y arc containing a replication stall site. The strength of the replication slowing was measured as the ratio between the maximum radioactive count at the stall site (peak) and that at the smooth arc (average between two flanking points on the Y arc, arc1 and arc2). (C) Quantitative analysis of the replication stalls shown in A.
Our vectors used for E. coli replication studies contain an SV40 origin that can support their episomal replication in T-antigen transformed cells, such as COS-1 monkey fibroblasts. The T-antigen functions both as an initiator and the replicative helicase, whereas other components of the replisome come from the host cell (reviewed in ref. 39). Replication intermediates from COS-1 cells were isolated 24 h after transfection, extensively digested with DpnI to eliminate unreplicated plasmid DNA, and analyzed by 2D gel-electrophoresis. Mammalian replication forks stalled at the Alu-IR, whereas direct repeats replicated smoothly (Fig. 2A). Note that the IR in this case spans nearly a quarter of the restriction fragment, yet the stalling is limited to a discrete spot, suggesting that the replication fork pauses consistently at the same site. Quantitatively, replication was slowed down 2.7-fold (Fig. 2C), a substantial inhibition, albeit less prominent than that observed in bacteria. The replication analysis of yeast pYES2 plasmids carrying direct and inverted Alu repeats showed a distinct replication stall site at the Alu IR, which was qualitatively and quantitatively similar to that in mammalian cells (Fig. 2 A and C). We conclude that replication fork pausing at long IRs is a universal phenomenon that occurs in prokaryotes and in lower and higher eukaryotes.
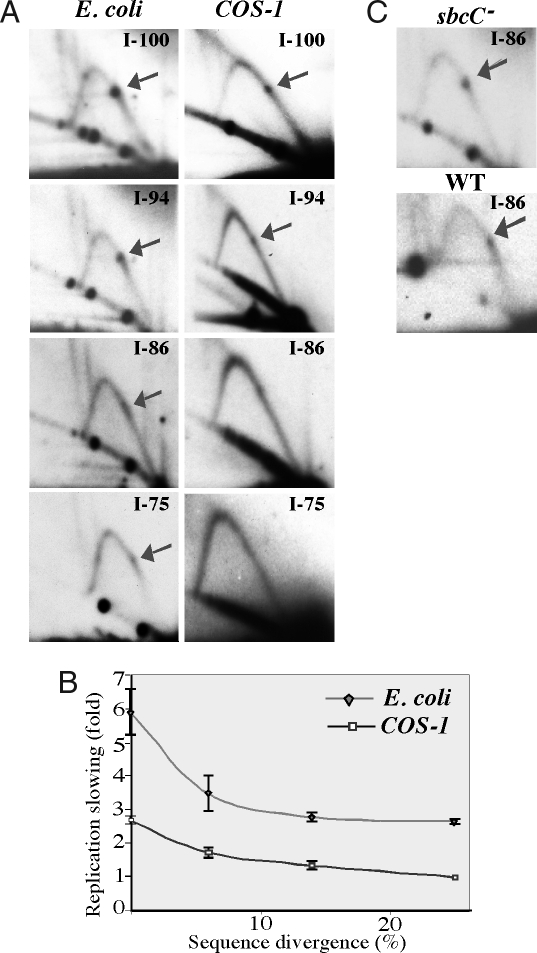
As mentioned above, IRs can form DNA hairpins and cruciforms. As these secondary structures are stabilized by intrastrand base-pairing, their formation and stability depend on the degree of sequence homology between the two repeated halves. To confirm that replication stalling at the Alu IR was caused by a DNA structure, we analyzed quasipalindromic repeats with different levels of sequence homology between their Alu halves. The diverged Alu IRs with 94%, 86%, and 75% homology contain base pair mismatches, small deletions, or insertions. In E. coli, the severity of the replication stalling at Alu IRs decreased markedly with the increase in sequence divergence (Fig. 3 A and B). As mentioned above, the replication slowdown caused by a 100% homologous IR is exceptionally strong (6-fold), thus mild replication blockage is still evident even at 75% sequence homology. Because our replication studies were performed in the SURE 2 strain deficient in the SbcC nuclease, which cleaves hairpin and cruciform DNA structures (27), we wondered whether the lack of this enzyme contributes to the profound replication stalling. Since imperfect IRs can be maintained in the WT E. coli strains, we looked at the replication stalling caused by the Alu IR with 86% sequence homology in the sbcC+ (DH5α) strain. We found no differences in the intensity of the IR-mediated replication stalling between the latter strain and the sbcC− (SURE 2) cells (Fig. 3C).
