Abstract
The ETS gene family is frequently involved in chromosome translocations that cause human cancer, including prostate cancer, leukemia, and sarcoma. However, the mechanisms by which oncogenic ETS proteins, which are DNA-binding transcription factors, target genes necessary for tumorigenesis is not well understood. Ewing's sarcoma serves as a paradigm for the entire class of ETS-associated tumors because nearly all cases harbor recurrent chromosomal translocations involving ETS genes. The most common translocation in Ewing's sarcoma encodes the EWS/FLI oncogenic transcription factor. We used whole genome localization (ChIP-chip) to identify target genes that are directly bound by EWS/FLI. Analysis of the promoters of these genes demonstrated a significant over-representation of highly repetitive GGAA-containing elements (microsatellites). In a parallel approach, we found that EWS/FLI uses GGAA microsatellites to regulate the expression of some of its target genes including NR0B1, a gene required for Ewing's sarcoma oncogenesis. The microsatellite in the NR0B1 promoter bound EWS/FLI in vitro and in vivo and was both necessary and sufficient to confer EWS/FLI regulation to a reporter gene. Genome wide computational studies demonstrated that GGAA microsatellites were enriched close to EWS/FLI-up-regulated genes but not down-regulated genes. Mechanistic studies demonstrated that the ability of EWS/FLI to bind DNA and modulate gene expression through these repetitive elements depended on the number of consecutive GGAA motifs. These findings illustrate an unprecedented route to specificity for ETS proteins and use of microsatellites in tumorigenesis.
Keywords: ChIP-chip, transcription, gene regulation, ETS, NR0B1
ETS proteins are extremely important in human tumor development. The first ETS gene, v-ets, was initially identified as part of the E26 avian erythroblastosis virus and corresponded to the human ETS1 protooncogene (1, 2). Based on the presence of a DNA-binding ETS domain, 27 unique human ETS family members have been identified (3). ETS family members are frequently dysregulated and/or mutated in human cancers through chromosomal rearrangements. Indeed, the observation that ≈70% of prostate cancers harbor translocations between ETS genes (ERG, ETV1, or ETV4) and androgen-responsive genes indicates that ETS gene rearrangements may be the most common chromosomal abnormalities in human cancer (4, 5).
Most members of the ETS family bind to DNA sequences containing a GGAA (or in some cases, GGAT) core motif, with sequences flanking the GGAA core contributing to the affinity and specificity of the interaction (3, 6, 7). Because most cell lines examined express multiple ETS family members simultaneously, and because ETS factors in many cases are not functionally redundant, there are likely mechanisms to allow for gene-specific regulation by different ETS proteins (8). Indeed, recent whole genome localization studies have supported this concept by demonstrating that in vivo ETS-binding sites may be grouped into two classes (9): (i) high-affinity binding sites found close to transcription start sites and (ii) lower-affinity binding sites found in close proximity to low affinity binding sites for other transcription factors that allow for cooperative DNA binding.
Ewing's sarcoma was the first tumor in which ETS family members were shown to be involved in chromosomal translocations and serves as a paradigm for ETS-driven cancers (10). Ewing's sarcoma is a highly malignant solid tumor of children and young adults that usually harbors a recurrent chromosomal translocation, t(11;22)(q24;q12), that encodes the EWS/FLI fusion oncoprotein (10). The oncoprotein consists of a transcriptional activation domain from EWS, joined, in frame, to a region of the ETS transcription factor FLI harboring a DNA-binding domain (10–12). EWS/FLI functions as an aberrant transcription factor that regulates genes involved in the tumorigenic phenotype of Ewing's sarcoma (11–15).
EWS/FLI plays a critical role in establishing and maintaining the tumorigenic phenotype of Ewing's sarcoma cells (13, 15–18). Thus, EWS/FLI regulates its downstream target genes nonredundantly with other coexpressed ETS factors in Ewing's sarcoma (13, 15, 18). EWS/FLI gene expression signatures include genes that are important for Ewing's sarcoma oncogenesis, such as NR0B1 (13). EWS/FLI up-regulates NR0B1 in Ewing's sarcoma cells, and this up-regulation is required for their transformed phenotype (13). Whether NR0B1 is regulated by EWS/FLI directly, or through other intermediary proteins, is unknown. Indeed, the transcriptional response elements that EWS/FLI uses to regulate its target genes are largely unknown.
