Abstract
Molecular clocks suggest that animals originated well before they first appear as macroscopic fossils, but geologic tests of these hypotheses have been elusive. A rare steroid hydrocarbon, 24-isopropylcholestane, has been hypothesized to be a biomarker for sponges or their immediate ancestors because of its relatively high abundance in pre-Ediacaran to Early Cambrian sedimentary rocks and oils. Biolipid precursors of this sterane have been reported to be prominent in several demosponges. Whether 24-isopropylcholestane can be interpreted as a sponge (and, hence, animal) biomarker, and so provide clues about early metazoan history, depends on an understanding of the distribution of sterol biosynthesis among animals and their protistan relatives. Accordingly, we characterized the sterol profile of the choanoflagellate Monosiga brevicollis, a representative of the unicellular sister group of animals. M. brevicollis does not produce a candidate sterol precursor for 24-isopropylcholestane under our experimental growth conditions. It does, however, produce a number of other sterols, and comparative genomics confirms its biosynthetic potential to produce the full suite of compounds recovered. Consistent with the phylogenetic position of choanoflagellates, the sterol profile and biosynthetic pathway of M. brevicollis display characteristics of both fungal and poriferan sterol biosynthesis. This is an example in which genomic and biochemical information have been used together to investigate the taxonomic specificity of a fossil biomarker.
Keywords: choanoflagellates, molecular fossils, origin of metazoans, Monosiga brevicollis
Sterols constitute a diverse class of triterpenoid lipids having wide ranging importance in biology. All eukaryotes require sterols; they serve important functions because membrane lipids play roles as developmental regulators and precursors to steroid hormones in multicellular organisms. Recently, sterols have emerged as an important investigative tool for paleontologists (1–4) because steranes, the hydrocarbon skeletons of sterols, resist microbial attack and remain stable over long periods of time. Thus, they are well represented as molecular fossils in sedimentary rocks. The discovery of diverse steranes in the geologic record has fueled a search for unique precursor sterols in many branches of the eukaryotic tree, because for a molecular fossil to be useful as a biomarker, it must have significant taxonomic and/or physiological specificity.
One candidate for a paleobiologically useful biomarker is 24-isopropylcholestane, the geologically stable derivative of the C30 sterol 24-isopropylcholesterol. Although much of eukaryotic sterol diversity is manifested in double bonds and functional groups that do not preserve in fossil hydrocarbon skeletons, the isopropyl moiety in the side chain of 24-isopropylcholesterol and related sterols results in a structure that is both unique and preservable. Particularly abundant as a molecular fossil in rocks deposited during the Ediacaran and Cambrian periods (3, 5) is 24-isopropylcholestane. Consistent with this stratigraphic distribution, its parent sterol is known to be synthesized by extant demosponges (6); 24-isopropylcholesterol has also been identified in small quantities in a stramenophile alga (7), but this is less likely to explain the abundance of 24-isopropylcholestane in Ediacaran and Cambrian, but not younger, rocks. As far as we know, 24-isopropylcholesterol is not synthesized by eumetazoans (cnidarians plus bilaterian animals). Recent molecular phylogenies indicate that sponges populate the basal branches of the animal tree, with eumetazoans forming a sister group to a specific class of sponges, the Calcarea (8–10). Thus, 24-isopropylcholestane potentially provides a powerful biomarker for early animal diversification.
The earliest mineralized sponge spicules occur just below the Proterozoic–Cambrian boundary, in ca. 544 Ma rocks from Mongolia (11), and probable sponge casts and molds have been found in Australian sandstones only a few million years older (12). The oldest potential animal macrofossils of any kind are 575 Ma problematica from Newfoundland (13), and the oldest morphologically preserved remains of any kind related to stem group animals are permineralized egg cysts in 632 Ma rocks from China (14). Nearly all molecular clock estimates for animal origins suggest that the kingdom originated earlier than this (15, 16). Thus, the discovery (17) of relatively abundant 24-isopropylcholestane in sedimentary rocks that lie stratigraphically below Marinoan glacial beds (>635 Ma) potentially brings molecular clocks and the geologic record into closer accord and further implies that animals originated in a unique environmental context, between two global glaciation events (18).
For the 24-isopropylcholestane skeleton to be interpreted in this way, however, it must be uniquely defined as a demosponge biomarker. It may be possible to assign this 24-isopropylcholestane confidently to the sponges within the Metazoa, because more derived metazoans use predominantly C27 sterols, either by de novo synthesis or by modification of dietary sterols to C27 molecules. No metazoan other than sponges has been shown to contain the biosynthetic capacity to methylate the sterol side chain, a requirement for formation of 24-isopropylcholestanes. As noted above, this molecule has been identified in small quantities in a stramenophile alga (7), but unambiguous body and molecular fossils of stramenopiles otherwise gain prominence in only Mesozoic and younger rocks (19).
To determine whether the 24-isopropylcholestane skeleton is uniquely associated with sponges within the opistokonts, the group that includes metazoa, fungi, and related unicellular protists, it is important to survey sterols in previously undescribed taxa within this group. To date, the sterol composition of choanoflagellates, the unicellular sister group of animals (20–22) has remained unknown. Therefore, to begin, we characterized the sterol composition of Monosiga brevicollis, a choanoflagellate recently developed as a model organism and used to represent the unicellular relatives of sponges and eumetazoans (22–24). We also used comparative bioinformatics to examine the sterol biosynthetic capacity of M. brevicollis from its complete genome sequence.