Fig. 3.
Replication stalling at the inverted Alu repeats depends on the level of sequence homology and is independent of the presence of the SbcC nuclease. (A) 2D gels showing replication through Alu IRs with different degrees of sequence homology in E. coli and COS-1 cells. (B) Quantitative analysis of the replication stalling caused by Alu IRs with various sequence divergence. ◆, E. coli; □, COS-1 cells. (C) Replication fork stalling at Alu IRs with 86% homology in the WT and sbcC− E. coli strains.
The replication fork slowdown at the Alu IRs strongly depended on their sequence homology in mammalian cells as well. The replication fork block was 1.6-fold weaker at 94% sequence homology, 2-fold weaker at 86% sequence homology, and virtually eliminated at 75% homology (Fig. 3 A and B). The dependence of the replication stalling on the repeat's sequence homology demonstrated the same pattern in eukaryotes as in bacteria. This tendency strongly indicates that a secondary structure formed by the IR is responsible for the replication blockage in both prokaryotes and eukaryotes.
Replication Fork Stalling at IRs Is Caused by DNA Hairpins.
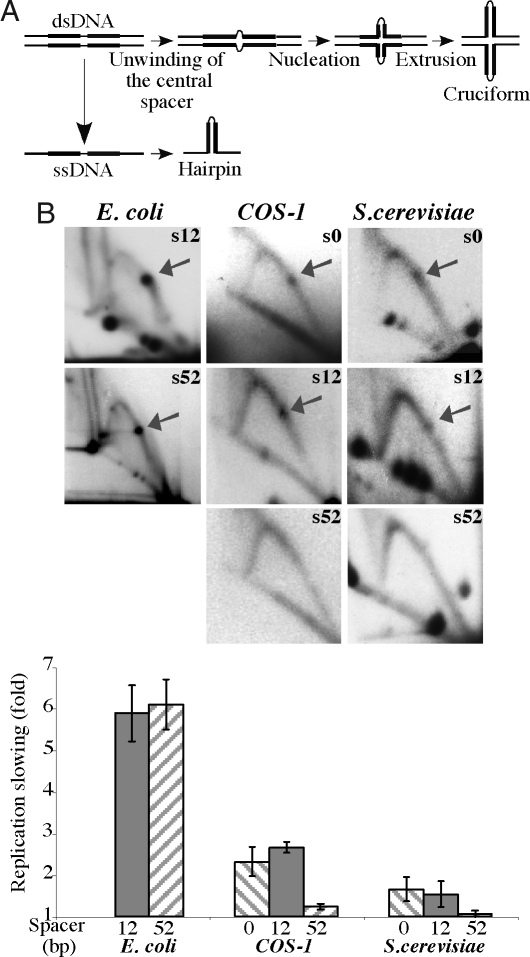
To determine whether the fork is stalled by preexisting DNA cruciforms or DNA hairpins (Fig. 4A) that could be formed during replication, we compared the replication-inhibition potentials of inverted Alu repeats that have 100% sequence identity but differ in the length of their central spacers. As the initial step of the cruciform extrusion requires complete unwinding of the dsDNA spacer at the center of the IR, the likelihood of the cruciform formation decreases drastically even with a modest increase in the spacer's length (40). At the same time, formation of the hairpin in the ssDNA is much less sensitive to the spacer's length. Thus, varying the length of the central spacer provides an opportunity to tune the rate of the cruciform extrusion in vivo without affecting the hairpin formation or stability. Based on the previous analysis (41), an increase in the spacer length from 12 to 52 bp should make cruciform extrusion virtually impossible. We observe, however, that in E. coli cells the intensities of the replication stalling caused by Alu IRs separated by these two spacers are indistinguishable (Fig. 4B). We conclude that replication blockage caused by IRs in bacteria is due to the formation of DNA hairpins.