One difficulty in the study of EWS/FLI is that the human cell of origin of Ewing's sarcoma is not currently known. Thus, some studies of EWS/FLI function have relied on heterologous cell types as model systems, but results from these systems may not be applicable to the human disease (19, 20). We recently developed a system that allows for the study of EWS/FLI in a relevant model system: in Ewing's sarcoma itself (13, 15, 17, 20). To understand the mechanisms by which EWS/FLI regulates its target genes in Ewing's sarcoma itself, we undertook two parallel approaches, including a genome wide analysis of fusion protein-binding sites in patient-derived Ewing's sarcoma cells and a directed analysis of EWS/FLI-regulated promoters. We found that EWS/FLI uses GGAA-containing microsatellites to regulate some of its target genes, including its key oncogenic target NR0B1. This demonstrates a new role for microsatellites in human cancer and suggests a unique mechanism for ETS transcription factor regulation of target genes.
Results
EWS/FLI functions as an aberrant ETS-type transcription factor in Ewing's sarcoma to regulate genes involved in tumorigenesis (11). To identify genes that are direct EWS/FLI targets, we performed genome wide localization studies (“ChIP-chip”) of the endogenous fusion protein in patient-derived A673 Ewing's sarcoma cells. Because wild-type FLI protein is not expressed in these cells, the anti-FLI antibody used for these studies only immunoprecipitates EWS/FLI in this context (15). Agilent 244k promoter microarrays were used, which interrogate ≈17,000 human promoters from −5.5 kb to + 2.5 kb relative to the transcriptional start site. Approximately 900 genes were identified that were directly bound by EWS/FLI [supporting information (SI) Dataset S1 (XLS)]. These included previously identified direct targets, such as TGFBR2, CAV1, and IGFBP3 (18, 21, 22).
Of importance, the gene whose promoter was most highly enriched in the ChIP-chip dataset was NR0B1. NR0B1 is regulated by EWS/FLI in Ewing's sarcoma cells and is absolutely required for the oncogenic phenotype of Ewing's sarcoma (13). This earlier study was not able to determine whether NR0B1 was a direct or indirect target of EWS/FLI. The ChIP-chip data demonstrate that NR0B1 is bound by EWS/FLI and suggest that it may be regulated directly by the fusion oncoprotein. Inspection of the promoter did not reveal any sequences matching the previously identified in vitro high-affinity ETS-binding site, which also binds FLI and EWS-FLI with high affinity (ACCGGAAG/aT/c; data not shown; ref. 23). We, therefore, sought to understand the regulation of this key target to understand how EWS/FLI might function in the absence of a high-affinity binding site.
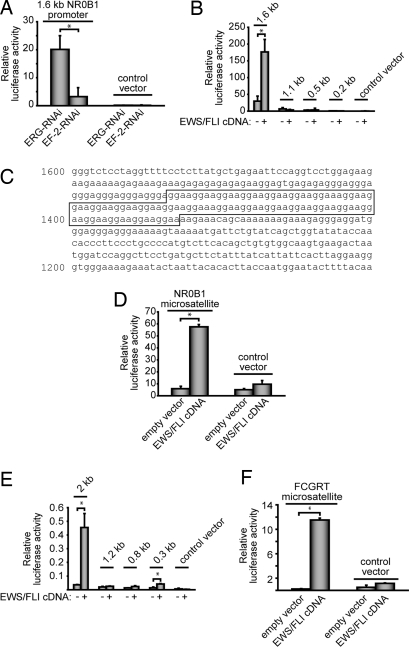
We cloned ≈1.6 kb of the NR0B1 promoter upstream of the luciferase cDNA and used this reporter construct to test for EWS/FLI responsiveness. Knock down of endogenous EWS/FLI (using the EF-2-RNAi retroviral construct) (15) showed that this promoter fragment was responsive to the fusion protein (Fig. 1A). This promoter fragment was also EWS/FLI-responsive in a heterologous cell type (293EBNA; Fig. 1B). These data support the notion that NR0B1 is a direct EWS/FLI target gene.
Fig. 1.
The GGAA microsatellites in the NR0B1 and FCGRT promoters are the EWS/FLI response elements. (A) TC71 Ewing's sarcoma cells cotransfected with a 1.6-kb NR0B1 promoter luciferase vector and either EF-2-RNAi (targeting EWS/FLI) or ERG-RNAi (negative control). The “control vector” does not contain NR0B1 promoter elements. The error bars the figure indicate SDs, and asterisks indicate P < 0.05. (B) 293EBNA cells cotransfected with the indicated NR0B1 promoter luciferase vectors (containing the indicated amount of promoter sequence upstream of the transcriptional start site) and an EWS/FLI (or empty control) cDNA expression vector. The control vector does not contain NR0B1 promoter elements. (C) NR0B1 promoter with GGAA microsatellite indicated. The microsatellite contains 25 GGAA repeats. (D) Luciferase assays in 293EBNA cells with the full 102-bp NR0B1 microsatellite upstream of a minimal promoter element. The control vector does not contain the microsatellite, but does contain the minimal promoter element. (E and F) Luciferase reporter assays by using 293EBNA cells and either the indicated FCGRT promoter deletion constructs or the isolated FCGRT GGAA microsatellite upstream of a minimal promoter, respectively. Of note, the FCGRT microsatellite is present at approximately −1.6 kb upstream of the transcriptional start site.