Results
Sterol Profile of M. brevicollis.
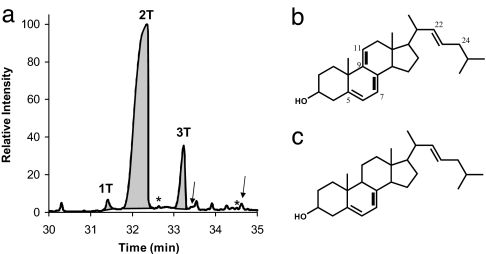
The profile of M. brevicollis ' sterol lipid fraction shows three dominant compounds (1, 2, and 3) by using the HCl-based extraction method described below. Fig. 1A shows the total ion chromatogram (TIC) of the trimethylsilyl ether (TMS) derivatives. These same three sterols also were obtained in identical proportions when extracted by using the Bligh–Dyer protocol (see Methods). Sterol identifications were made by comparison of mass spectra for both acetate and TMS derivatives with those from the literature. Mass spectral fragments of 1–3 are provided in Table 1, and reference fragments of other C27 trienes and tetraenes are given in supporting information (SI) Table S1.
Fig. 1.
Sterol profile of M. brevicollis: (a) Total ion chromatogram of the sterol fraction of M. brevicollis (as TMS ethers), showing choanoflagellate sterols (1–3). Other minor components may be derived from growth medium (arrows) or may be of choanoflagellate source (asterisks), as discussed in the text. (b) Proposed structure of 1. (c) Proposed structure of 2.
Table 1.
Mass spectra (70eV fragments) of major sterol components of M. brevicollis
| Molecule | TIC peak no.* | m/z(relative abundance, %) |
|---|---|---|
| C27:4 Cholesta-5,7,9(11),22-tetraene-3β-ol | 1T | 452 (13), 437 (5), 362 (34), 347 (13), 325 (8), 251 (100), 249 (20), 235 (9), 209 (18), 211 (8), 195 (15), 73 (25), 69 (24), 55 (21) |
| 1A | 422 (10), 362 (100), 347 (18), 251 (45), 209 (30), 69 (25), 55 (30) | |
| C27:3 Cholesta-5,7,22-triene-3β-ol | 2T | 454 (29), 439 (2), 364 (17), 349 (100), 323 (71), 253 (20), 237 (7), 211 (17), 197 (11), 143 (17), 111 (8), 73 (16), 69 (14), 55 (13) |
| 2A | 424 (10), 364 (100), 394 (35), 323 (8), 235 (30), 211 (12), 69 (20), 55 (30) | |
| C28:3 Ergosta-5,7,22-triene-3β-ol | 3T | 468 (29), 453 (3), 378 (16), 363 (100), 337 (64), 253 (21), 237 (6), 211 (19), 197 (11), 73 (22), 69 (24), 55 (17) |
| 3A | 438 (10), 378 (100), 363 (30), 253 (25), 211 (12), 157 (28), 69 (21), 55 (30) |
*1T, 2T, and 3T are the trimethylsilyl ethers, and 1A, 2A, and 3A are the corresponding acetates.
Mass spectral fragmentation patterns of 1A and 1T are typical of C27 tetraene sterol derivatives (25–31), based on comparison with literature spectra for C27 sterol acetates and TMS ethers. The molecular ions (m/z 422 for 1A and m/z 452 for 1T) suggest that this molecule has four unsaturations. The diagnostic base peak resulting from side-chain cleavage of the TMS ether yields the tetracyclic m/z 251 fragment ion. This is consistent with the presence of three unsaturations in the ring system and one in the side chain.
We identify 1 as cholesta-5,7,9 (11),22-tetraen-3β-ol (Fig. 1B) based on the published spectrum of an acetate derivative (32). Comparison of the M. brevicollis compound (1A; Table 1) with this spectrum reveals identical fragment masses and relative abundances, supporting the identification. The reference molecule was synthesized in an enzymatic study by a yeast mutant deficient in a sterol methyl transferase enzyme essential for the synthesis of C28 sterols (32). These mutants were fed exogenous cholesterol and were able to desaturate at C-22, with the resulting (synthetic) compound being cholesta-5,7,9 (11),22-tetraen-3β-ol. Naturally occurring cholesta-5,7,9 (11),22-tetraen-3β-ols have been reported as minor sterols in the demosponges Axinella cannabina (33, 34) and taxa within the orders Dendroceratida and Dictyoceratida (34). Here, we assign the structure of M. brevicollis sterol (1) tentatively to C27Δ5,7,9(11),22.