Fig. 4.
Replication fork stalling at inverted Alu repeats with varying lengths of the central spacer. (A) Schematic representation of hairpin and cruciform structure formation by an IR. (B) 2D gels and quantitative analysis of replication stalling at Alu IRs with 100% sequence homology and either 0 bp (s0), 12 bp (s12) or 52 bp (s52) spacers in E. coli, COS-1 cells, and S. cerevisiae
DNA hairpins likely form on the lagging strand DNA template, when an IR within the single-stranded Okazaki initiation zone (OIZ) snaps back into the stem-loop structure (42). Thus, the size of the OIZ might be a limiting factor in the process of hairpin formation. In prokaryotes, the OIZ is at least 1.5 kb long, making it possible for the entire 650-bp Alu IR to fold into a hairpin. In eukaryotes, the OIZ is ≈200 bp long; thus, only a third of the Alu IR can fold into a hairpin during DNA replication (43, 44), and only if OIZ overlaps with the center of the repeat. Therefore, increasing the length of the IR's central spacer by 40 bp would abolish both its cruciform- and hairpin-forming potential in eukaryotes. Alternatively, we completely eliminated the central spacer, which would markedly increase the rate of cruciform extrusion (40), without changing the rate of the hairpin formation.
Our data show that changing the length of the repeat's central spacer from 12 to 0 bp has no effect on the replication fork stalling in either primate cells or yeast (Fig. 4B). As expected, we did not detect the replication blockage at the IR with a 52-bp-long spacer in eukaryotic cells (Fig. 4B), which supports the hypothesis that hairpin structures are formed on the lagging strand template. Overall, our data indicate that the replication fork pausing at IRs is caused by the formation of the DNA hairpins, likely during lagging strand synthesis in both prokaryotes and eukaryotes.
Mrc1 and Tof1 Stabilize Replication Forks Stalled at DNA Hairpins in Yeast.
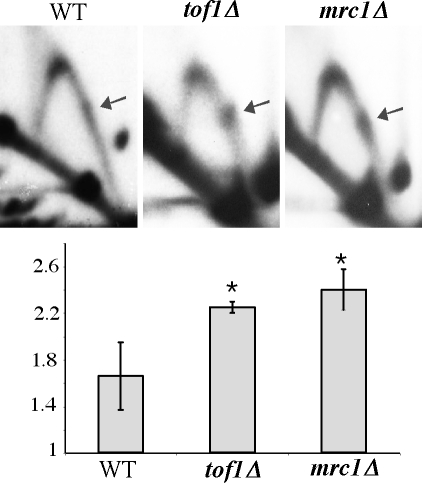
The Saccharomyces cerevisiae, Mrc1, and Tof1 proteins, as well as their Schizosaccharomyces pombe and mammalian homologues, are involved in stabilizing stalled replication forks and in replication fork progression under normal conditions (reviewed in ref. 45). We thus compared the replication fork progression through a perfect Alu IR in the WT, tof1Δ, and mrc1Δ strains of S. cerevisiae. The IR-mediated fork stalling was markedly increased in the tof1Δ strain compared with the WT (Fig. 5). This result was somewhat unexpected, as it was previously reported that protein-mediated replication stalling at replication fork barriers decreases significantly in a Tof1 knockout strain (34, 35). Therefore, our results indicate that Tof1p functions primarily as a fork stabilizer at DNA structure-mediated stalls, while it facilitates the replication fork pausing at protein–DNA complexes.
Fig. 5.
Tof1 and Mrc1 proteins counteract replication fork stalling at inverted Alu repeats. (Upper) 2D analysis of replication of perfect inverted Alu repeats (I-100-s0) in the WT, tof1Δ, and mrc1Δ strains of S. cerevisiae. (Lower) Quantitative analysis of replication stalls shown in A. P values are 0.025 for tof1Δ and 0.018 for mrc1Δ.