To identify the EWS/FLI response element in the NR0B1 promoter, a series of deletion constructs were tested in the luciferase reporter assay. We found that EWS/FLI responsiveness was contained within the −1.6 to −1.1 kb promoter region (Fig. 1B). This 500-bp region was also sufficient to confer EWS/FLI responsiveness to a minimal promoter derived from SV40 (data not shown).
Whereas the 500-bp NR0B1 promoter region did not contain high-affinity ETS-binding elements (ACCGGAAG/aT/c) (23), it did contain a 102-bp microsatellite consisting of 25 GGAA repeats (as well as two single base insertions; Fig. 1C). Because ETS family members, including EWS/FLI, bind with high affinity to consensus sequences containing a GGAA core element (9), we hypothesized that the GGAA microsatellite might represent a previously unrecognized in vivo EWS/FLI-binding element. We cloned the 102-bp GGAA microsatellite into a luciferase reporter construct that contained a minimal promoter derived from SV40. The microsatellite was sufficient to mediate EWS/FLI responsiveness in heterologous 293EBNA cells (Fig. 1D). Thus, the GGAA microsatellite is the EWS/FLI response element in the NR0B1 promoter.
In a parallel approach to identify potential EWS/FLI binding and response elements, we used an unbiased computational approach [multiple Em for motif elicitation (MEME)] (24) to identify sequence motifs that were enriched near the most highly EWS/FLI-bound DNA fragments identified in the ChIP-chip experiment. The MEME analysis identified a sequence (ACCGGAAGTG; E value = 1.4 × 10−40) that perfectly matched the previously described high-affinity ETS-binding element (23). However, the most highly enriched sequence identified by MEME was a repetitive GGAA motif (that corresponded to the microsatellite sequence in the NR0B1 promoter; GGAAGGAAGGAAGGAA; E value = 1 × 10−173). The GGAA-repetitive sequence element was identified in 12 of the 134 top EWS/FLI-bound gene promoters (9%) that were used for this analysis. There was virtually no overlap between promoter elements that harbored GGAA microsatellites vs. those that contained the high-affinity ETS-binding elements (data not shown).
To test whether other genes use their GGAA microsatellites as EWS/FLI response elements, we analyzed the FCGRT promoter. FCGRT was also identified as an EWS/FLI-bound target in the ChIP-chip analysis and contains a GGAA microsatellite at approximately −1.6 kb relative to its transcriptional start site. A 2-kb region of the FCGRT promoter was EWS/FLI-responsive in luciferase reporter assays (Fig. 1E). Deletion analysis of the promoter demonstrated that the GGAA microsatellite containing region was necessary for EWS/FLI responsiveness (Fig. 1E). Furthermore, the GGAA microsatellite was sufficient to confer EWS/FLI responsiveness to a reporter containing a minimal SV40 promoter (Fig. 1F). These data provide an independent confirmation of the role of GGAA microsatellites as EWS/FLI response elements.
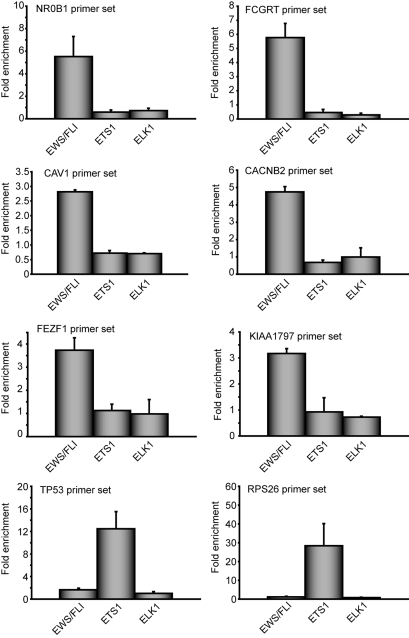
To further validate in vivo occupancy of GGAA microsatellites by EWS/FLI, directed ChIP experiments were performed at six microsatellite-containing genes: NR0B1, FCGRT, CAV1, CACNB2, FEZF1, and KIAA1797. Each of these genes contain GGAA microsatellites within 5 kb of their transcriptional start sites. We found that EWS/FLI bound to each of these GGAA microsatellite-containing promoters in vivo but not to control TP53 or RPS26 promoters, neither of which contain GGAA microsatellites (Fig. 2). None of the six microsatellite-containing promoters was significantly occupied by two other members of the ETS family, ETS1 or ELK1 (Fig. 2). Thus, binding of these promoters appears to be specific to EWS/FLI.