Although mass spectrometry often does not afford a unique structural assignment, the fragmentation patterns observed here are consistent with specific structural features and support the identification proposed above. Cleavage in the D ring results in the observed 209-Da ion, suggesting that 1 has a B/C ring triene (35, 36). Also consistent with this diagnosis is the ratio of relative abundances of the [M+] to the [M–AcOH] fragment, which is 1:10 in 5,7,9(11) triene acetates (35). The ring system, after loss of the side chain, will be at m/z 251. The significant 111-Da fragment ion is often attributed to the loss of a side chain with a single unsaturation. The position of this double bond, however, is difficult to deduce from fragmentation patterns alone (32, 37). The base peak of 1A is a m/z 362 fragment that commonly results from cleavage of the side chain favored by an unsaturation in the C-22 position. The published spectra of cholesta-5,7,9(11),24-tetraen-3β-ol show fragment ions similar to those found in the spectra of 1, but the C-24(25)-ene displays different relative abundances for each fragment (30).
Compound 2, the dominant product, is a C27:3 compound. It was confirmed as cholesta-5,7,22 trien-3β-ol (Fig. 1C) by comparison with cholesta-5,7,22-trien-3β-ol acetates. This molecule has the same unsaturation pattern as ergosterol, but lacks the methyl group at position C-24. Although C27 trienes are fairly well known, and, in some cases have been shown to be intermediates in the cholesterol biosynthetic pathway, they are still rare compared with their C28 counterparts, such as ergosterol (25–27, 38). Mass spectral comparisons were made with all known trienes (Table S1), and identification was made based on best fit to C27Δ5,7,22. Compound 3 was identified by TMS ethers and acetate mass spectra as ergosta-trien-3β-ol (36, 39), or ergosterol.
Two of the minor peaks in the sterol fraction can be attributed to sterols in the choanoflagellate media (labeled with arrows; Fig. 1A). M. brevicollis is cultured with Flavobacter sp. bacteria in media of seawater brewed with Ward's cereal grass. The cereal grass is a mixture of various plant materials and contains phytosterols. We extracted a standard mass of the dry cereal grass used for the culture media by using the Bligh–Dyer method (40). Analysis of the cereal grass yielded a TIC in which two sterol products could be identified: a C28:1 and a C29:1, both most consistent with the single unsaturation (Δ5), probably campesterol and sitosterol, respectively. The presence of these two sterols in the choanoflagellate profiles (left arrow, C28:1 and right arrow, C29:1) indicates they were either incorporated into or adsorbed onto M. brevicollis biomass without being altered. Alternatively, they were extracted directly from the small amount of residual media collected during cell pelleting. A third, cleaner extraction of cultured biomass did not yield any of these minor components but still recovered the suite of sterols (1, 2, 3) originating from M. brevicollis. Control extractions also were performed on cultures of Flavobacter, the single food source for M. brevicollis. Flavobacter was grown under the same culture conditions as used for the M. brevicollis experiments, and an equal volume of biomass was extracted by using the Bligh–Dyer method. This control did not yield detectable sterols, and Flavobacter is not among the small number of bacteria expected to synthesize sterols (41). These controls are consistent with our assertion that 1, 2, and 3 are synthesized de novo by M. brevicollis and are not modified dietary sterols.
Other minor products (labeled with asterisks; Fig. 1A) detected in the Monosiga extracts but not found in the cereal grass control include a C28:4 sterol and lanosterol (C30:2). The C28:4 compound (left asterisk; Fig. 1A) appears to have the same fragmentation pattern as 1 but with one additional carbon atom on the side-chain. This is likely a methyl group at C-24 as found in ergosterol, making it a 24-methyl-C28Δ5,7,9(11),22-tetraene; this is the C28 counterpart of 1. Other C28 tetraenes such as 13 in Fig. 2 are known intermediates in sponge and fungal sterol biosynthesis. However, compound 13 is a typical C28 tetraene (24-methyl-C28Δ5,7,22,24(28)-tetraene), which would be formed as the penultimate step in ergosterol biosynthesis. The M. brevicollis C28-tetraene is not 13, but again appears to be another sterol unique to the choanoflagellate. Because the C28:4 compound was only seen in one of the three extractions of choanoflagellate biomass, and in trace quantities, it may be a biosynthetic intermediate. It is unlikely to be a minor modification of dietary sterol, given the configuration of the double bonds in the tetraene.
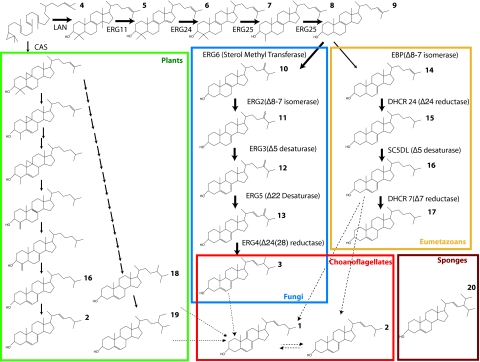
Fig. 2.
Canonical sterol biosynthetic pathways, leading to plant, fungal, or eumetazoan sterols. Choanoflagellates and sponges are shown separately. Dotted lines represent possible pathways to choanoflagellate sterols. Multiple arrows represent steps not described in the text. Gene names are given by using fungal notation, e.g., ERG4.