In the mrc1Δ background, we also observed an increase in the replication fork arrest at the IR, consistent with a role of Mrc1 in stabilizing stalled replication forks at hairpin structures (Fig. 5). The increase over WT was very similar in the Mrc1 and Tof1 knockout strains. These results show that both Tof1 and Mrc1 counteract replication fork stalling at DNA secondary structures, and that they are equally important for fork stabilization at these stall sites.
Discussion
We show here that long IRs are replication stall sites in bacteria, yeast, and primate cells. The replication inhibition at Alu IRs could be caused by the repeat's secondary structure or a protein bound to it. Sequence-specific protein binding can be ruled out, as the replication forks progress smoothly through direct Alu repeats (Fig. 2). One could also envision a protein that binds specifically to the secondary structure formed by the repeat, inhibiting replication. Such a protein should stabilize this secondary structure, however, which is inconsistent with a profound decrease in the replication stalling upon a modest 6% decrease in sequence homology between the repeat halves (Fig. 3). We conclude, therefore, that the replication blockage at the IR is caused by its secondary DNA structure.
IRs can adopt two types of secondary structures: DNA hairpins and DNA cruciforms (46), which were both implicated in genomic instability (37). We show that replication stalling at long IRs is mediated by DNA hairpins rather than cruciforms. By modifying the length of the spacer region, we drastically changed the likelihood of cruciform extrusion without affecting the hairpin formation (40). Yet decreasing the spacer's length from 12 to 0 bp did not alter repeat-mediated replication blockage in any of our systems. Furthermore, the spacer can be increased up to 52 bp in bacteria without consequence for the replication blockage. Consistent with our conclusions, the SbcC nuclease induces double-stranded breaks at long IRs in E. coli, by acting primarily on DNA hairpins (47). Because the presence of the SbcC nuclease did not affect the severity of replication stalling at long IRs (Fig. 3), the cleavage must occur after the replication fork escapes the stall site (Fig. 6Left).
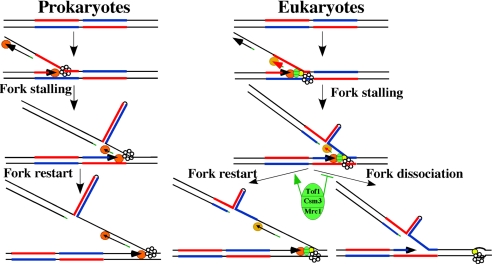
Fig. 6.
A model for the hairpin-mediated replication fork stalling in prokaryotes and eukaryotes. Blue and red lines, halves of an IR; orange circles, DNA polymerases; hexameric rings, DNA helicases, green oval, Tof1/Csm3/Mrc1 complex; yellow circle, Cdc45p.
The transition of long IRs into hairpin-like structures requires significant unwinding of the dsDNA. This event occurs during DNA replication (Fig. 6), where a portion of the lagging strand template stays single-stranded to allow priming of the Okazaki fragments (reviewed in ref. 44). The hairpin can thus be formed when an IR overlaps with the OIZ and stalls lagging strand synthesis (Fig. 6). How could this effect translate into an arrest of the whole replisome? When a lesion on the lagging strand template arrests the lagging strand polymerase, the leading strand synthesis continues and the lagging strand polymerase is eventually released and reprimed downstream of the lesion. The polymerase release is triggered by the accumulation of ssDNA resulting from the uncoupling between the helicase and polymerase (48). In the case of repeat-mediated blockage, this restart mechanism could be impaired, as ssDNA exposed by the advancing helicase might continue folding into a hairpin (Fig. 6).
Our data support the idea that the length of the OIZ is a limiting factor in the formation of DNA hairpins during DNA replication. First, in prokaryotes, where the OIZ is much longer than in eukaryotes, IRs cause a much stronger fork arrest (Fig. 2), likely because of the formation of longer and more stable hairpins (Fig. 6 Left). Second, increasing the length of the central spacer of the IRs up to 52 bp abolished the replication fork stalling in eukaryotes (Fig. 4). As the OIZ in eukaryotes is only ≈200 nt long (43), the longer central spacer would significantly decrease both the likelihood and the area of overlap between the IR and OIZ. This might explain the tolerance of eukaryotes to long palindromes, as only relatively short hairpins can extrude during DNA replication (Fig. 6 Right).