Fig. 2.
EWS/FLI occupies GGAA microsatellite containing promoters in vivo. Chromatin immunoprecipitation of the indicated promoters from A673 Ewing's sarcoma cells by using antibodies against FLI (which recognizes EWS/FLI), ETS1, or ELK1. Data are plotted as fold enrichment for each region compared to the average enrichment of two negative control genes. The error bars indicate SEMs of two to five independent experiments.
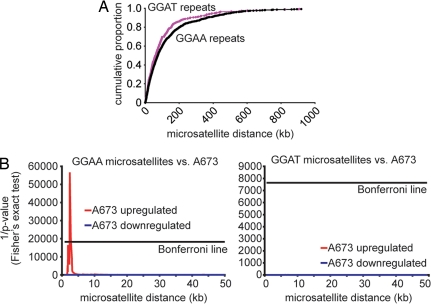
To determine whether EWS/FLI regulation through microsatellites is a generalized phenomenon in Ewing's sarcoma, we asked whether there was a correlation between the presence of a GGAA-containing microsatellite and EWS/FLI responsiveness. All 2,577 GGAA microsatellite-containing genes in the human genome were mapped, and the distances between the microsatellites and the transcriptional start sites were determined. As a control, all 942 GGAT microsatellite-containing genes were similarly identified and mapped. The distribution of GGAA and GGAT microsatellites relative to adjacent genes was similar (Fig. 3A). Genes were rank-ordered by distance between the microsatellite and the transcriptional start site. Fisher's exact test was then performed reiteratively to determine whether there was an over-representation of EWS/FLI-up- or -down-regulated genes at each rank position. The very conservative Bonferroni correction was applied to control for multiple hypothesis testing. A dataset derived from A673 cells showed significant enrichment of EWS/FLI-up-regulated genes within 5 kb of GGAA-containing microsatellites (Fig. 3B). EWS/FLI-down-regulated genes were not enriched, and there was no enrichment of EWS/FLI-regulated genes when compared to GGAT-containing microsatellites (Fig. 3B). A second independent EWS/FLI dataset derived from two other Ewing's sarcoma cell lines (TC71 and EWS502) showed a similar pattern of enrichment of EWS/FLI-regulated genes close to GGAA, but not GGAT, microsatellites (data not shown).
Fig. 3.
Enrichment of GGAA microsatellites in the promoters of EWS/FLI-up-regulated genes. (A) Cumulative portion of microsatellites (GGAA or GGAT) plotted as a function of distance between the microsatellites and the closest 5′ gene edge. (B) Correlation between EWS/FLI-up- and -down-regulated genes (in red and blue, respectively) and microsatellite distance analyzed by Fisher's exact test in the A673 Ewing's sarcoma cell line. Significant correlations cross over the “Bonferroni line” (see Methods). N.B., only up-regulated genes vs. GGAA microsatellites can be seen at the scales used.
A series of controls that included either randomly sampled gene sets or published “cancer gene neighborhood” gene sets [group C4 from the Molecular Signature Database (MsigDB version 2, January 2007 release; http://www.broad.mit.edu/gsea/msigdb/msigdb_index.html)] were tested in the same analyses. The A673 and TC71/EWS502 Ewing's sarcoma datasets were significantly enriched over the randomly sampled gene sets (P = 0.0005 and P = 0.0001, respectively) and were also significantly enriched as compared to the “cancer gene neighborhood” gene sets (P = 0.005 and P = 0.007, respectively). These data strongly suggest that the use of GGAA microsatellites as EWS/FLI response elements for gene up-regulation is not limited to NR0B1 and FCGRT but is more widespread.