Lanosterol (right asterisk; Fig. 1A) was detected in trace amounts and is the initial biosynthetic sterol in the pathway to both fungal and metazoan sterols and, therefore, not unexpected. In one of the three M. brevicollis extracts, an additional, unusual and trace C31 sterol was cyclolaudenol (24-methyl-5α-cycloart-25-en-3β-ol). The identification is based on the close similarity of its mass spectrum to those published for this compound (36). This compound is less expected, because it is a derivative of cycloartenol; it may have been a minor component, or a modification product, of a sterol associated with the growth medium. Importantly, none of the sterols from any of the extractions was found to have an isopropyl group in the side chain. The only C30 molecule detected was the biosynthetic intermediate, lanosterol. Thus, no sterols identified in M. brevicollis are candidate precursors of the sterane biomarker, 24-isopropylcholestane.
The sterol profile from M. brevicollis has several distinguishing factors. First, it resembles the sterol composition of metazoans by having a dominant C27 sterol. Indeed compound 1 is otherwise known from demosponges. Second, it shows features characteristic of fungal sterol biosynthesis by displaying the same unsaturation pattern found in the C28 sterol, ergosterol, the dominant sterol of fungi. Unlike fungi, however, Monosiga expresses the Δ5,7,22 in its C27 sterol product. Consistent with this preference for ergosterol-like products, the choanoflagellate also produces ergosterol as its second most abundant sterol. The combination of fungal and animal sterol characters seen in choanoflagellates is consistent with their phylogenetic position (8–10,20,22; Fig. 1). Choanoflagellate sterol biosynthesis, as suggested by the suite of genes in its genome, includes a pathway similar to the fungal pathway to ergosterol, with methylation on the C-24 position via a sterol methyl transferase enzyme (SMT) and double bonds inserted with C-5 and C-7 desaturases. This suggests that this pathway may be plesiomorphic for the opisthokont clade, i.e., present in the last common ancestor of fungi, choanoflagellates, and animals, but subsequently lost in eumetazoans. No eumetazons have yet been shown to contain the SMT genes necessary for the synthesis of ergosterol; therefore, a sterol profile dominated by C27 products synthesized de novo can be considered a derived feature of eumetazoans within the opistokonts.
Because 1-3 are not found in the choanoflagellate media or its bacterial prey, they must be synthesized by M. brevicollis. Monosiga might, in principle, convert C28:1 and C29:1 sterols from its diet into 1-3, but this would require significant molecular modification. Furthermore, the available dietary sterols do not contain the important C-22 unsaturation, so this feature must have been introduced by the choanoflagellate. Genomic evidence discussed below suggests that M. brevicollis possesses the necessary genes for complete de novo synthesis of all three.
Genomic Investigation of Sterol Biosynthesis.
The biosynthetic pathways for sterols in plants, fungi, and derived metazoans are well characterized as are the corresponding enzymes and their gene sequences (Fig. 2). To evaluate the sterol biosynthetic capabilities of M. brevicollis, we used protein sequence information obtained from its recently completed whole-genome sequence. Traditionally, labeling studies have been used to verify biosynthetic activity in heterotrophic organisms such as choanoflagellates. Instead, we used genomic information to identify homologs for enzymes involved in sterol biosynthesis, mapping out a putative biosynthetic capacity. A full set of biosynthetic genes demonstrates that an organism may be able to synthesize the molecules of interest. One advantage to this approach is that it may also suggest or confirm unique abilities based on enzymatic variations and could help explain the origin of unusual products such as a dominance of C-22 unsaturations.
Protein–protein BLAST (Basic Local Alignment Search Tool) searches of the catalog of predicted M. brevicollis proteins by using fungal and metazoan query sequences indicate that this choanoflagellate contains homologs for a full suite of sterol biosynthesis enzymes and, therefore, could synthesize sterols de novo (Table 2). Candidate sequences were identified by using a cut-off expectation (E) value of <10−15 relative to at least one empirically characterized fungal or metazoan sequence. Each candidate protein sequence was then analyzed phylogenetically to determine homology (Fig. S1, sequence information Table S2 and Table S3). Interestingly, a gene for the C-22 desaturase enzyme was the only step in the sterol biosynthetic pathway for which a predicted homolog from the M. brevicollis genome was not recovered. Homologs of this gene can be found easily (and can be distinguished from the aforementioned C-14 demethylases) in fungi and plants, but C-22 desaturases cannot be similarly found in animals. To date, the enzymatic products of this homologous family of C-22 desaturases represent the only known method for inserting a C-22 double bond into a sterol side chain, yet in M. brevicollis, no sequence with high homology to known C-22 desaturases was recovered, despite the apparent requirement for this functionality. Sequences with less stringent BLAST scores (expectation values 10−9 to 10−15) were recovered by searching with a representative fungal sequence, whereas none were recovered by searching with a plant sequence. Furthermore, the phylogenetic analysis shows that these sequences with lower scoring BLAST hits do not group with known C-22 desaturases. Therefore, it must be assumed that the enzyme operating in M. brevicollis is different or highly divergent from previously known C-22 desaturases.
Table 2.