The fork-stabilizing proteins Mrc1, Tof1, and Csm3 are required to prevent dissociation of replisomes stalled upon cell treatment with hydroxyurea (32). Mrc1 is also required to maintain the optimal replication speed under normal growth conditions (49). Fork pausing at replication fork barriers, in contrast, is facilitated by the Tof1 protein (33–35), which was attributed to the Tof1p ability to counteract the Rrm3 helicase (35). We show that the severity of the replication stalling at DNA hairpins increases significantly in the tof1Δ and mrc1Δ backgrounds. The two proteins seem to be equally potent in stabilizing the replisome (Fig. 5). Thus, both Tof1 and Mrc1 proteins may act as fork stabilizers at DNA structure-mediated stalls. Maintaining the normal architecture of the replisome at the stall site by fork-stabilizing protein would allow the efficient restart of the lagging strand synthesis (Fig. 6 Right). At the same time, the replication-pausing function of Tof1 protein did not apply to hairpin-mediated replication stalls, implying that the Rrm3 helicase is not involved in fork progression through DNA hairpins, consistent with our model of hairpin structure formation behind the advancing helicase.
Materials and Methods
Strains and Plasmids.
Cloning was carried out in the E. coli SURE 2 strain (Stratagene). Replication studies were performed in E. coli SURE 2 strain, S. cerevisiae CH1585 strain (MATa leu2-Δ1, trp1-Δ63, ura3–52, his3–200), and COS-1 fibroblasts (ATCC CRL-1650). The tof1Δ and mrc1Δ strains were obtained by one-step gene disruption using a kanamycin cassette PCR-amplified from the pFA6A-KanMX4 plasmid (50). Various Alu repeats were cloned from pHS plasmids (20) into pSV2neo and pYES2 [see Fig. 1 and supporting information (SI) Text and Table S1].
Isolation of Replication Intermediates and 2D Electrophoresis.
Replication intermediates from E. coli were isolated as described (51). Yeast replication intermediates were isolated according to ref. 52 with minor modifications (see SI Text). Replication intermediates from COS-1 cells were isolated by using Hirt's protocol (53) (see SI Text). 2D electrophoresis was carried out as described (54).
Quantitative Analysis of 2D Gels.
Quantitative analysis of 2D gels was performed on either a Storm 860 PhosphorImager using Imagequant software or a BioRad Pharos FX PhosphorImager using Quantity One software. The severity of replication fork slowdown was calculated as the ratio between the maximum radioactive count of the bulge (Fig. 4A) and an average between the radioactive counts of two points on the adjacent arc (Fig. 4A). The values for each construct are averages of at least three independent experiments with corresponding standard deviations. The comparison of replication slowing in WT and mutant yeast strains were carried out by t test. Differences were considered significant for P < 0.05.
Supplementary Material
Acknowledgments.
We thank George Samadashwily and Michael Resnick for initial input to this study; Vera Egorova, Maria Krasilnikova, and Alexander Shishkin for experimental help; Catherine Freudenreich, Pat Higgins, David Leach, Susan Lovett, David Lilley, Bernardo Schvartzman, and anonymous reviewers for useful comments and suggestions; Ranjith Anand, Nicole Cherng, Rangapriya Sundararajan, and Christine Surka for proofreading this manuscript; and John and Penny White for their generous contribution. This study was supported by National Institutes of Health Grant GM60987 (to S.M.) and National Science Foundation Grant MCB-0417088 (to K.S.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804510105/DCSupplemental.