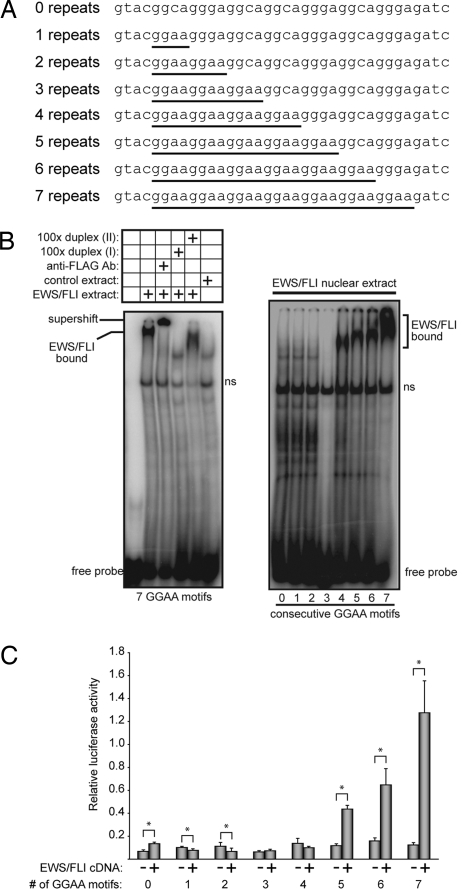
The in vivo occupancy and direct ChIP experiments suggested direct binding between the ETS domain of EWS/FLI and the microsatellite repeat. However, the GGAA tandem repeats are spaced too closely for each to be used as a binding site for an ETS domain. Site size requirement experiments indicate that an ETS domain requires at least 15 bp of DNA duplex, although only 9–10 bp show sequence preference (25). Furthermore, the GGAA flanks surrounding the GGAA core of a microsatellite/repetitive element do not create a sequence similar to the selected consensus site for FLI (23). To evaluate the binding properties of the repetitive elements, we performed electrophoretic mobility shift assays with DNA probes bearing variable number of repeats (Fig. 4A). Initially, nuclear extract from cells expressing recombinant 3xFLAG-EWS/FLI were used. EWS/FLI-specific DNA complexes were detected as confirmed by supershifts with anti-FLAG antibodies and competition experiments (Fig. 4B). A minimum of four repeats was necessary to detect binding (Fig. 4B). This result indicated that the 9- to 10-bp sequence centered within three repeats, GAAGGAAGGA, does not create a strong binding site and suggested that the affinity of EWS/FLI for the microsatellite might be enhanced by multiple binding events possible with additional repeats. Indeed, the mobility of the shifted complex was reduced with added number of repeats suggesting multiple binding events on the fragments bearing five, six, or seven repeats. Similar results were obtained with highly purified, recombinant FLI-derived protein (data not shown). These in vitro binding studies demonstrated direct binding and strongly suggested that in vivo occupancy detected by ChIP experiments was attributable to sequence-specific DNA binding between EWS/FLI and the microsatellite repeats and did not require other cellular proteins. Interestingly, nuclear extract containing 3xFLAG-FLI also showed a similar binding pattern with these variable repeats oligonucleotides (Fig. S1A).
Fig. 4.
Ability of EWS/FLI to bind and activate via GGAA repetitive regions depends on the number of consecutive GGAA motifs. (A) Sequences of the oligonucleotides used for these analyses. The GGAA repeats are underlined. (B) Left shows an EMSA with a DNA duplex containing seven consecutive GGAA motifs. A specific EWS/FLI band is present when 3xFLAG-EWS/FLI from nuclear extracts is included. This specific band is supershifted with anti-FLAG antibody and competed with DNA duplex (I) containing a high-affinity ETS site but is not competed with DNA duplex (II) containing a PU.1 site that does not bind EWS/FLI (14). A control extract that does not contain EWS/FLI produces only nonspecific binding (indicated by “ns”). Right shows EMSA with DNA duplexes containing the indicated number of consecutive GGAA motifs and 3xFLAG-EWS/FLI. The positions of specific EWS/FLI-bound complexes are indicated. (C) Luciferase assays in 293EBNA cells with 36-bp sequence containing the indicated number of consecutive GGAA motifs (as indicated in A) upstream of a minimal promoter. The error bars indicate SDs, and asterisks indicate P < 0.05.
Promoter fragments with the same set of synthetic GGAA repeats were tested for transcriptional activity in the context of the minimal SV40 promoter. Transcriptional activity required at least five consecutive GGAA repeats and exhibited increased activity with six and seven repeats (Fig. 4C). Similar results were observed with DNA probes representing the endogenous NR0B1 microsatellite (data not shown). Wild-type FLI, on the other hand, was not able to regulate reporter gene activity via the NR0B1 microsatellite (Fig. S1B). Taken together, although both EWS/FLI and FLI are capable of binding GGAA microsatellites, only EWS/FLI is able to transcriptionally activate via these elements.