Sterol biosynthesis gene in M. brevicollis
| Gene description (fungal gene name) | Protein ID no. | Best hit to known sequence/percent similarity | Homology |
|---|---|---|---|
| OSC | 18210 | gi∣47933397∣ref∣NP001001438.1∣lanosterol synthase [Homo sapiens] (hit %: 97, score: 1975, % id: 46) | Yes |
| C-14 demethylase (ERG 11) | 38433 | gi∣83287777∣sp∣Q5RE72∣Cytochrome P450 (P45014DM) [Pongo pygmaeus] (model %: 89, hit %: 88, score: 1205, % id: 48) | Yes |
| C-4 demethylase (ERG 25) | 27469 | gi∣18390767∣ref∣NP_563789.1∣SMO2–2 [Arabidopsis thaliana] (hit %: 78, score: 294, % id: 31) | Yes |
| C-14 reductase (ERG 24) | 38824 (fungal/animal-like) | gi∣33320186∣gb∣AAQ05836.1∣AF480070_1 [Mus musculus] (hit %: 63, score: 979, % id:50) | Yes |
| 29161 (plant-like) | gi∣9714052∣emb∣CAC01296.1∣(FACKEL) [Arabidopsis thaliana]; gi∣8917585∣gb∣AAF81279.1∣C-14 sterol reductase [Arabidopsis thaliana] (model %: 63, hit %: 66, score: 522, % id: 32) | Yes | |
| C-24 methyltransferase (ERG 6) | 33702 | gi∣73761691∣gb∣AAZ83345.1∣[Gossypium hirsutum] (model %: 76, hit %: 87, score: 800, % id: 46) | Yes |
| Δ8-7 isomerase (ERG 2) | 25184 | gi∣70799755∣gb∣AAZ09671.1∣[Leishmania major strain Friedlin] (hit %: 60, score: 532, % id: 71) | Yes |
| Δ5 desaturase (ERG 3) | 32318 | gi∣52219112∣ref∣NP_001004630.1∣[Danio rerio] (model %: 98, hit %: 94, score: 853, % id: 55) | Yes |
| 39006* | gi∣89298745∣gb∣EAR96733.1∣Sterol desaturase family protein [Tetrahymena thermophila SB210] (model %: 70, hit %: 60, score: 524, % id: 46) | Yes | |
| C-24 reductase (ERG 4) | 37477 | gi∣6321426∣ref∣NP_011503.1∣[Saccharomyces cerevisiae] (model %: 94, hit %: 94, score: 902, % id: 41) | Yes |
| C-7 reductase** | 26465 | gi∣145336583∣ref∣NM_103926.4∣[Arabidopsis thaliana] (DWARF 5); (model %: 47, hit %: 60, score: 476, % id: 38) | Yes |
| Δ22 desaturase (ERG 5) | 33214 | gi∣71411448∣ref∣XP_807973.1∣cytochrome P450 [Trypanosoma cruzi strain CL Brener] (model %: 80, hit %: 82, score: 941, % id: 32) | No |
Sterol biosynthesis genes beginning with the first, cyclized product 2,3 sterol epoxide and extending to fungal or animal end products. SQMO, squalene monooxygenase; OSC, oxidosqualene cyclase. ERG notation for gene names in fungi. Δ5 desautrase and C-14 reductrase have two potential homologs in M. brevicollis. ERG 25 and ERG 3 have two potential homologs.
*, undefined sterol desaturase;
**, no fungal analog exists.
We also extracted some information on sterol biosynthetic capacity for demosponges from the whole-genome sequence of the genomic model sponge, Amphamedon queenslandica. This genome is also sequenced by the Joint Genome Institute. Unlike the M. brevicollis genome, this genome is not yet assembled so determination of specific gene sequences and numbers of genes copies in the genome is not possible. The raw sequence data in the form of trace archives is publicly available from National Center for Biotechnology Information and was used to search for all sterol biosynthetic genes by using tBLASTn searches. Recovered sequences were assembled by hand and included in the phylogenetic analyses to determine homology (Fig. S1 and Table S3). It appears that A. queenslandica has all biosynthetic genes necessary for sterol biosynthesis, including one or more SMT genes. It also appears to be lacking a C-22 desaturase, like M. brevicollis, although this must remain speculative until a fully assembled version of the genome can be searched.
Evolution of Sterol Biosynthesis in Choanoflagellates.
Both the sterols and the suite of genes present in the genome of M. brevicollis indicate the intermediate nature of choanoflagellate sterols relative to those of fungi and animals. The presence in M. brevicollis of putative homologs for enzymes found in the fungal sterol biosynthetic pathway but not found in known metazoan biosynthetic pathways (e.g., C-24 reductase) and an enzyme previously found in the metazoan, but not in the fungal, biosynthetic pathway (C-7 reductase), suggests a combination of the features previously thought to be unique to fungi or to animals. The M. brevicollis pathway thus may represent an evolutionary intermediate between fungi and animals. C-24 reductase typically acts in the penultimate step in ergosterol biosynthesis and is not found in the cholesterol biosynthetic pathway. Likewise, C-7 reductase removes the C-7 unsaturation in intermediates leading to cholesterol, but is not found in the ergosterol biosynthetic pathway. This genomic survey of sterol biosynthetic enzymes suggests that unique aspects of pathways can indeed be predicted, based on analysis of gene homologs in conjunction with knowledge of an organism's sterol products.