References
- 1.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Wells RD. Non-B DNA conformations, mutagenesis, and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Cox R, Mirkin SM. Characteristic enrichment of DNA repeats in different genomes. Proc Natl Acad Sci USA. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA-forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley DMJ. The inverted repeat as a recognizable structural feature in supercoiled DNA. Proc Natl Acad Sci USA. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panayotatos N, Wells RD. Cruciform structures in supercoiled DNA. Nature. 1981;289:466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- 7.Chalberg MD, Englund PT. The effect of template secondary structure on vaccinia DNA polymerase. J Biol Chem. 1979;254:7820–7826. [PubMed] [Google Scholar]
- 8.Kaguni LS, Clayton DA. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci USA. 1982;79:983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman LA, Gefter ML. Studies of the mechanism of enzymatic DNA elongation by Escherichia coli DNA polymerase II. J Mol Biol. 1976;103:61–76. doi: 10.1016/0022-2836(76)90052-8. [DOI] [PubMed] [Google Scholar]
- 10.Weaver DT, DePamphilis ML. Specific sequences in native DNA that arrest synthesis by DNA polymerase alpha. J Biol Chem. 1982;257:2075–2086. [PubMed] [Google Scholar]
- 11.Kang S, Ohshima K, Shimizu M, Amirhaeri S, Wells RD. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J Biol Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 12.Lindsey JC, Leach DR. Slow replication of palindrome-containing DNA. J Mol Biol. 1989;206:779–782. doi: 10.1016/0022-2836(89)90584-6. [DOI] [PubMed] [Google Scholar]
- 13.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 14.Nag DK, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasar F, Jankowski C, Nag DK. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol Cell Biol. 2000;20:3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobachev KS, et al. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farah JA, Hartsuiker E, Mizuno K, Ohta K, Smith GR. A 160-bp palindrome is a Rad50. Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobachev KS, et al. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 22.Nag DK, Suri M, Stenson EK. Both CAG repeats and inverted DNA repeats stimulate spontaneous unequal sister-chromatid exchange in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5677–5684. doi: 10.1093/nar/gkh901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 25.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 26.Kurahashi H, et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair. 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leach DR. Long DNA palindromes, cruciform structures, genetic instability, and secondary structure repair. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 29.Lebofsky R, Bensimon A. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol. 2005;25:6789–6797. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Exp Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Paulsen RD, Cimprich KA. The ATR pathway: Fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 33.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 34.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Leung FC. Long inverted repeats in eukaryotic genomes: Recombinogenic motifs determine genomic plasticity. FEBS Lett. 2006;580:1277–1284. doi: 10.1016/j.febslet.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 37.Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- 38.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 39.Borowiec JA, Dean FB, Bullock PA, Hurwitz J. Binding and unwinding–how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 40.Sinden RR, Zheng GX, Brankamp RG, Allen KN. On the deletion of inverted repeated DNA in Escherichia coli: Effects of length, thermal stability, and cruciform formation in vivo. Genetics. 1991;129:991–1005. doi: 10.1093/genetics/129.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vologodskii AV, Frank-Kamenetskii MD. The relaxation time for a cruciform structure in superhelical DNA. FEBS Lett. 1983;160:173–176. doi: 10.1016/0014-5793(83)80960-0. [DOI] [PubMed] [Google Scholar]
- 42.Mirkin SM. DNA structures, repeat expansions, and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.DePamphilis ML. Eukaryotic DNA replication fork. Chemtracts Biochem Mol Biol. 2002;15:313–325. [Google Scholar]
- 44.DePamphilis ML, Wassarman PM. Replication of eukaryotic chromosomes: A close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- 45.Labib K, Hodgson B. Replication fork barriers: Pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinden RR. DNA Structure and Function. San Diego: Academic; 1994. [Google Scholar]
- 47.Eykelenboom JK, Blackwood JK, Okely E, Leach DR. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 48.McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279:21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 49.Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 51.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu JR, Gilbert DM. Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res. 1995;23:3997–3998. doi: 10.1093/nar/23.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 54.Krasilnikova MM, Mirkin SM. Analysis of triplet repeat replication by two-dimensional gel electrophoresis. Methods Mol Biol. 2004;277:19–28. doi: 10.1385/1-59259-804-8:019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.