Discussion
Our findings demonstrate that EWS/FLI uses GGAA-containing microsatellites as specific response elements for a subset of fusion protein-up-regulated genes. The use of microsatellites as cancer-relevant genetic elements has not been previously demonstrated. Up-regulation of the EWS/FLI target gene NR0B1 has been shown previously to be necessary for the transformed phenotype of patient-derived Ewing's sarcoma cells (13). The present report demonstrates that the regulation of NR0B1 by EWS/FLI depends on the GGAA microsatellite in the NR0B1 promoter. Similarly, a second EWS/FLI-regulated gene, CAV1, which also contains a GGAA microsatellite in its promoter, has also been shown to be involved in the tumorigenic phenotype of Ewing's sarcoma (22). Although the cancer-relevant protein p53 has been shown to use a microsatellite as a response element at one of its target genes, PIG3, no functional role for PIG3 has been defined in tumorigenesis (26). Thus, the data presented in this report suggest a new role for microsatellites in human cancer development.
Experimental analysis of DNA probes and promoter fragments with variable numbers of repeats indicated that at least four to five consecutive GGAA motifs are required for DNA binding and gene activation and that the efficiency of these processes increased with increasing numbers of repeats. These results suggest interactions between multiple binding events. We speculate that protein–protein interactions may mediate cooperative DNA binding or that the presence of multiple sites affects the local effective concentration of active protein. Both phenomena could explain the use of suboptimal binding sequences within the GGAA-repetitive elements. The relatively high ChIP signal from the in vivo occupancy study and direct ChIP experiment is consistent with a mechanism that enhances the binding affinity of EWS/FLI to the microsatellites. Whereas eukaryotic promoters are often characterized by multiple transcription factor binding sites in close proximity, these findings indicate that the GGAA repetitive elements have emergent properties and do not simply represent a collection of independent binding sites.
In addition to the length-dependent interaction between EWS/FLI and GGAA microsatellites, other features may contribute to the selection of certain microsatellites as EWS/FLI-binding targets. Indeed, only ≈30% of GGAA microsatellites that could be detected by the Agilent promoter microarray used in our studies were bound by EWS/FLI (S.C.H., K.G., and S.S., unpublished observations). Furthermore, we have been unable to detect binding of endogenous wild-type FLI to GGAA microsatellites in Jurkat T cells (K.G., unpublished observation), nor have we observed enrichment of GGAA microsatellite binding in previously published ChIP-chip data of three other ETS transcription factors, ETS1, ELF, and GABPα (P.C.H., unpublished observation; ref. 9). Additional features that may contribute to ETS protein binding to microsatellites in vivo include local chromatin structure, nucleosome positioning, and the presence of other bound proteins at these sites, all of which may modulate the accessibility of the microsatellite for ETS protein binding. Minor sequence variations could also play a role in binding affinity to these sites. Additionally, although EWS/FLI can bind to microsatellite sequences in vitro without the assistance of other proteins (K.G., unpublished observations), it is possible that other proteins are required for binding in vivo. We demonstrated that both EWS/FLI and wild-type FLI bind to GGAA microsatellites with similar efficiency in vitro, but only EWS/FLI is capable of activating a microsatellite-containing reporter construct (Fig. S1). This suggests that activation of genes through these microsatellites, but not microsatellite binding, is a neomorphic function of the EWS/FLI fusion protein. Whereas these data suggest that the use of microsatellites by EWS/FLI may be a unique function of the fusion protein in Ewing's sarcoma, additional work will be required to fully address this question.
One interesting implication of this work is that differences in microsatellites (e.g., size, sequence, or location) could result in differences in Ewing's sarcoma susceptibility. Ewing's sarcoma is ≈10 times more prevalent in Caucasian populations than in African American populations (27). It may be that some microsatellites that regulate cancer-relevant EWS/FLI target genes are polymorphic between populations. Five microsatellites (including the NR0B1 microsatellite) were analyzed for length polymorphisms in genomic DNA from groups of individuals of defined ethnic backgrounds (CEPH reference panel) (28). Whereas we did not detect any significant length polymorphism differences between African and European populations in this small series (W.S.W. and L.B.J., unpublished observations), a more comprehensive analysis is needed to fully evaluate this hypothesis.