Overall, the dominance of C27 sterols is similar to modern eumetazoan sterol profiles, and according to preliminary phylogenetic analyses of biosynthetic genes, the choanoflagellate genes always appear more closely related to metazoan genes (and closely related to A. queenslandica, where a potential homolog for the demosponge was assembled). The eumetazoan pathway (Fig. 2) always leads to C27 sterols in the animals that have been studied, because they lack the ability to methylate the sterol side chain. However, in contrast to animals, the choanoflagellate also makes minor quantities of C28 sterols. This is somewhat unusual, because organisms with the ability to methylate the C-24 position tend to make large quantities of sterols with methylated side chains. This is seen in fungi, where the fungal pathway (Fig. 2) always produces C28 sterols in the fungi that have been well studied. Fungi also produce dominant Δ5,7,22 triene sterols with side chain methylations at the C-24 position and side chain unsaturations at C-22. These features are similar to the choanoflagellate sterol composition; thus it appears that choanoflagellates employ aspects of both pathways.
Without further information on the physiological role of methylated sterols, we are unable to speculate why this particular choanoflagellate mainly produces C27 sterols. However, a similar trend can be seen in organisms like red algae that produce dominant C27 sterols but have the ability to make C28 and C29 molecules. The hypothetical pathways shown by dotted lines in Fig. 2 only suggest the possible paths to the choanoflagellate sterol products. Compound 1 and 2 could be synthesized directly from a C27 precursor but could conceivably be modified from a C28 or C29 sterol taken from the media (as shown from compound 18 and 19) or from compound 3. The most logical path to biosynthesis of compound 3 would be by following the typical fungal pathway, although it is possible it arises from methylation of compound 2, or modification of compound 18 or 19.
Although the biosynthetic pathway to 24-isopropylcholesterol is not known, it has been suggested that an errant C-22 desaturase could be the enzyme responsible for the isopropyl moiety (42). The lack of a homolog for the C-22 desaturase in M. brevicollis and the presence of C-22 unsaturations in its sterols suggests that they may also be employing an unusual C-22 desaturase in the biosynthetic pathway. Much more work is needed on the method for insertion of the C-22 unsaturation in the M. brevicollis sterols, and further work on the biosynthesis of 24-isopropylcholesterol if any connection is to be made between these two modifications.
Implications.
Our studies indicate that M. brevicollis does not produce a candidate isopropyl sterol precursor for the 24-isopropylcholestanes. Such a conclusion is consistent with the hypothesis that 24-isopropyl steranes in pre-Ediacaran sedimentary rocks are molecular proxies for demosponges (17), although it will be necessary to analyze a greater diversity of choanoflagellates to provide more definitive support. Exploration of biosynthetic pathways although comparative genomics provides an approach to investigating the uniqueness and utility of lipid biomarkers. Genomic characterization of enzymatic pathways can provide insight into the biosynthetic capabilities of an organism. Because the observed molecular composition of a given sample of biomass can be influenced by physiological condition, diet, or the presence of symbionts, genomics presents an independent assessment of an organism's ability to produce molecules of interest. Comparative genomics also provides support for the uniqueness of the overall choanoflagellate sterol pathway, because it appears that enzymes or enzyme combinations lead here to unique biomarkers. Further understanding of the genetic and enzymatic controls on the synthesis of the 24-isopropyl group in sponges, and an improved understanding of the side-chain (C-22) unsaturation in choanoflagellates, promises to provide a definitive test of the hypothesis that 24-isopropylcholestane diagnostically records sponges in the geologic record. This, in turn, points the way toward strong geologic tests of molecular clock estimates for the origin of metazoans.
Methods
Cultures.
Cultures of M. brevicollis (50154; American Type Culture Collection) were grown in natural seawater media brewed with Wards cereal grass media, filtered, and inoculated with a bacterial monoculture of Flavobacter as a food source (cat. no.9448606; Scholar Chemistry). Cultures were maintained at room temperature and split to a 1:15 dilution every 3 days. Cultures were monitored for contamination by using microscopy and to ensure that Flavobacter was the sole food source for M. brevicollis. Three separate rounds of cultures were grown over the course of 1 year from the same frozen stock. Control cultures of axenic Flavobacter were grown under the same conditions as experimental samples, and both uninoculated media and the cereal grass source for the media were also used as controls.
Lipid Extractions.
Total lipid extracts were generated by using two different techniques to verify complete sterol extraction. In the first, lyophilized biomass was extracted with ultrasonication using a 10:5:4 methanol/chloroform/water (Bligh–Dyer) solvent (40). Phase separation was performed with addition of an equal volume of chloroform, and then water followed by centrifugation. Neutral lipids were separated from the organic phase with a cold acetone precipitation and separated into fractions by using silica TLC plates with elution in 2× dichloromethane to a 12-cm mark followed by 1× hexane. The sterol fraction was collected and analyzed as acetate derivatives.
The second method extracted wet biomass for 4 hours in a 10% aqueous HCl solution at 70°C. The total lipid extract was collected by liquid–liquid extraction in a separatory funnel with dichloromethane as solvent. The organic phase was separated into fractions by using silica flash column chromatography. Sterol fractions were analyzed as TMS and acetate derivatives. Controls of Flavobacter cultures and extractions of cereal grass used in media were conducted by using the first extraction technique and analyzed as TMS derivatives.
Mass Spectrometry.