Ewing's sarcoma has only been observed in humans. Thus, no other organism develops Ewing's sarcoma spontaneously, nor have genetically engineered mouse models of Ewing's sarcoma been reported. We speculate that at least part of this difference in susceptibility is related to differences in GGAA microsatellites between organisms. For example, there is no GGAA-containing microsatellite in the Mus musculus Nr0b1 promoter, even though there are thousands of such microsatellites in the murine genome (data not shown). Murine Nr0b1 is not induced by EWS/FLI in NIH 3T3 cells (M.K., unpublished observation). Similarly, the GGAA microsatellites found in the EWS/FLI target genes FCGRT and CAV1 are also absent in the murine orthologs (data not shown). This suggests that even if the EWS/FLI fusion were expressed in mice, either spontaneously or via genetic engineering, it would not up-regulate critical genes required for oncogenic transformation, and thus tumors would not form (13, 22). This has important implications for the development of a genetically engineered mouse model of Ewing's sarcoma and may explain why Ewing's sarcoma development is limited to humans.
Methods
DNA Cloning.
The 3xFLAG-EWS/FLI MSCV-hygro retroviral expression construct and the EF-2-RNAi retroviral construct have been described previously (15, 19). 3xFLAG-FLI was prepared by using standard molecular biology approaches and was cloned into the MSCV-neo retroviral plasmid. Full-length and 5′ deleted NR0B1 and FCGRT promoters were cloned into the pGL3-Basic luciferase reporter vector (Promega Corporation). Constructs without promoter elements (e.g., the 500 bp NR0B1 microsatellite region, isolated microsatellites, and constructs containing varying numbers of GGAA motifs) were cloned into the pGL3-Promoter vector (Promega Corporation).
Cell Culture.
TC71, A673, and 293EBNA cells were cultured as described previously (13, 15, 29).
Luciferase Assays.
293EBNA or TC71 cells were transfected with each firefly reporter, Renilla plasmid, and cDNA or RNAi plasmids. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Two-tailed Student's t tests were used for statistical comparisons.
EMSA.
Nuclear extracts were prepared from 293EBNA cells transfected with 3xFLAG-EWS/FLI, 3xFLAG-FLI, or empty vector control expression plasmids. Twenty milligrams of nuclear extract protein, 5 nM [32P]-labeled probes, and 1× Gel Shift Binding Buffer (Promega Corporation) were used in each reaction. DNA duplex (I) (500 nM) (containing a high-affinity EWS/FLI-binding site, called “ETS2 probe” in ref. 12) and 500 nM DNA duplex (II) (bearing a variant ETS motif, which binds the ETS protein PU.1, but not EWS/FLI, called “PU.1 probe” in ref. 12) were used as specific and nonspecific unlabeled competitors for protein binding, respectively.
ChIP and Whole Genome Localization Studies (ChIP-chip).
ChIP from A673 cells was performed as previously described (9), by using anti-ETS1, anti-ELK1, or anti-FLI-1 antibodies (sc-350, sc-355, or sc-356, respectively; Santa Cruz Biotechnology, Inc.). Quantitative PCR was performed with NR0B1, FCGRT, CAV1, CACNB2, FEZF1 (LOC389549), KIAA1797 (hsa-mir-491), RPS26, or TP53 primers and with ALB and BCL2L1 primers (as normalization controls). See Table S1 for sequences. For ChIP-chip, anti-FLI immunoprecipitated genomic DNA samples from A673 cells (two independent biological replicates) were processed and hybridized to Agilent 244k promoter microarrays, as described (9). These microarrays interrogate ≈17-kb human promoters from −5.5 to +2.5 kb relative to the transcriptional start site. Initial analysis of the datasets was performed by using the Agilent ChIP Analytics software (version 1.3.1) to average both replicates as previously described (9).
MEME.
After processing the ChIP-chip data via the Agilent ChIP Analytics software, the most highly enriched DNA probe for each gene was identified. In some cases, the software package identified two enriched segments, suggesting that there were two separate EWS/FLI-binding sites in that gene. In each case, the genomic DNA sequence for the region surrounding the most enriched probe(s) (including the adjacent proximal and distal probes) were downloaded from the University of California at Santa Cruz Genome Browser (http://genome.ucsc.edu) by using the May 2004 Human genome assembly. Because of input data size limitations of the web-based MEME application (http://meme.sdsc.edu/meme/meme.html; version 3.5.7), only 60 kb of sequence could be used as input data (24). This corresponded to sequences from the 134 most highly enriched promoter fragments. The data were analyzed with the following parameters: any number of repetitions, minimum width of eight bases, maximum width of 16 bases, and identify two motifs.
Genome Wide in Silico Analysis.
For the purposes of this study, microsatellites were defined as sequence elements that contained at least 20 GGAA (or GGAT) motifs in a window of 120 bp and were identified from Homo sapiens genome data (Ensembl version 35), and the distance between the closest microsatellite “edge” and the transcriptional start site of the two closest genes in either direction was determined. There were 2,577 GGAA microsatellite-containing genes, and 942 GGAT microsatellite-containing genes identified.
For A673 cells, previously published “stable knockdown” data consisting of 320 EWS/FLI-up-regulated and 1,151 EWS/FLI-down-regulated genes were used (15). For TC71 and EWS502, the 1,610 EWS/FLI-up-regulated and 436 EWS/FLI-down-regulated gene sets derived from a similar stable knockdown experiment were used (13). ProbeIDs were mapped to their Ensembl identifiers by using the HG-U133A.na21.annot.csv and HG-U133A_2_annot.csv (September 2005 release) annotation files from Affymetrix. Probes without associated Ensembl identifiers were masked from further analysis.
For the Fisher's exact test analyses, the microsatellite-neighboring genes were rank-ordered based on distance between the microsatellite and the gene. Fisher's exact test was performed at each position of the rank-ordered list by using the following two-by-two table: genes with microsatellites at or closer than a particular distance (where the distance increases with each iteration of the analysis) vs. those greater than that particular distance and genes that are EWS/FLI-regulated vs. those that are not. EWS/FLI-up- or -down-regulated genes from A673 cells or TC71/EWS502 cells were considered in separate analyses (13, 15). The total number of genes analyzed was the intersection between genes containing microsatellites that were also present on the U133A microarray (Affymetrix). The Bonferroni correction was calculated by dividing 0.05 by the total number of genes in each analysis.
Supplementary Material
Acknowledgments.
We thank Don Ayer for critical reading of the manuscript, Bradley Cairns and members of the laboratory of S.L.L. for helpful discussions, and Whitney Tolpinrud for technical assistance. This work was supported by funds awarded to S.L.L. from the National Cancer Institute (K08 CA96755), Hope Street Kids, the Liddy Shriver Sarcoma Initiative, Primary Children's Medical Center Foundation, the Terri Anna Perine Sarcoma Fund, and Huntsman Cancer Institute/Huntsman Cancer Foundation. We also acknowledge support from the National Institutes of Health to the Huntsman Cancer Institute (Grant P30 CA 42014), to B.J.G. (Grant R01 GM38663), and to L.B.J. (Grant R01 GM59290 and R01 HL070048).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801073105/DCSupplemental.
References
- 1.Nunn MF, Seeburg PH, Moscovici C, Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- 2.de Taisne C, Gegonne A, Stehelin D, Bernheim A, Berger R. Chromosomal localization of the human proto-oncogene c-ets. Nature. 1984;310:581–583. doi: 10.1038/310581a0. [DOI] [PubMed] [Google Scholar]
- 3.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 7.Szymczyna BR, Arrowsmith CH. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J Biol Chem. 2000;275:28363–28370. doi: 10.1074/jbc.M004294200. [DOI] [PubMed] [Google Scholar]
- 8.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 11.May WA, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci USA. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May WA, et al. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res. 2006;4:851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 14.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing's sarcoma EWS/FLI- 1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 15.Smith R, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11:1049–1054. [PubMed] [Google Scholar]
- 17.Owen LA, Lessnick SL. Identification of target genes in their native cellular context: An analysis of EWS/FLI in Ewing's sarcoma. Cell Cycle. 2006;5:2049–2053. doi: 10.4161/cc.5.18.3213. [DOI] [PubMed] [Google Scholar]
- 18.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing's sarcoma. Cell Cycle. 2006;5:2753–2759. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 20.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7:250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 21.Hahm KB, et al. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 22.Tirado OM, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing's sarcoma cells. Cancer Res. 2006;66:9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 23.Mao X, Miesfeldt S, Yang H, Leiden JM, Thompson CB. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J Biol Chem. 1994;269:18216–18222. [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 25.Gillespie ME. Salt Lake City: University of Utah; 1998. A structural and genetic investigation of DNA binding by the murine Ets-1 Ets domain, a winged HTH transcription factor. PhD thesis. [Google Scholar]
- 26.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 27.Gurney JG, Swensen AR, Bulterys M. Malignant bone tumors. In: Ries LAG, et al., editors. Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program 1975–1995. Bethesda, MD: Natl Cancer Inst, SEER Program; 1999. pp. 99–110. DHHS Publ No (NIH) 99-4649. [Google Scholar]
- 28.Dausset J, et al. Centre d'etude du polymorphisme humain (CEPH): Collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 29.Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.