Acetate derivatives of choanoflagellate biomass were analyzed by using a HP 6890 gas chromatograph with a Varian CP-Sil-5 column (60 m, 0.32 mm ID, 0.25-μm film thickness) fused silica capillary column, coupled to HP 5973 mass-selective detector and operated over 50–650 Da at 70 eV. TMS derivatives of choanoflagellate TLE, bacterial biomass TLE, and cereal grass TLE were analyzed with an Agilent 6890N gas chromatograph with an Agilent HP-5MS column (30 m, 0.25-mm ID, 0.25micron film thickness; Agilent part number 19091S-433) coupled to an Agilent 5973 inert MSD.
Identification of Sterol Biosynthesis Genes.
The whole-genomic genome sequence (WGS) of M. brevicollis is available from the Joint Genome Institute (JGI) (genome.jgi-psf.org). We identified potential homologs of genes for these enzymes from the M. brevicollis WGS through translated nucleotide BLAST comparisons [tBLASTn; (36)] with empirically defined protein sequences, with a cutoff expectation value (E value) of <10−15 (see Table 2 for GenBank accession numbers). Potential partial gene sequences for A. queenslandica were acquired with local tBLASTn searches of the WGS trace archives. Homology was further established by reciprocal BLAST hits to other similar proteins and by phylogenetic tree constructions.
Protein alignments for phylogenetic analysis were made with Geneious 2.0.1 (www.geneious.com) with Blosum 62 cost matrix, free end gaps, and two refinement iterations. Alignments were realigned by using Jalview (43) access to Mafft, Muscle, or Clustal W alignment algorithms for similarity comparison. Maximum-likelihood trees were made with the Phylip package using Seqboot and Proml (44) and using PHYML online server (45) using WAG, Dayhoff, and JTT models with four rate substitution categories and 100 bootstrap datasets. Tree topologies were identical with each model and bootstrap values very similar if not identical. Results from analyses using the WAG model are shown as Fig. S1.
Supplementary Material
Acknowledgments.
We thank Mark Martindale, Dave Matus, and Scott Nichols for BLAST help with the A. queenslandica trace archive and Tracy LaGrassa, Jess Lyons, and Andrea Morris (University of California, Berkeley, CA) for providing choanoflagellate biomass for analysis. Gordon Love provided constructive comments, as did reviewers J. Brocks, J. Giner, K. Peterson, and J. Volkman. This work was supported in part by National Science Foundation (NSF) Grant EAR-0420592 (to R.E.S. and A.H.K.) and NSF grant EAR-0641899 (to A.P.) and funding from the Gordon and Betty Moore Foundation Marine Microbiology Initiative (to N.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Annotations for all sequences in Table 2 have been deposited in the GenBank database (accession nos. XP_001744097.1, XP_001748747.1, XP_001747965.1, XP_001749711.1, XP_001749698.1, XP_001748534.1, XP_001745538.1, XP_001748534.1, XP_001245522.1, XP_001745538.1, XP_001750121.1, XP_001746631.1, XP_001746961.1, and XP_001747487.1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803975105/DCSupplemental.
References
- 1.Ourisson G, Rohmer M, Poralla K. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Ann Rev Microbiol. 1987;41:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 2.Brocks JJ, Summons RE. In: Treatise on Geochemistry. Holland HD, Turekian KK, editors. Amsterdam: Elsevier; 2003. pp. 63–115. [Google Scholar]
- 3.Peters KE, Walters CC, Moldowan JM. The Biomarker Guide. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 4.Brocks JJ, Pearson AP. Building the biomarker tree of life. Rev Mineral Geochem. 2005;59:233–258. [Google Scholar]
- 5.McCaffrey MA, et al. Paleoenvironmental implications of novel C30 steranes in Precambrian to Cenozoic age petroleum and bitumen. Geochem Cosmochim Acta. 1994;58:529–532. [Google Scholar]
- 6.Hofheinz W, Oesterhelt G. 24-isopropylcholesterol and 22-dehydro-24-isopropylcholesterol, novel sterols from a sponge. Helv Chim Acta. 1979;58:529–532. [Google Scholar]
- 7.Giner J, Li X, Boyer GL. Sterol composition of Aureoumbra lagunensis, the Texas brown tide alga. Phytochemistry. 2001;57:787–789. doi: 10.1016/s0031-9422(01)00135-2. [DOI] [PubMed] [Google Scholar]
- 8.Sperling EA, Pisani D, Peterson KJ. Poriferan paraphyly and its implications for Precambrian paleobiology. Geol Soc London Spec Publ. 2007;286:355–368. [Google Scholar]
- 9.Erpenbeck D, Wörheide G. On the molecular phylogeny of sponges (Porifera) Zootaxa. 2007;1668:107–126. [Google Scholar]
- 10.Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasier M, Green O, Shields G. Ediacarian sponge spicule clusters from southwestern Mongolia and the origins of the Cambrian fauna. Geology. 1997;25:303–306. [Google Scholar]
- 12.Gehling JG, Rigby JK. Long expected sponges from the Neoproterozoic Ediacara fauna of South Australia. J Paleontol. 1996;70:185–195. [Google Scholar]
- 13.Narbonne GM. The Ediacara biota: Neoproterozoic origin of animals and their ecosystems. Ann Rev Earth Planet Sci. 2005;33:421–442. [Google Scholar]
- 14.Yin L, et al. Doushantuo embryos preserved inside diapause egg cysts. Nature. 2007;466:661–663. doi: 10.1038/nature05682. [DOI] [PubMed] [Google Scholar]
- 15.Peterson KJ, et al. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci USA. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roger AJ, Hug LA. The origin and diversification of eukaryotes: Problems with molecular phylogenetics and molecular clock estimation. Phil Trans R Soc London Ser B. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love GD, et al. Constraining the timing of basal metazoan radiation using molecular biomarkers and U-Pb isotope dating. Geochem Cosmochim Acta. 2006;70:A625. [Google Scholar]
- 18.Hoffman PF, Schrag DP. The snowball Earth hypothesis: testing the limits of global change. Terra Nova. 2002;14:129–155. [Google Scholar]
- 19.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 20.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 22.Lang BF, O'Kelly C, Nerad T, Gray MW, Berger G. The closest unicellular relatives of animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 23.King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 24.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulheirn LJ, Aberhart DJ, Caspi E. Dehydrogenation of sterols by protozoan Tetrahymena pyriformis. J Biol Chem. 1971;246:6556–6559. [PubMed] [Google Scholar]
- 26.Scallen T, Dean WJ, Laughran ED, Vora BV. Isolation and chemical characterization of Δ5,7,24-cholestatrien-3beta-ol from pig tissues. J Lipid Res. 1969;10:121–127. [PubMed] [Google Scholar]
- 27.Gerst N, Ruan B, Pang J, Wilson WK, Schroepfer GJ. An updated look at the analysis of unsaturated C27 sterols by gas chromatography and mass spectrometry. J Lipid Res. 1997;38:1685–1701. [PubMed] [Google Scholar]
- 28.Ruan B, et al. Sterols in blood of normal and Smith–Lemli–Opitz subjects. J Lipid Res. 2001;42:799–811. [PubMed] [Google Scholar]
- 29.Longley RP, Rose AH, Knights BA. Composition of protoplast membrane from Saccharomyces cerevisiae. Biochem J. 1968;108:401–412. doi: 10.1042/bj1080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabiani D, Caruso D, Cavaleri M, Kienle MG, Galli G. Cholesta-5,7,9(11)-trien-3 beta-ol found in plasma of patients with Smith–Lemli–Opitz syndrome indicates formation of sterol hydroperoxide. J Lipid Res. 1996;37:2280–2287. [PubMed] [Google Scholar]
- 31.Galli G, Maroni S. Mass spectrometric investigations of some unsaturated sterols biosynthetically related to cholesterol. Steroids. 1967;10:189–197. doi: 10.1016/0039-128x(67)90046-3. [DOI] [PubMed] [Google Scholar]
- 32.Pelillo M, Galletti G, Lercker G. Mass spectral fragmentations of cholesterol acetate oxidation products. Rapid Com Mass Spectr. 2000;14:1275–1279. doi: 10.1002/1097-0231(20000730)14:14<1275::AID-RCM25>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 33.Itoh T, Sica D, Djerassi C. Minor and trace sterols in marine-invertebrates: 35. Isolation and structure elucidation of 74 sterols from the sponge Axinella cannabina. J Chem Soc Perkins Trans. 1983;1:147–153. [Google Scholar]
- 34.Bergquist PR, Karuso P, Cambie RC, Smith DJ. Sterol composition and classification of the Porifera: 3. Biochem Syst Ecol. 1991;19:17–24. [Google Scholar]
- 35.Goad LJ. In: Methods in Plant Biochemistry. Day PM, Harborne JB, editors. Vol 7. London: Academic; 1991. pp. 369–434. [Google Scholar]
- 36.Goad LJ, Akihisa T, editors. Analysis of Sterols. Berlin: Springer; 1997. [Google Scholar]
- 37.Wyllie SG, Djerassi C. Mass spectrometry in structural and stereochemical problems: 146. Mass spectrometric fragmentations typical of sterols with unsaturated side chains. J Org Chem. 1968;33:305–313. [Google Scholar]
- 38.Conner RL, Mallory FB, Landrey JR, Iyengar CWL. Conversion of cholesterol to Δ5,7,22-cholestatrien-3beta-ol by Tetrahymena pyriformis. J Biol Chem. 1969;244:2325–2333. [PubMed] [Google Scholar]
- 39.Raederstorff D, Rohmer M. Sterol biosynthesis de-novo via cycloartenol by the soil ameba Acanthamoeba polyphaga. Biochem J. 1985;231:609–613. doi: 10.1042/bj2310609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 41.Pearson A, Budin M, Brocks JJ. Phylogenetic biochemical evidence for sterol synthesis in the bacterium Gemmata obsuriglobus. Proc Nat Acad Sci. 2003;100:15352–15357. doi: 10.1073/pnas.2536559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva CJ, Djerassi C. Biosynthetic-studies of marine lipids: 36. The origin of common sterol side-chains in 11 sponges using [3-h-3]-squalene. Comp Biochem Biophys. 1992;101:255–268. doi: 10.1016/0305-0491(92)90188-w. [DOI] [PubMed] [Google Scholar]
- 43.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Department of Genome Sciences, University of Washington; 2005. http://evolution.genetics.washington.edu/phylip. [Google Scholar]
- 45.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online - a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.